ABSTRACT
The S. cerevisiae kinesin Kip2 stabilises astral microtubules (MTs) and facilitates spindle positioning through transport of MT-associated proteins, such as the yeast CLIP-170 homologue Bik1, dynein and the adenomatous-polyposis-coli-related protein Kar9 to the plus ends of astral MTs. Here, we show that Kip2 associates with its processivity factor Bim1, the yeast homologue of the plus-end-tracking protein EB1. This interaction requires an EB1-binding motif in the N-terminal extension of Kip2 and is negatively regulated by phosphorylation through Mck1, the yeast glycogen synthase kinase 3. In addition, Mck1-dependent phosphorylation decreases the intrinsic MT affinity of Kip2. Reduction in Kip2 phosphorylation leads to stabilisation of astral MTs, and accumulation of Kip2, dynein and Kar9 at MT plus ends, whereas loss of Mck1 function leads to defects in spindle positioning. Furthermore, we provide evidence that a subpopulation of Mck1 at the bud-cortex phosphorylates Kip2. We propose that yeast GSK-3 spatially controls astral MT dynamics and the loading of dynein and Kar9 on astral MT plus ends by regulating Kip2 interactions with Bim1 and MTs.
KEY WORDS: GSK-3, Kip2, EB1, CLIP-170, Yeast
Summary: The yeast GSK-3 kinase controls astral microtubule functions by regulating the interaction of the microtubule-stabilising kinesin Kip2 with microtubules and its processivity factor Bim1/EB1.
INTRODUCTION
The microtubule (MT) plus end is a site of remarkable versatility: owing to its dynamicity it easily explores cellular space, but it can also mediate and maintain stable interactions with other cytoskeletal factors. Functionality of microtubule plus ends can be modified by a set of specialised proteins termed MT plus-end tracking proteins (+TIPs) that are able to stably associate with the growing end of the MT (Akhmanova and Steinmetz, 2008; Perez et al., 1999). Among +TIPs, MAPRE1 (hereafter referred to as EB1) and cytoplasmic linker protein CLIP-170 play a central role: EB1 is required for the plus-end-tracking of other proteins (Akhmanova and Steinmetz, 2008; Honnappa et al., 2009; Vaughan, 2005), whereas members of the CLIP-170 family are positive regulators of MT growth (Brunner and Nurse, 2000; Komarova et al., 2002; Steinmetz and Akhmanova, 2008).
Yeast homologues of EB1 (Bim1) and CLIP-170 (Bik1) are both involved in spindle positioning during mitosis of S. cerevisiae. Spindle positioning in budding yeast ensures that the spindle elongates along the mother-bud axis during anaphase and depends on two redundant pathways, the dynein pathway and the Kar9 pathway. The plus-end-directed kinesin motor Kip2 participates in both pathways by transporting +TIPs to the plus ends of astral microtubules (aMTs). In the Kar9 pathway, Kip2 is required for efficient accumulation of the +TIP Kar9 at aMT plus ends (Maekawa et al., 2003). Similar to adenomatous polyposis coli (APC) – a tumour suppressor that links MTs to actin, Kar9 mediates interactions of aMTs with cortical actin that are required for pre-anaphase spindle positioning and nuclear migration close to the bud (Bienz, 2001; Miller et al., 2000; Miller and Rose, 1998). As part of the dynein pathway, Kip2 transports Bik1 and cytoplasmic dynein from the spindle poles to the plus ends of aMTs (Sheeman et al., 2003; Carvalho et al., 2004; Roberts et al., 2014). Dynein is subsequently offloaded from aMTs and immobilised at the cell cortex, where it pulls on aMTs and facilitates correct positioning of the mitotic spindle in anaphase (Moore et al., 2009).
Besides its role in spindle positioning, Kip2 has an intriguing property in budding yeast: it mediates MT stabilisation (Carvalho et al., 2004; Cottingham and Hoyt, 1997; Huyett et al., 1998). Deletion of KIP2 results in extremely short aMTs, whereas KIP2 overexpression leads to cells with abnormally long aMTs. Stabilisation of aMTs by Kip2 seems to be coupled to the transport of Bik1 to aMT plus ends (Carvalho et al., 2004).
Glycogen synthase kinase 3 (GSK-3) is a highly conserved kinase with a key role in signalling during development (Doble and Woodgett, 2003, 2007; Kim et al., 2009; Wu and Pan, 2010), as well as in regulation of MT function and chromosome segregation (Wakefield et al., 2003; Tighe et al., 2007; Buttrick and Wakefield, 2008). In migrating cells and developing neurons, GSK-3 regulates cell polarisation by phosphorylating several +TIPs including APC and CLASP2 (Etienne-Manneville and Hall, 2003; Watanabe et al., 2009). However, the role of GSK-3 in MT regulation within other systems, including yeast cells, is poorly defined.
Here, we show that Kip2 physically interacts with Bim1 through its N-terminal extension, which precedes the kinesin motor domain. This extension is heavily phosphorylated by the yeast GSK-3 kinase homologue Mck1 in a cell-cycle dependent manner, and probably requires a priming phosphorylation by the LATS-related kinase Dbf2. We provide evidence that the N-terminal extension is a regulatory hot spot because phosphorylation not only interferes with Bim1 binding, but also reduces the MT affinity of Kip2. We propose that Mck1 and, possibly, Dbf2 control spindle positioning through spatial regulation of aMT dynamics, and the deposition of dynein and Kar9 at aMT plus ends through phosphorylation of the kinesin Kip2.
RESULTS
Kip2 is phosphorylated by budding yeast GSK-3/Mck1
Mitotic Cdc28 (the budding yeast Cdk1) phosphorylates Kip2 in vitro (Ubersax et al., 2003). In line with this, we identified two potential phosphorylation sites in Kip2 – residues S63 and T275 – that fit the Cdc28 consensus [S/T]PxR sequence, in which x represents any amino acid (aa) (Fig. 1A). Indeed, in western blot analysis of cell extracts, Kip2 C-terminally tagged with 13 Myc epitopes (Kip213myc) displayed a complex migration pattern that collapsed after treatment with alkaline phosphatase (Fig. S1A). This suggested that part of Kip2 is present within cells as a number of phosphoisoforms that display different electrophoretic mobilities on SDS PAGE. Furthermore, replacing S at position 63 with A (Kip2-AT13myc) largely abrogated Kip2 phosphorylation (Fig. 1B, Fig. S1A). The single T275A replacement did not show any significant effect in vivo (Fig. S1A), whereas combination of S63A and T275A mutations (Kip2-AA13myc) displayed similar reductions when compared with Kip2-AT13myc (Fig. 1B, Fig. S1A). We next tested whether Cdc28 phosphorylates Kip2 in vivo by inhibiting Cdc28 over time using the cdc28-as1 strain (Ubersax et al., 2003). In this experiment, the bona fide Cdc28 substrate Kar9 served as an internal positive control (Liakopoulos et al., 2003). Whereas phosphorylation of Kar9TAP decreased rapidly within 20 min after Cdc28 inhibition, Kip213myc phosphorylation remained fairly stable over 50 min (Fig. S1B). In addition, pairwise deletion of the yeast cyclins Clb1–6, which are essential for Cdc28 activity did not significantly decrease Kip2 phosphorylation (Fig. S1C). Hence, we conclude that Cdc28 is not the main kinase involved in Kip2 phosphorylation.
Fig. 1.
Mck1 phosphorylates Kip2 in vivo and in vitro. (A) Details of Kip2 variants described in this paper and abbreviations used in labels, as well as graphical representation of Kip2. Positions of the GSK-3, Cdc28 and Dbf2 consensus sites, residues S63 and T275 are shown in red. The SxIP Bim1/EB1-binding motif is marked in yellow. (B) Comparison of phosphorylation and expression levels between unphosphorylatable Kip213myc variants and Kip213myc in mck1Δ and rim11Δ cells. Phosphorylation of Kip213myc is reduced upon introduction of S63A mutation, in mck1Δ cells and marginally in rim11Δ cells. Shown is a western blot against the Myc epitope in whole-cell extracts. Variant labelling see (A), AA corresponds to Kip213myc-S63A T275A. Arc1 serves as loading control. (C) Depletion of Mck1 leads to decrease in Kip2 phosphorylation. Overexpression of Mck1 does not increase phosphorylation of hypo-phosphorylated Kip213myc variants mutated in the GSK-3 consensus site(s). HAMck1 expression from the GAL1-10 promoter (PGAL) was induced by addition of galactose (+), depletion (−) by addition of glucose to the medium. Western blot shows Kip213myc phosphorylation and HAMck1 expression levels. Arc1 serves as loading control. (D,E) In vivo 32P-phosphate labelling of Kip213myc variants and Kip213myc in different kinase mutants. Kip2 loading: western blot of the Kip213myc variants after immunoprecipitation from cells labelled with 32P-phosphate, the same blot was used for the autoradiography shown. (F) Mck1 phosphorylates Kip2 in vitro. MBPKip2ΔCGFP and displayed variants were pre-incubated with Cdk1 and cyclin B and/or ATP followed by incubation with Mck1TAP and γ32P-ATP as indicated. Autoradiography of the subsequent SDS-PAGE is shown. Mck1TAP phosphorylates Kip2 only after pre-incubation with Cdk1 and phosphorylation is abolished after introduction of the S63A and S275A mutations. (G) Sequential phosphorylation of Kip2 N-terminus by Mck1. A fusion of the N-terminal 80 aa of Kip2 to GST (GST-Kip21-80) is phosphorylated only after pre-incubation with Cdk1. Mutation of S63 but also of the upstream T59 abrogated phosphorylation.
We took a systematic approach to identify the kinase mainly responsible for Kip2 phosphorylation. We did not detect any reduction in Kip213myc phosphorylation in cells with mutations in the yeast Polo kinase (cdc5-2; Charles et al., 1998), the mitotic exit network kinase Cdc15 (cdc15-1; Shirayama et al., 1998) or the yeast kinase Aurora (ipl1-321; Biggins et al., 1999; data not shown). Next, we examined Kip2 phosphorylation in 60 strains each of which was deficient for a non-essential kinase. We observed the most prominent reduction in Kip2 phosphorylation in cells deleted for MCK1, a kinase that encodes one of four homologues of the GSK-3 kinase in budding yeast (Fig. 1B, Fig. S1D; the other three GSK-3 homologues are Rim11, Mrk1 and Ygk3). Phosphorylation of Kip2 in rim11Δ cells is only marginally reduced (Fig. 1B), whereas deletion of Mrk1 or Ygk3 had no significant negative effect on Kip2 phosphorylation levels (Fig. S1D). Moreover, phosphorylation of Kip2 in vivo decreased upon downregulation of MCK1 (expressed under the regulatable PGAL1-10 promoter) and hyperphosphorylation was restored upon overexpression of the kinase – the latter effect was absent upon introduction of the S63A mutation (Fig. 1C). Thus, we conclude that Mck1 is the main kinase responsible for the S63-dependent Kip2 phosphorylation.
Mck1 phosphorylates the unstructured N-terminus of Kip2
We next searched for regions of Kip2 that are targeted by Mck1. GSK-3 is known to sequentially phosphorylate multiple residues within a continuous S/TxxxS/T pattern upstream of an initial phosphorylation site that is often (but not always) phosphorylated by a priming kinase (Roach, 1991). Such a pattern extends throughout the entire S/T-rich N-terminal region of Kip2 (32 S and T residues within the first 80 aa) and is predicted to be heavily phosphorylated by GSK-3 at three S/T clusters according to several algorithms (clusters A, B and C; Fig. 1A; for site prediction see Materials and Methods). Furthermore, T275 is also part of a GSK-3 consensus site. Thus, mutation of S63 and T275 disrupts not only the Cdc28 but also the GSK-3 consensus site and may abrogate phosphorylation either by inhibiting priming phosphorylation or by stopping the sequential phosphorylation cascade. In line with this, overexpression of MCK1 failed to induce hyperphosphorylation of Kip213myc variants with mutations of S63 or in the N-terminal GSK-3 clusters (Fig. 1C). Since a single aa replacement at S63 has the same effect as replacing the S and T residues in the entire N-terminal clusters with D residues, we conclude that S63 is, indeed, crucial for Mck1-dependent Kip2 phosphorylation.
To investigate differences in phosphorylation of the different N-terminal variants of Kip2 in vitro (Fig. 1A), we labelled living cells with 32P-orthophosphate and examined phosphorylation by autoradiography after immunoprecipitation (Fig. 1D,E). Consistent with our previous results, phosphorylation of Kip213myc in mck1Δ cells was severely reduced, as was that of Kip2-AT13myc (and, to a similar extent, that of Kip2-AA13myc). Deletion of the clusters A and B (Δ37-Kip213myc) severely reduced phosphorylation, whereas Kip2 deleted for the whole N-terminus before aa residue 70 (Δ70-Kip213myc) did not exhibit any detectable phosphorylation (Fig. 1D), even when considering that Δ70-Kip213myc protein levels were clearly lower compared to wild-type Kip2. These data suggest that the S/T-rich terminus of Kip2 bears the main phosphorylation sites and that phosphorylation extends through all three clusters of the GSK-3 consensus site.
We next tested phosphorylation of Kip2 by Mck1 in vitro. We, thus, purified from bacteria an MBP-fusion of Kip2 lacking the C-terminal tail (Kip2 1–560) followed by GFP (MBPKip2ΔCGFP; this fusion was more stable than full-length Kip2 isolated from yeast and from insect cells) and Mck1TAP from yeast cells. In line with the requirement for a priming-phosphorylation by GSK-3 kinases, Mck1TAP alone was not able to phosphorylate MBPKip2ΔC (Fig. 1F). However, pre-incubation with recombinant Cdk1 and cyclin B allowed MBPKip2ΔC to become efficiently phosphorylated after Mck1TAP addition. Introducing a S63A mutation reduced phosphorylation, whereas a double S63A T275A mutation nearly abrogated in vitro phosphorylation. These results suggested that priming phosphorylation of S63 by using Cdk11 and cyclin B can support sequential phosphorylation by Mck1 of sites further upstream. Accordingly, mutation of T59 to an unphosphorylatable residue should prevent phosphorylation of the upstream cluster and phenocopy of the S63A mutation. To test this, we constructed GSTKip21–80 fusion proteins bearing the S63A or T59A mutation and examined their phosphorylation in vitro. As predicted, both constructs were only poorly phosphorylated compared to the wild-type control (Fig. 1G). Taken together, our results suggest that Mck1 sequentially phosphorylates Kip2 at the GSK-3 clusters within its N-terminus in vivo and in vitro. It is also likely that the GSK-3 consensus site that comprises T275 is phosphorylated with T275 acting as a priming site. However, we cannot exclude that T275 acts as a priming site only in the presence of the S63A mutation.
Dbf2 might act as priming kinase for Mck1
Although our in vitro experiments suggest that Cdc28 is a priming kinase for Mck1, inhibition of Cdc28 did not affect Kip2 phosphorylation in vivo and deactivation of Cdc28 did not decrease Kip2 phosphorylation in absence of Mck1 (Fig. S1B,C,D). Furthermore, S63 is part of cluster C but is not the most downstream Ser within that cluster. We, therefore, reasoned that S69 or S72 might act as a priming site in vivo.
Indeed, we observed a clear reduction of Kip213myc phosphorylation upon introduction of S69A and the highest reduction upon introduction of the S72A mutation, suggesting that these sites control phosphorylation of the upstream cluster in vivo (Fig. 2A). A consensus site for protein kinase C comprises S69, whereas the aa sites 69–75 are also predicted CKI sites (ELM; http:/elm.eu.org/). However, we did not observe a significant reduction in Kip213myc phosphorylation in yeast stt1-1 (PKC) or yck1Δ yck2-2ts (CKI) mutants (Fig. S1E). Therefore, the (priming) kinases that phosphorylate these sites remain unknown.
Fig. 2.
Dbf2 and additional kinases act on Kip2 as a priming kinases for GSK-3. (A) Amino acids at positions S69 and S72, downstream of S63 act presumably as phosphorylation priming sites for Mck1. Western blots of the indicated Kip213myc variants. Kip213myc phosphorylation is reduced upon mutation of the indicated sites. (B) Kip213myc phosphorylation is clearly reduced in dbf2-2 dbf20Δ mutants. (C) Kip2 phosphorylation is cell-cycle regulated and increases in anaphase (western blot against Kip213myc of cell extracts from cells synchronized with α-factor for 3 h and released in fresh medium lacking the pheromone, at 30°C).
However, during the analysis of the yeast-kinase deletion strains, we observed that Kip2 phosphorylation is diminished in dbf20Δ-null mutants as well as in dbf2Δ strains (Fig. S1F). Dbf20 is a close homologue of the LATS-related Dbf2 kinase in yeast. Dbf2 kinase is a part of the mitotic exit network that is required in metaphase for correct spindle orientation (Hotz et al., 2012) and is further activated following anaphase to promote exit from mitosis (Bardin and Amon, 2001). Upon analysis of the dbf2-2 dbf20Δ double mutant, we observed that phosphorylation of Kip213myc was, indeed, clearly reduced (Fig. 2B). Furthermore, we examined phosphorylation of Kip2 during the cell cycle (Fig. 2C, Fig. S1G). Phosphorylation levels of Kip213myc were lowest during metaphase, followed by increase in Kip213myc phosphorylation immediately after. Therefore, the pattern of Kip2 phosphorylation fits well with the timing of principle Dbf2 activation. Although some phosphorylation was still evident, the increase of phosphorylated Kip213myc upon anaphase entry was less pronounced in the dbf2-2 dbf20Δ mutant (and the mck1Δ mutant) when compared to wild type (Fig. S1G), in agreement with the hypothesis that Dbf2 acts on Kip2 as a priming kinase for Mck1.
Upon closer inspection of the Kip2 sequence, we noticed that residues S13, S63 and S69 within the N-terminal clusters are preceded by an R and, thus, match the preferred sequence for the Dbf2 kinases [RxxS/T] (Mah et al., 2005; Fig. 1A). Furthermore, T14, S18 and S33 are part of such consensus sites although not part of the GSK-3 [SxxxS] pattern. Intriguingly, neither an R60A nor an R66A single mutation (Fig. S1H and Fig. 2A, respectively) caused significant difference in the Kip213myc phosphorylation pattern. We refrained from mutating more R residues in the N-terminal Dbf2 consensus sites because this would have drastically changed the pI of this domain and might have perturbed Kip2-MT interactions (see below). Our interpretation of these data so far is that priming for Mck1 phosphorylation is mediated by Dbf2 and/or Dbf20 within the Kip2 N-terminus, and possibly by additional kinases that act at S69 and S72.
Regulation of MT stability through phosphorylation of Kip2
GSK-3 is an important regulator of MT function in mammalian cells (Buttrick and Wakefield, 2008), but its role in MT regulation in budding yeast is unknown. Kip2 localises exclusively on aMTs and has been reported to promote aMT stability by transporting Bik1 to aMTs plus ends (Carvalho et al., 2004). This prompted us to examine the aMT phenotypes of hypo-phosphorylated Kip2 variants that bear mutations within the GSK-3 consensus site in cells at G2/M phase. For this, we created kip2Δ cells that genomically express different Kip2 variants under the control of the native KIP2 promoter. Interestingly, cells that express Kip2 variants that are defective in Mck1 phosphorylation (Kip2-AT) displayed an increased number as well as abnormally long astral MTs (Fig. 3A,B,C). This phenotype is exacerbated in Kip2-AA-expressing cells, in which the numbers of long aMTs were increased. In addition, these cells were growth resistant to the MT-destabilising drug Benomyl (Fig. 3D). Cells that express Kip2 with point mutations at T275 (S→A or S→E) within the GSK-3 consensus site alone did not display a significant aMT phenotype (Fig. S2A). Consistently, reduction of Kip2 phosphorylation through deletion of MCK1 also results in aMT stabilisation and increase in aMT number (Fig. 3A,B,C), whereas these aMT phenotypes are even more pronounced in mck1Δ rim11Δ mutant cells, but not after combining the mck1Δ deletion with mrk1Δ or ygk3Δ (Fig. S2B). Importantly, we also observed the same aMT phenotypes in dbf2-2 dbf20Δ cells in all phases of the cell cycle (Fig. 3A,B,C), consistent with the role of Dbf2 as a priming kinase for Mck1. In conclusion, these experiments show that expression of the hypophosphorylated form of Kip2 stabilizes MTs.
Fig. 3.
Mck1 and Dbf2 regulate aMT stability through Kip2 phosphorylation. (A) Lack of Mck1- and Dbf2-dependent Kip2 phosphorylation stabilises aMTs. Images of MTs from kip2Δ cells expressing unphosphorylated Kip2 variants and from depicted kinase mutants. The MTs shown here do not represent the mean MT length but are characteristic for each strain. (B) Length distribution and corresponding statistics for bud aMTs in G2/M-phase cells of depicted strains. Number of counted aMTs are shown in brackets, P , statistical significance of the difference between the distribution of the strain on the left of the graph and the MT distribution of the strain denoted in brackets. Imaging of all strains was performed in the same experiment. See Fig. S2B for additional aMT length measurements. (C) The number of aMTs is increased upon a reduction in Kip2 phosphorylation. The number of aMTs growing from either spindle pole was counted; n>100 cells for each strain. (D) kip2Δ cells show growth sensitivity, and Kip2-AA-expressing cells resistance to the MT-destabilising drug Benomyl. See Fig. S2A for images of Kip2-SA and -SE (T275E mutation) variants.
Phosphorylation inhibits the binding of Kip2 to MTs
We next set out to elucidate the molecular mechanism through which Mck1-dependent Kip2 phosphorylation might affect aMT stability. We first reasoned that phosphorylation simply might control the stability of Kip2. In line with this idea, cells expressing a Kip2-AT–YFP variant display a significantly higher plus-end accumulation at aMTs when compared to cells expressing wild-type Kip2–YFP (Fig. 4A). This effect was not specific only for the plus ends but is, rather, a result of the overall increased amounts of Kip2-AT–YFP on aMTs. Consistently, depleting cells of Mck1 also results in increased aMT- and plus-end accumulation of wild-type Kip2–YFP, whereas overexpression of Mck1 had the opposite effect (Fig. 4A). We, therefore, set up a cycloheximide-chase experiment to follow degradation kinetics of different Kip2 variants, and co-expressed Kip213myc-AA and wild-type Kip2TAP as an internal control (Fig. 4B). Both proteins, however, displayed indistinguishable turnover kinetics, suggesting that Mck1-dependent Kip2 phosphorylation does not regulate Kip2 stability and that the increased plus-end accumulation of the hypo-phosphorylated Kip2 must have a different cause.
Fig. 4.
Phosphorylation regulates association of Kip2 with MTs. (A) Mck1-dependent phosphorylation regulates Kip2 load on aMTs. Fluorescence intensity of Kip2–YFP, Kip2-AT–YFP and allD-Kip2–YFP in wild-type cells and of Kip2–YFP in HAMck1-overexpressing or Mck1-depleted cells. Examples of quantified images are given above each graph. Numbers of counted aMTs in brackets, arbitrary intensity units ±s.e.m. are shown. Scale bars: 3 µm. (B) Cycloheximide chase of plasmid-borne Kip213mycAA under control of the KIP2 promoter in cells expressing Kip2TAP from the KIP2 locus as an internal control. Kip2 phosphoisoforms were not resolved here in order to facilitate comparison between different Kip2 variants. (C) A variant of Kip2 with a phosphorylation-mimicking N-terminus (allD-MBPKip2ΔCGFP) binds less efficiently to MTs in vitro, compared to the unchanged variant. Shown are Coomassie-Blue-stained gels of microtubule sedimentation assays, after incubation of in vitro polymerised, taxol-stabilized MTs with recombinant and MBPKip2ΔCGFP or allD-MBPKip2ΔCGFP. S, supernatant; P, pellet. Refer also to Fig. S2D.
Increase in affinity for MTs or in processivity of hypo-phosphorylated Kip2 isoforms might also account for increased accumulation at aMT plus ends. In the first case, more Kip2 would be loaded onto aMTs, in the second case loaded Kip2 would be more likely to reach the plus end. We first addressed the possibility that phosphorylation of the Kip2 N-terminus increases the MT affinity of the motor. This unstructured, S/T-rich region is highly basic (with pI=13) and is reminiscent of the Ndc80 N-terminal extension that contributes to binding of Ndc80 to MTs (Ciferri et al., 2008). Interestingly, introducing negative charges by phosphorylation of the N-terminal extension in Ndc80 weakens its interaction with the MT lattice (Ciferri et al., 2008). We, therefore, asked whether phosphorylation of the N-terminal sequence of Kip2 has an analogous effect on MT binding. Recombinant GFPKip2 purified from insect cells is hyperphosphorylated in an S63-dependent manner in western blots – similar to that from yeast cells – and we verified by using mass-spectrometry that S63 is, indeed, phosphorylated (Fig. S2C,D; see also Roberts et al., 2014). We, thus, asked which forms of purified GFPKip2 interact stronger with MTs. In MT- sedimentation assays, the affinity of the hyperphosphorylated GFPKip2 isoforms was reduced compared to hypo-phosphorylated GFPKip2 (Fig. S2D). Furthermore, to mimic a constitutively phosphorylated Kip2, we mutated all S or T residues to D within the N-terminal cluster of our MBPKip2ΔCGFP construct (allD-MBPKip2ΔCGFP) and repeated the co-sedimentation experiments. Again, wild-type MBPKip2ΔCGFP co-sedimented with increasing tubulin concentrations, the majority of allD-MBPKip2ΔCGFP remained in the supernatant (Fig. 4C). Consistent with these findings, introduction of the same mutations to the Kip2–YFP construct (allD-Kip2–YFP) reduced Kip2 MT load by ∼50% in vivo (Fig. 4A). Therefore, we propose that Mck1 controls aMT stability by phosphorylating the N-terminus of Kip2 and by regulating the affinity of Kip2 for MTs.
Mck1 controls interaction of Kip2 with the yeast EB1
During sequence analysis of the N-terminal extension, we also identified a putative EB1/Bim1-binding (SxIP) motif (Honnappa et al., 2009) at aa 21-24, with S21 being part of the GSK-3 cluster A (SNIP1; Fig. 5A; see also Roberts et al., 2014). The site might be conserved in the proposed fission yeast Kip2 homologue Tea2 (Fig. 5A). A second motif could be identified starting at aa 415 (SNIP2). The SNIP1 Bim1 binding site at aa 21–24 is of particular interest because Bim1 (and also Bik1) act as a processivity factor for Kip2 (Bieling et al., 2007; Roberts et al., 2014). Phosphorylation next to EB1 binding motifs has been shown to disrupt interactions with EB1 (Honnappa et al., 2009; Roberts et al., 2014; Zumbrunn et al., 2001). In this case, the SNIP1 motif is flanked by the Mck1 clusters A and B, the first of which includes S21 of SNIP1. This suggested that Mck1-dependent phosphorylation interferes with the Kip2–Bim1 interaction. Therefore, Mck1-dependent Kip2 phosphorylation might control plus-end accumulation of Kip2 by regulating Kip2 interaction to its processivity factor(s), in addition to its interaction with MTs.
Fig. 5.
Kip2 interacts with Bim1 over its N-terminal extension in a phosphorylation-dependent manner. (A) Alignment of the N-terminal extensions of Kip2 and fission yeast Tea2. The proposed EB1-interacting motifs are shown in green, positively charged residues in yellow. The GSK-3 consensus site comprising S63 is shown in bold. (B,C) Bim1 interacts with the EB1-binding motif at the Kip2 N-terminus, independently of Bik1. Immunoprecipitations of HABim1 from kip2Δ or kip2Δ bik1Δ cells expressing depicted plasmid-borne Kip213myc variants. Note that only non- (or hypo)-phosphorylated Kip2 isoforms co-precipitate with HABim1 (seen in C). PGAL3HA-BIM1 expression was induced for 3 h prior to immunoprecipitation. Immunoprecipitated proteins (IP) and extracts after the immunoprecipitation (input) were probed in western blots with anti-HA and anti-Myc antibodies. Asterisk indicates antibody heavy chain. SS refers to the Kip213myc variant with the SNIP motif mutated to SNSS. (D) Localisation of the Kip2–YFP variants which show decreased interaction with Bim1 (C and D). Representative images of cells expressing CFP–Tub1 and integrated Kip2–YFP variants from the KIP2 promoter as the sole Kip2 source. (E) The two EB1-binding motifs are required for Kip2–Bim1 interaction in vitro. Shown are coomassie stained gels INPUT: amounts of the recombinant proteins used in the pulldown assay, the EB1 domain of Bim1 (EB1Bim1) as a GST-fusion was bound to beads (bait). SxIP1 and SxIP2 denote the recombinant Kip2 variants with respective mutations (SNIP to SNNN) in the two EB1-binding motifs of Kip2 (see main text). Mutation of these motifs does not abrogate but weakens the Kip2-interaction in an additive manner. The Kip2–Bim1 interaction is almost lost upon deletion of the N-terminus (Δ70) or upon mutation of the N-terminal GSK-3 clusters to aspartates (allD, refer also Fig. 1A).
We, thus, first tested whether the identified motifs are required for Kip2 interaction with Bim1 in vivo. Indeed, HABim1 co-immunoprecipitated Kip213myc from cell extracts (Fig. 5B), whereas Bik1 was dispensable for this interaction in vivo [Bim1 and Bik1 physically interact with each other (Blake-Hodek et al., 2010; Wolyniak et al., 2006]. The truncated Kip2 variants that lack either the first 37aa (Δ37-Kip213myc, lacking also SNIP1) or the entire N-terminal Kip2 extension (Δ70-Kip213myc) almost completely lost interaction with HABim1 (Fig. 5B), although the effect was difficult to assess because the cellular levels of these Kip2 variants were also decreased. However, mutation of the N-terminal SNIP1 to SNSS abrogated the interaction between Kip2 and Bim1 (Fig. 5C, for localisation of the Kip2-variants see Fig. 5D).
In vitro, recombinant MBPKip2ΔCGFP protein interacted with the EB1 domain of Bim1 (GST-EB1Bim1) and this interaction was abrogated after deletion of the N-terminal sequences that included the SxIP1 motif (Fig. 5E). Mutation of the N-terminal SxIP1 motif reduced, but did not abolish, the interaction with GST-EB1Bim1, whereas additional mutation of the second SxIP2 motif weakened the interaction further (Fig. 5E). Importantly, deletion of the first 70 N-terminal aa of Kip2 essentially abrogated binding. Hence, we concluded that the N-terminal SNIP1 is, indeed, a Bim1-binding site in Kip2.
Does Mck1 regulate the interaction between Kip2 and Bim1? In support of this notion, we found that only the hypo-phosphorylated form of Kip2 co-immunoprecipitated with HABim1 in vivo (Fig. 5C). Importantly, the recombinant allD-MBPKip2ΔCGFP variant failed to interact with the EB1-binding domain of Bim1 in vitro (Fig. 5E). Therefore, these data suggest that the N-terminal Kip2 domain contains a functional EB1-interaction motif that mediates interaction of Kip2 with Bim1, and that phosphorylation of Kip2 by Mck1 close to the EB1-binding motif inhibits the interaction between Kip2 and Bim1. Thus, Mck1-dependent phosphorylation interferes with the cellular activity of Kip2 through two different mechanisms; first, by reducing its overall affinity to MTs and, second, by inhibiting Kip2 binding to its processivity factor Bim1.
Kip2 phosphorylation controls the amount of Kar9 and dynein on astral MT plus ends
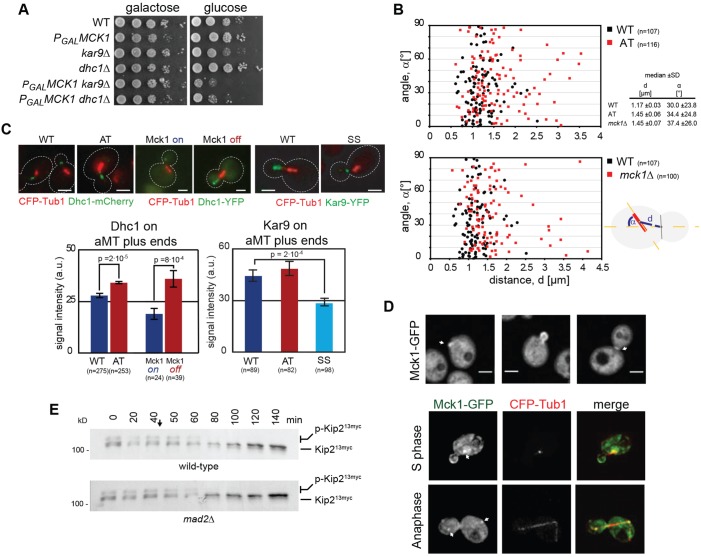
We examined the implications of Mck1-dependent Kip2 regulation in spindle positioning. As mentioned, Kip2 is required for deployment of dynein and Kar9 to aMT plus ends (Maekawa et al., 2003; Sheeman et al., 2003). Thus, we first analysed genetic interactions of MCK1 with the dynein and the Kar9 pathways. Both pathways require aMTs for their function, and their simultaneous genetic deactivation is lethal for yeast cells. Depletion of Mck1 resulted in growth defects of cells deleted for either DYN1 (the dynein heavy chain) or KAR9 (Fig. 6A), suggesting that Mck1 is, indeed, required for the correct function of both pathways and consistent with the role of Mck1 in regulation of aMT function by Kip2 phosphorylation. In agreement with this idea, we found that a fraction of spindles were mispositioned in mck1Δ cells as well as in cells expressing the hypo-phosphorylated Kip2-AT variant (Fig. 6B) – mainly because of an increase in the distance of the spindle from the cleavage apparatus at the bud neck.
Fig. 6.
Kip2 phosphorylation controls the amount of Kar9 and dynein on aMT plus ends and spindle positioning. (A) Genetic interactions of MCK1 with genes required for spindle positioning. PGALHAMCK1 kar9Δ and PGALHAMCK1 dyn1Δ cells display slow growth upon deactivation of MCK1 expression on glucose-containing plates. (B) Spindles are mispositioned in mck1Δ cells and cells expressing Kip2-AT. Plotted is the angle of the spindle to the mother-bud axis (α, y-axis), against the distance from the bud neck (d, x-axis), the vertical line denotes the bud neck. Number of spindles counted in brackets. (C) Fluorescence intensity of Dyn1–YFP and Kar9–YFP on aMT plus ends in cells expressing the Kip2 variants indicated or in cells expressing wild-type Kip2 in HAMck1 overexpressing or depleted cells. Numbers of counted aMTs in brackets, arbitrary intensity units ±s.e.m. are shown. Scale bar: 3 µm. (D) Localisation of Mck1–GFP. A Mck1 sub-pool localises to the incipient bud, to the bud cortex until G2/M phase, and the bud neck. Mck1 also localises to the spindle poles (possibly SPBs or kinetochores) from S phase until anaphase. Scale bar: 2.5 µm. (E) Phosphorylation of Kip2 requires MTs. Time course showing Kip213myc phosphorylation in extracts of wild type- and spindle assembly checkpoint-defective mad2Δ cells, after MT depolymerisation. Nocodazole was added at 40 min (arrow). Equal number of cells were used to generate the extracts for each time point.
As Kip2 is able to transport dynein to the MT plus end (Roberts et al., 2014), we next examined whether lack of Kip2 phosphorylation by Mck1 would affect deployment of dynein to aMT plus ends. For this, we quantified localisation of dynein on aMT plus ends in cells expressing the hypo-phosphorylated Kip2-AT variant. Dynein accumulation at aMT ends increased in these cells (Fig. 6C). Depletion of Mck1 (which was expressed under the repressible PGAL1-10 promoter) also resulted in increased amounts of dynein at aMT ends, whereas Mck1 overexpression led to the opposite effect (Fig. 6C). Thus, we propose that Mck1 controls plus-end localisation of dynein by adjusting the MT affinity and processivity of Kip2.
The protein Kar9 requires Bim1 for aMT localisation (Miller et al., 2000) and also depends on Kip2 to be transported to the plus ends of aMTs (Carvalho et al., 2004; Maekawa et al., 2003). We, therefore, also examined whether the interaction between Kip2 and Bim1 is required for localisation of Kar9 complexes to MT plus ends. Intensity of Kar9 at aMT plus ends was clearly reduced in cells expressing Kip2-SS as a sole Kip2 source (Fig. 6C). Intriguingly, expression of the hypo-phosphorylated Kip2-AT variant did not increase the amount of Kar9 at bud MT plus ends significantly. However, because Mck1 phosphorylation inhibits binding of Kip2 to Bim1 and the Kip2-SS variant showed reduced targeting of Kar9 to aMT plus ends we, nevertheless, think that Mck1 might control the plus-end deployment of Kar9 as well as dynein.
Phosphorylation of Kip2 requires MTs and might take place at cortical sites
Mammalian GSK-3 is spatially regulated at cortical sites (Etienne-Manneville and Hall, 2003; Wittmann and Waterman-Storer, 2005). We, thus, asked whether phosphorylation of Kip2 by Mck1 takes place at specific cellular sites, and closer examined the localisation of Mck1 and Rim11 by using spinning disc confocal microscopy and subsequent deconvolution. Mck1–GFP is located throughout the cytoplasm and the nucleus, as previously reported (Huh et al., 2003). In addition, Mck1–GFP accumulates at the cortex of emerging and small buds, at the bud neck, as well as at the spindle poles – either at the spindle pole bodies (SPBs – the yeast MT-organising centres) or at kinetochores (Fig. 6D). Rim11–GFP displays similar localisation but does not localise to the spindle poles (data not shown). The localisation of Mck1 suggests that Kip2 phosphorylation takes place either in the cytoplasm at the cortex or – because Kip2 localises exclusively on aMTs – at the spindle poles.
To further clarify this, we asked whether aMTs are required for Kip2 phosphorylation. After depolymerisation of aMTs with nocodazole, phosphorylation of Kip2 was lost within 20 min (Fig. 6E). Loss of phosphorylation is independent of the nocodazole-induced cell cycle arrest because it still occurred in mad2Δ cells. In view of the cortical localisation of Mck1 and Rim11, these data suggest that Mck1 does not phosphorylate Kip2 in the cytoplasm or at the SPB but, possibly, as aMTs reach the cell cortex. It is, therefore, possible that Mck1 controls aMT function by phosphorylating Kip2, thereby regulating dynein-plus-end and Kar9-plus-end turnover at cortical sites (see discussion).
DISCUSSION
In this work we show that Mck1 phosphorylates Kip2 by regulating aMT length, and the load of dynein and Kar9 complexes at aMT plus ends. We demonstrate that phosphorylation occurs mainly at the entire S/T-rich N-terminal part of Kip2. Phosphorylation of Kip2 by Mck1 occurs in a sequential manner and requires the S63. Mck1 has been shown to prefer substrates that are pre-phosphorylated in close vicinity to the GSK-3 consensus site (Tan et al., 2014), similar to its mammalian counterpart (Fiol et al., 1988). In vitro, replacing GSK-3β sites with D or E residues supports the phosphorylation of S/T residues that are located further upstream (Roach, 1991). However, mutation of S63 in Kip2 to D or E failed to mimic and, rather, abrogated phosphorylation (data not shown).
Two lines of evidence suggests that phosphorylation of T275 within the GSK-3 consensus site that is found in the motor domain of Kip2 contributes to Kip2 regulation. First, the double S63A T275A mutation caused the strongest effects regarding aMT stabilisation and resistance of cells to the MT destabilising effect of Benomyl (Fig. 3D). Second, phosphorylation by Mck1 was reduced after introduction of the T275A mutation in vitro (Fig. 1F). We, thus, think that phosphorylation of Kip2 at T275 contributes to Kip2 regulation through Mck1, but how exactly T275 phosphorylation affects Kip2 function remains unclear.
It is also not clear which kinases prime Kip2 for phosphorylation downstream of the GSK-3 consensus sites. Our in vivo experimental data do not support the in vitro data, which suggests that Cdc28 acts as a priming kinase at S63 (Ubersax et al., 2003). In fact, later proteomic studies did not identify Kip2 among the in vivo Cdc28 substrates (Holt et al., 2009). In contrast, Dbf2/Dbf20 is a strong candidate for a priming kinase: phosphorylation of Kip2 was reduced in dbf2-2dbf20Δ cells and deactivation of Dbf2 kinases led to aMT stabilisation. Moreover, the N-terminal extension of Kip2 contains nine sequences that each match the Dbf2 consensus site. In addition, Kip2 phosphorylation is cell cycle regulated and increases in anaphase – at the time of Dbf2 activation, whereas this regulation is lost upon Dbf2 deactivation. Further experiments are required to address this issue, which is complicated by the fact that other kinases seem to act on the Kip2 N-terminus, mainly at aa positions 69 and 72.
Importantly, lack of Kip2 phosphorylation by Mck1 stabilizes aMTs in a manner that is similar to the overexpression of Kip2 (Carvalho et al., 2004). Therefore, aMT stabilisation in cells with reduced Kip2 phosphorylation could be due to increased amount of Kip2 within the cell. Several lines of evidence argue against this idea: first, cellular steady-state levels of the aMT-stabilising Kip2-AT and Kip2-AA variants did not significantly differ from wild type (for example Fig. 1B or Fig. S1A). Second, cellular Kip2 levels did not increase upon Mck1 downregulation or decrease upon Mck1 overexpression (Fig. 1C). And, third, turnover of Kip2-AA was indistinguishable from wild-type Kip2 (Fig. 4B). In contrast, the stabilising effect of Kip2-AT on aMTs can be adequately explained by the finding that loss of Kip2 phosphorylation increases the amount of the Kip2 that is able to bind aMTs, a prediction that we confirmed in vivo (Fig. 4A) and in vitro (Fig. 4C). This, in turn, suggests that aMT stabilisation in Kip2-AT-expressing cells is due to increased transport of the Kip2 cargo Bik1 to aMT plus ends. Intriguingly, however, we did not detect any significant increase of Bik1 at aMT plus ends in Kip2-AT-expressing cells, thus the molecular mechanisms that cause aMT stabilisation in these cells remain to be determined.
We found that Kip2 interacts with Bim1 through an EB1-interaction motif that is present inside the Kip2 N-terminal domain, close to the Mck1 and Dbf2 phosphorylation sites. Although we can efficiently detect the Kip2–Bim1 interaction in pull-down assays, the complex is probably not very stable because it cannot withstand gel filtration (Roberts et al., 2014). Furthermore, we propose that a similar N-terminal extension of Tea2 (the fission yeast Kip2 homologue) also contains two EB1-binding sites. The fission yeast EB1 homologue Mal3 binds to the N-terminus of the kinesin-like protein Tea2 acting as a processivity factor, because Tea2 does not efficiently track MTs in the absence of Mal3 (Bieling et al., 2007; Browning and Hackney, 2005). Similarly, Bim1 and Bik1 act as processivity factors for Kip2; they are required for Kip2 in order to counteract the minus-end force when it transports dynein in vitro (Roberts et al., 2014). This evidence is consistent with our observation that abrogation of Kip2 phosphorylation leads to Kip2- and dynein accumulation at aMT plus ends, and that the Bim1–Kip2 association is required for efficient transport of Kar9 to the aMT plus ends. However, it is not clear how Kip2 transports Kar9 to the ends of MTs. Acting as a processivity factor for Kip2, Bim1 might indirectly affect Kar9 transport to aMT plus ends (Browning and Hackney, 2005). Alternatively, the Bim1-Kar9 complex might be transported by ‘hitchhiking’ on Kip2, although it is difficult to envisage how Bim1 would bind simultaneously to both proteins. A third possibility is that Kip2 indirectly alters plus-end localisation of Kar9 by affecting deployment of Bim1 to aMT ends.
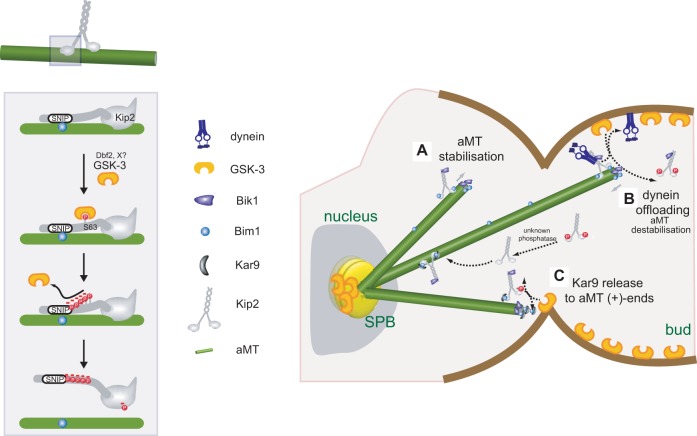
Our data suggest that Kip2 phosphorylation by Mck1 weakens the association of Kip2 with Bim1 and aMTs. In this manner, Mck1 and Dbf2 could control aMT dynamics at the cell cortex (Fig. 7). According to this model, Kip2 becomes phosphorylated by Mck1 as it reaches the cortex. Phosphorylated Kip2 dissociates from aMTs, causing aMT destabilisation (Fig. 7A,B). Dissociation of the Kip2–Bik1 complex from aMTs upon phosphorylation by Mck1/Dbf2 might also be part of the mechanism that is used to offload dynein at the cell cortex (Fig. 7B). This idea is also supported by the finding that overexpression of Mck1 results in reduction of dynein at aMT plus ends, whereas depletion of Mck1 has the opposite effect. By contrast, when Mck1/Dbf2 phosphorylates Kip2, it disrupts not only the interaction between Kip2 and aMTs but also the interaction between Kip2 and Bim1. This would lead to dissociation of Bim1 (and possibly of Kar9) from Kip2, and its deployment to aMT plus ends (Fig. 7C). Our model predicts that both dynein- and Kar9-dependent spindle positioning are defective in cells that lack Mck1, as supported by the genetic interactions of mck1Δ with both dyn1Δ and kar9Δ. In vitro assays are certainly required to test these ideas in detail.
Fig. 7.
Model on how Mck1 regulates aMT functions. Left: Mck1 phosphorylates Kip2 at the N-terminal extension, including the GSK-3 consensus site that comprises S63, close to the EB1-binding motif (SNIP1). Phosphorylation disrupts interaction between Kip2 and Bim1. In addition, phosphorylation (involving possibly also T275) causes dissociation of Kip2 from aMTs. Dbf2 and unknown kinase(s) act as priming kinases for Mck1. Right: Mck1 might regulate aMT function through Kip2 phosphorylation in three ways. (A) Unphosphorylated Kip2 binds and stabilises aMTs. (B) When aMTs reach the cortex, Mck1 phosphorylates Kip2, leading to Kip2 dissociation from aMTs and offloading of dynein at cortical sites. The process may be enhanced upon anaphase onset, the time of Dbf2 activation. (C) Phosphorylation by Mck1 also disrupts interaction between Kip2 and Bim1, leading to release of transported Bim1–Kar9 complexes at aMT plus ends. Dephosphorylation of Kip2 by a phosphatase restarts the cycle.
To this date, the role of yeast GSK-3 in the control of aMT dynamics was largely unclear. This study fills the gap concerning GSK-3 function between yeast and mammalian cells. Kip2 stabilises aMTs, binds to Bim1 and promotes transport of APC-related Kar9 to aMT plus ends in budding yeast. This resembles the role of kinesin KIF17 – which was recently found to stabilise MTs, interact with EB1 and localise APC to MT plus ends – in mammalian cells (Jaulin and Kreitzer, 2010). We found that Mck1 accumulates at cortical sites of polarised growth in yeast cells, regulates aMT dynamics, and the amount of Kip2 and dynein at aMT plus ends. Moreover, Mck1 regulates the interaction between Kip2 and Bim1. We also showed that the Kip2–Bim1 interaction is required for efficient localisation of Kar9 to aMT plus ends. In mammalian cells, GSK-3 phosphorylates +TIPs, such as CLASP2 or APC, and weakens their interaction with MTs or other +TIPs (Rubinfeld et al., 1996; Zumbrunn et al., 2001), whereas local inhibition of GSK-3 stabilises MTs and promotes their interaction with the cortical cytoskeleton during cell migration (Akhmanova et al., 2001; Etienne-Manneville and Hall, 2003; Kumar et al., 2009; Watanabe et al., 2009). Regulation of Kip2 through Mck1 in budding yeast might be another example for a GSK-3-dependent mechanism to couple the regulation of MT dynamics with the control of aMT-cortical interactions.
MATERIALS AND METHODS
Yeast growth and medium
All strains were derivates of S288C (ura3-52 lys2-801 ade2-101 trp1-Δ63 his1-Δ200 leu2-Δ1) or BY4743 (for details see Table S1) and grown on standard medium.
Sequence analysis and generation of Kip2 variants
Sequence analysis to identify phosphorylation sites was performed by NetPhosK (Technical University of Denmark, http://www.cbs.dtu.dk/services/NetPhosK/) and the Eukaryotic Linear Motif resource (Gould et al., 2010) (ELM; http:/elm.eu.org/). All Kip2 variants were generated by site-directed mutagenesis. Untagged versions as well as Venus-YFP-tagged Kip2 variants were cloned into the integrative pRS305 plasmid under control of the endogenous KIP2 promoter and integrated into the LEU2 locus. 13×Myc-tagged (13myc) Kip2 variants expressed under control of the KIP2 promoter were sub-cloned into the pRS314 plasmid. For full plasmid list see Table S2.
Microscopy
Microscopy was carried out by using an Olympus IX81 microscope equipped with the Cell R software (Olympus Germany, Hamburg). MTs were visualised by integrating a GFP–Tub1- or CFP–Tub1-encoding plasmid into the URA3, LYS2 or TRP1 locus and analyzed in small-budded cells with spindles no longer than 2 µm by using ImageJ. Length measurements were made using ImageJ. Fluorescence intensities were measured with ImageJ in an area of 0.3 µm (a circle with a radius of 0.32 µm) in the relevant focal plane of a nine-image z-stack after background subtraction. Only bud aMTs of metaphase cells were examined. This restriction takes into account the temporal and spatial localisation pattern of Mck1 and Rim11 (see Fig. 6D). Mck1–GFP and Rim11–GFP imaging was performed using a Perkin Elmer spinning disc on a Nikon TE2000 inverted microscope. Deconvolution was performed with Huygens Essential 3.4 (Scientific Volume Imaging, Hilversum, The Netherlands).
Statistical analysis
Statistical comparison of datasets was made by using a two-tailed t-test of unequal variance. The number of measurements was large enough to allow generation of histograms. aMT lengths are stochastic and expected to follow a normal distribution.
Protein expression and purification
His6eGFP–Kip2, His6eGFP–Kip2-AA, His6mCherry–Bik1
All proteins were expressed in SF9 cells from a pFastBacM13 vector integrated in its corresponding Bacmid system carried by DH10Bac cells (BAC-TO-BAC™ expression system; Invitrogen, Paisley, UK) according to Wasilko et al. (2009). Cells were lysed in 50 mM HEPES pH 7.5, 150 mM NaCl, 3 mM EGTA, 1.5 mM MgCl2, 0.5 mM ATP, 5% glycerol, 0.1% Tween-20, 15 mM sodium pyrophosphate plus protease inhibitors. Purification was carried out in a two steps by using Blue Sepharose (6 Fast Flow) and Ni2+-Sepharose (HisTrap HP, both GE Healthcare Bio-Sciences AB, Uppsala, Sweden).
Mck1-CBP
Yeast cells expressing Mck1-TAP expressed from its genomic locus were lysed in 50 mM Tris pH 7.5, 300 mM NaCl, 1.5 mM MgCl, 0.5 mM DTT, 0.1 mM ATP, 10% glycerol, 0.01% NP-40, complete protease inhibitor and 1 mM β-phosphoglycerate. Bait protein was allowed to bind to IgG-Sepharose (6 Fast Flow, GE Healthcare Bio-Sciences AB, Uppsala, Sweden) and bound protein was eluted by incubating with TEV-protease.
His6Bim1, GST-Kip21-80, GST-EB1Bim1, and MBPKip2ΔC constructs
Cells expressing the corresponding proteins from the pET28 vector in the Rosetta host strain (Merck, Darmstadt, Germany) were lysed in 50 mM Tris pH 7.5, 200 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 0.01% NP-40 plus protease inhibitors (Complete™, Boehringer) and purified using standard conditions. After elution, the proteins were dialysed against the lysis buffer, aliquoted and snap-frozen in liquid nitrogen.
Western blots, cycloheximide-chase, immunoprecipitation and immunostaining, metabolic labelling
Western blots
Protein extraction was performed as described in (Knop et al., 1999) or with glass beads in 50 mM Tris pH 7.6, 150 mM NaCl, 1 mM MgCl2, protease- and phosphatase-inhibitors and 0.1% NP-40 (better resolution of phosphospecies). For visualisation of Kip2 phosphoisoforms cell lysates were analyzed by SDS-PAGE (6%) and western blotting. Antibodies used for western blots were primary antibodies anti-Myc (1:2000, Rabbit polyclonal, Sigma) anti-HA (1:2000, 12CA5, ABGENT, San Diego, CA), anti-GFP (Rabbit polyclonal, gift from Johannes Lechner, Univ. Heidelberg), anti-Arc1 (1:40,000, Rabbit polyclonal, gift from Ed Hurt, (Univ. Heidelberg), anti-His (1:1000, 6-His, Covance, Emeryville, CA), rabbit PAP, polyclonal (1:1000, DakoCytomation) and anti-tubulin (1:1000, DM1A, Sigma-Aldrich, St Louis, MO), and secondary antibodies anti-mouse IgG peroxidase (Fc-part) and anti-rabbit IgG peroxidase (entire molecule) (both 1:10,000, goat, Sigma-Aldrich). For cycloheximide-chase cells were grown in selective medium, protein translation was stopped by addition of 500 µg/ml cycloheximide and samples were analysed by western blotting. For CIP-treatment lysates were treated with 0.5 u/µg protein CIP or phosphatase inhibitor cocktail II (Sigma-Aldrich). The reaction-mix was incubated for 30 min at 37°C.
Immunoprecipitations of HA-Bim1 and HA-Bik1
PGAL1-dependent expression was induced at OD0.5 for 4 h by adding galactose to 2% final concentration. Cells were lysed in 50 mM Tris pH 7.6, 150 mM NaCl, 1 mM MgCl2, protease- and phosphatase-inhibitors and 0.1% NP-40 and proteins precipitated overnight with 2 µg anti-haemagglutinin (HA) antibody (clone 7HA, Sigma-Aldrich) and protein-A–Sepharose.
Immunostaining
Infected SF9 cells were harvested, washed twice with PBS and fixed with 4% paraformaldehyde. After washing with PBS, permeabilisation with 1% Triton X-100 in PBS for 10 min and additional washing with PBS, cells were blocked in PBS+0.1% Tween+1% BSA for 1 h at 4°C. Cells were stained overnight at 4°C with anti-tubulin antibody (1:250 in blocking buffer, DM1A, Sigma-Aldrich), washed three times for 10 min with PBS and incubated for 1 h at 4°C with secondary antibody (anti-mouse Alexa-Fluor-680, 1:1000 in blocking buffer; Molecular Probes/Invitrogen, Invitrogen, Paisley, UK). Cells were again washed three times with PBS and mounted for microscopy.
Metabolic labelling
Cells were grown to an OD0.5 in low phosphate YPD; 50 ml were harvested and re-suspended in 1 ml low-phosphate YPD with 4% glucose. Cells were then grown for an additional hour in the presence of 500 µCi/ml inorganic 32P phosphate. After cell lysis, Kip213myc was immunoprecipitated (see above) using anti-Myc (1:2000, Rabbit polyclonal) antibody for 2 h at 4°C and analyzed by western blot.
In vitro kinase assay and in vitro binding
The His6-tag of eGFP–Kip2 was cleaved off by His6-TEV protease. Uncleaved His6–eGFP as well as the TEV-protease itself was removed with metal affinity beads (Talon IMAC, Clonetech Laboratories, Mountain View, CA). Six pmol of purified eGFP–Kip2 were incubated with 2.5 pmol purified Mck1-CBP for 30 min at 30°C in kinase-buffer (50 mM Tris pH 7.0, 10 mM MgCl2, 0.1 mM EGTA, 1 mM DTT, 100 µM unlabelled ATP, 0.3-0.4 µM 32P y-ATP). For the subsequent in vitro binding assay, an excess of His6mCherry–Bik1 (120 pmol) or His6Bim1 (300 pmol) was added and the buffer adjusted to 150 mM NaCl (plus complete protease inhibitor). Proteins were allowed to bind to metal affinity beads for 1 h at 4°C and beads were washed five times with IP-Buffer (see above). Supernatants and beads were analyzed by SDS-Page followed by western blotting or autoradiography.
MT-sedimentation assay
MT polymerisation of 200 mM tubulin was induced by 5% glycerol and 1 mM GTP for 30 min at 37°C. Tubulin was diluted to 10 mM final concentration in 50 mM Tris pH 7.0, 150 mM KCl supplemented with 10 mM Taxol or 30 µM nocodazole. Polymerised and non-polymerised tubulin was separated by centrifugation at 67,000 g at 28°C through a 40% glycerol cushion. His6eGFP–Kip2 was cleared for 15 min at 67,000 g and 28°C. 7 pmol Kip2 and ∼40 nmol tubulin were incubated for 20 min at 28°C. Bound Kip2 was again separated by centrifugation through a glycerol cushion. Supernatants and MT-containing pellets were analyzed by SDS-PAGE and western blotting as described above.
Acknowledgements
We are grateful to Joe Howard and Vladimir Varga (MPI-CBG Dresden) for their support on the project and Régis Lemaitre (MPI-CBG Dresden) for help with protein expression; Lea Stevermann for the Bim1 protein and critical reading of the manuscript; to J. Reichert for conducting the mass-spectrometry; Yves Barral (ETH Zurich), Jennifer Ortiz (Univ. Heidelberg) and Johannes Lechner (Univ. Heidelberg) for sharing information, strains, plasmids and antibodies; to Ed Hurt and Thomas Söllner (Univ. Heidelberg) for technical support; and to Ulrike Engel, Peter Bankhead and the Nikon Imaging Center (Univ. Heidelberg) for support with microscopy and deconvolution. The Nikon Imaging Center is supported by CellNetworks-Cluster of Excellence (EXC81).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
H.D. and D.L. designed the study and performed experiments, A.N.T. performed experiments, H.D. prepared the figures and D.L. wrote the manuscript.
Funding
H.D. is supported by the Boehringer Ingelheim Foundation, A.N.T. and D.L. were supported by the German Research Foundation (DFG). Deposited in PMC for immediate release.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.166686/-/DC1
References
- Akhmanova A. and Steinmetz M. O. (2008). Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9, 309-322. 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- Akhmanova A., Hoogenraad C. C., Drabek K., Stepanova T., Dortland B., Verkerk T., Vermeulen W., Burgering B. M., De Zeeuw C. I., Grosveld F. et al. (2001). Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 104, 923-935. 10.1016/S0092-8674(01)00288-4 [DOI] [PubMed] [Google Scholar]
- Bardin A. J. and Amon A. (2001). Men and sin: what's the difference? Nat. Rev. Mol. Cell Biol. 2, 815-826. 10.1038/35099020 [DOI] [PubMed] [Google Scholar]
- Bieling P., Laan L., Schek H., Munteanu E. L., Sandblad L., Dogterom M., Brunner D. and Surrey T. (2007). Reconstitution of a microtubule plus-end tracking system in vitro. Nature 450, 1100-1105. 10.1038/nature06386 [DOI] [PubMed] [Google Scholar]
- Bienz M. (2001). Spindles cotton on to junctions, APC and EB1. Nat. Cell Biol. 3, E67-E68. 10.1038/35060140 [DOI] [PubMed] [Google Scholar]
- Biggins S., Severin F. F., Bhalla N., Sasson I., Hyman A. A. and Murray W. A. (1999). The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13, 532-544. 10.1101/gad.13.5.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake-Hodek K. A., Cassimeris L. and Huffaker T. C. (2010). Regulation of microtubule dynamics by Bim1 and Bik1, the budding yeast members of the EB1 and CLIP-170 families of plus-end tracking proteins. Mol. Biol. Cell. 21, 2013-2023. 10.1091/mbc.E10-02-0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning H. and Hackney D. D. (2005). The EB1 homolog Mal3 stimulates the ATPase of the kinesin Tea2 by recruiting it to the microtubule. J. Biol. Chem. 280, 12299-12304. 10.1074/jbc.M413620200 [DOI] [PubMed] [Google Scholar]
- Brunner D. and Nurse P. (2000). CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102, 695-704. 10.1016/S0092-8674(00)00091-X [DOI] [PubMed] [Google Scholar]
- Buttrick G. J. and Wakefield J. G. (2008). PI3-K and GSK-3: Akt-ing together with microtubules. Cell Cycle 7, 2621-2625. 10.4161/cc.7.17.6514 [DOI] [PubMed] [Google Scholar]
- Carvalho P., Gupta M. L. Jr, Hoyt M. A. and Pellman D. (2004). Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev. Cell 6, 815-829. 10.1016/j.devcel.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Charles J. F., Jaspersen S. L., Tinker-Kulberg R. L., Hwang L., Szidon A. and Morgan D. O. (1998). The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr. Biol. 8, 497-507. 10.1016/S0960-9822(98)70201-5 [DOI] [PubMed] [Google Scholar]
- Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J. G., De Wulf P. et al. (2008). Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133, 427-439. 10.1016/j.cell.2008.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham F. R. and Hoyt M. A. (1997). Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 138, 1041-1053. 10.1083/jcb.138.5.1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doble B. W. and Woodgett J. R. (2003). GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 116, 1175-1186. 10.1242/jcs.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doble B. W. and Woodgett J. R. (2007). Role of glycogen synthase kinase-3 in cell fate and epithelial-mesenchymal transitions. Cells Tissues Organs 185, 73-84. 10.1159/000101306 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. and Hall A. (2003). Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature 421, 753-756. 10.1038/nature01423 [DOI] [PubMed] [Google Scholar]
- Fiol C. J., Haseman J. H., Wang Y., Roach P. J., Roeske R. W., Kowalczuk M. and DePaoli-Roach A. A. (1988). Phosphoserine as a recognition determinant for glycogen synthase kinase-3: phosphorylation of a synthetic peptide based on the G-component of protein phosphatase-1. Arch. Biochem. Biophys. 267, 797-802. 10.1016/0003-9861(88)90089-6 [DOI] [PubMed] [Google Scholar]
- Gould C. M., Diella F., Via A., Puntervoll P., Gemund C., Chabanis-Davidson S., Michael S., Sayadi A., Bryne J. C., Chica C. et al. (2010). ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 38, D167-D180. 10.1093/nar/gkp1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt L. J., Tuch B. B., Villen J., Johnson A. D., Gygi S. P. and Morgan D. O. (2009). Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682-1686. 10.1126/science.1172867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., Gouveia S. M., Weisbrich A., Damberger F. F., Bhavesh N. S., Jawhari H., Grigoriev I., van Rijssel F. J. A., Buey R. M., Lawera A. et al. (2009). An EB1-binding motif acts as a microtubule tip localization signal. Cell 138, 366-376. 10.1016/j.cell.2009.04.065 [DOI] [PubMed] [Google Scholar]
- Hotz M., Leisner C., Chen D., Manatschal C., Wegleiter T., Ouellet J., Lindstrom D., Gottschling D. E., Vogel J., Barral Y. (2012). Spindle pole bodies exploit the mitotic exit network in metaphase to drive their age-dependent segregation. Cell 148, 958-972. 10.1016/j.cell.2012.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.-K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S. and O'Shea E. K. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686-691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Huyett A., Kahana J., Silver P., Zeng X. and Saunders W. S. (1998). The Kar3p and Kip2p motors function antagonistically at the spindle poles to influence cytoplasmic microtubule numbers. J. Cell Sci. 111, 295-301. [DOI] [PubMed] [Google Scholar]
- Jaulin F. and Kreitzer G. (2010). KIF17 stabilizes microtubules and contributes to epithelial morphogenesis by acting at MT plus ends with EB1 and APC. J. Cell Biol. 190, 443-460. 10.1083/jcb.201006044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.-Y., Wang X., Wu Y., Doble B. W., Patel S., Woodgett J. R. and Snider W. D. (2009). GSK-3 is a master regulator of neural progenitor homeostasis. Nat. Neurosci. 12, 1390-1397. 10.1038/nn.2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K. and Schiebel E. (1999). Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15, 963-972. [DOI] [PubMed] [Google Scholar]
- Komarova Y. A., Akhmanova A. S., Kojima S.-I., Galjart N. and Borisy G. G. (2002). Cytoplasmic linker proteins promote microtubule rescue in vivo. J. Cell Biol. 159, 589-599. 10.1083/jcb.200208058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Lyle K. S., Gierke S., Matov A., Danuser G. and Wittmann T. (2009). GSK3beta phosphorylation modulates CLASP-microtubule association and lamella microtubule attachment. J. Cell Biol. 184, 895-908. 10.1083/jcb.200901042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D., Kusch J., Grava S., Vogel J. and Barral Y. (2003). Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112, 561-574. 10.1016/S0092-8674(03)00119-3 [DOI] [PubMed] [Google Scholar]
- Maekawa H., Usui T., Knop M. and Schiebel E. (2003). Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 22, 438-449. 10.1093/emboj/cdg063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah A. S., Elia A. E. H., Devgan G., Ptacek J., Schutkowski M., Snyder M., Yaffe M. B. and Deshaies R. J. (2005). Substrate specificity analysis of protein kinase complex Dbf2-Mob1 by peptide library and proteome array screening. BMC Biochem. 6, 22 10.1186/1471-2091-6-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. K. and Rose M. D. (1998). Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140, 377-390. 10.1083/jcb.140.2.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. K., Cheng S.-C. and Rose M. D. (2000). Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Cell 11, 2949-2959. 10.1091/mbc.11.9.2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. K., Stuchell-Brereton M. D. and Cooper J. A. (2009). Function of dynein in budding yeast: mitotic spindle positioning in a polarized cell. Cell Motil. Cytoskeleton 66, 546-555. 10.1002/cm.20364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F., Diamantopoulos G. S., Stalder R. and Kreis T. E. (1999). CLIP-170 highlights growing microtubule ends in vivo. Cell 96, 517-527. 10.1016/S0092-8674(00)80656-X [DOI] [PubMed] [Google Scholar]
- Roach P. J. (1991). Multisite and hierarchal protein phosphorylation. J. Biol. Chem. 266, 14139-14142. [PubMed] [Google Scholar]
- Roberts A. J., Goodman B. S. and Reck-Peterson S. L. (2014). Reconstitution of dynein transport to the microtubule plus end by kinesin. Elife 3, e02641 10.7554/eLife.02641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinfeld B., Albert I., Porfiri E., Fiol C., Munemitsu S. and Polakis P. (1996). Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly. Science 272, 1023-1026. 10.1126/science.272.5264.1023 [DOI] [PubMed] [Google Scholar]
- Sheeman B., Carvalho P., Sagot I., Geiser J., Kho D., Hoyt M. A. and Pellman D. (2003). Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 13, 364-372. 10.1016/S0960-9822(03)00013-7 [DOI] [PubMed] [Google Scholar]
- Shirayama M., Zachariae W., Ciosk R. and Nasmyth K. (1998). The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 17, 1336-1349. 10.1093/emboj/17.5.1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M. O. and Akhmanova A. (2008). Capturing protein tails by CAP-Gly domains. Trends Biochem. Sci. 33, 535-545. 10.1016/j.tibs.2008.08.006 [DOI] [PubMed] [Google Scholar]
- Tan K., Roberts A. J., Chonofsky M., Egan M. J. and Reck-Peterson S. L. (2014). A microscopy-based screen employing multiplex genome sequencing identifies cargo-specific requirements for dynein velocity. Mol. Biol. Cell 25, 669-678. 10.1091/mbc.E13-09-0557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe A., Ray-Sinha A., Staples O. D. and Taylor S. S. (2007). GSK-3 inhibitors induce chromosome instability. BMC Cell Biol. 8, 34 10.1186/1471-2121-8-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., Shah K., Shokat K. M. and Morgan D. O. (2003). Targets of the cyclin-dependent kinase Cdk1. Nature 425, 859-864. 10.1038/nature02062 [DOI] [PubMed] [Google Scholar]
- Vaughan K. T. (2005). TIP maker and TIP marker; EB1 as a master controller of microtubule plus ends. J. Cell Biol. 171, 197-200. 10.1083/jcb.200509150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield G. J., Stephens J. D. and Tavaré M. J. (2003). A role for glycogen synthase kinase-3 in mitotic spindle dynamics and chromosome alignment. J. Cell Sci. 116, 637-646. 10.1242/jcs.00273 [DOI] [PubMed] [Google Scholar]
- Wasilko D. J., Lee S. E., Stutzman-Engwall K. J., Reitz B. A., Emmons T. L., Mathis K. J., Bienkowski M. J., Tomasselli A. G. and Fischer H. D. (2009). The titerless infected-cells preservation and scale-up (TIPS) method for large-scale production of NO-sensitive human soluble guanylate cyclase (sGC) from insect cells infected with recombinant baculovirus. Protein Expr. Purif. 65, 122-132. 10.1016/j.pep.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Noritake J., Kakeno M., Matsui T., Harada T., Wang S., Itoh N., Sato K., Matsuzawa K., Iwamatsu A. et al. (2009). Phosphorylation of CLASP2 by GSK-3{beta} regulates its interaction with IQGAP1, EB1 and microtubules. J. Cell Sci. 122, 2969-2979. 10.1242/jcs.046649 [DOI] [PubMed] [Google Scholar]
- Wittmann T. and Waterman-Storer C. M. (2005). Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3beta in migrating epithelial cells. J. Cell Biol. 169, 929-939. 10.1083/jcb.200412114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolyniak M. J., Blake-Hodek K., Kosco K., Hwang E., You L. and Huffaker T. C. (2006). The regulation of microtubule dynamics in Saccharomyces cerevisiae by three interacting plus-end tracking proteins. Mol. Biol. Cell 17, 2789-2798. 10.1091/mbc.E05-09-0892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D. and Pan W. (2010). GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem. Sci. 35, 161-168. 10.1016/j.tibs.2009.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumbrunn J., Kinoshita K., Hyman A. A. and Nathke I. S. (2001). Binding of the adenomatous polyposis coli protein to microtubules increases microtubule stability and is regulated by GSK3 beta phosphorylation. Curr. Biol. 11, 44-49. 10.1016/S0960-9822(01)00002-1 [DOI] [PubMed] [Google Scholar]