Abstract
Janthinobacterium lividum is a Gram-negative bacterium able to produce violacein, a pigment with antimicrobial and antitumor properties. Janthinobacterium lividum colonizes the skin of some amphibians and confers protection against fungal pathogens. The mechanisms underlying this association are not well understood. In order to identify the advantages for the bacterium to colonize amphibian skin we sequenced Janthinobacterium lividum strain MTR, a strain isolated from Cajón del Maipo, Chile. The strain has capnophilic behavior, with growth favored by high concentrations (5 %) of carbon dioxide. Its genome is 6,535,606 bp in size, with 5,362 coding sequences and a G + C content of 62.37 %. The presence of genes encoding for products that participate in the carbon fixation pathways (dark CAM pathways), and the entire set of genes encoding for the enzymes of the glyoxylate cycle may explain the capnophilic behavior and allow us to propose that the CO2 secreted by the skin of amphibians is the signal molecule that guides colonization by Janthinobacterium lividum.
Electronic supplementary material
The online version of this article (doi:10.1186/s40793-015-0104-z) contains supplementary material, which is available to authorized users.
Keywords: Violacein, Janthinobacterium lividum, Carbon dioxide, Capnophilic, Carbon fixation
Introduction
Janthinobacterium lividum is a betaproteobacterium common in soil and water in cold-temperate regions [1, 2]. This bacterium produces violacein, a compound with antibiotic and antiviral properties that is toxic for eukaryotic cells [3, 4]. Although the biological function of violacein for Janthinobacterium lividum remains unknown, it has been proposed that the compound provides protection against bacterial predators [5, 6]. Janthinobacterium lividum is also present in the skin microbiome of some amphibians, where its presence and production of violacein confers protection against the fungal pathogen Batrachochytrium endrobatidis [7–9]. This bacterial-amphibian relationship has been classified as symbiotic (mutualism) [7, 10, 11], although the benefit for the bacteria has not been clearly established. Carbon dioxide is secreted by amphibian skin alveoli. Although carbon dioxide is a highly abundant, its role as a signaling molecule in bacteria has not been well characterized. Janthinobacterium lividum strain MTR is an isolate from our laboratory that grows best in carbon dioxide concentrations higher than 1 % (with an optimum of 5 %). These carbon dioxide concentrations are close to the ranges (1.2–2.5 % CO2) observed in amphibian skin alveoli [12]. Because this property has not been described before in other strains of Janthinobacterium lividum, we focused on sequencing the genome of this bacterial strain to elucidate the function of the carbon dioxide as a signal molecule.
Organism information
Classification and features
Janthinobacterium lividum is a rod-shaped, Gram-negative, motile, aerobic bacterium, commonly isolated from of soil and water of cold regions, such as mountains or glaciers. It is a heterotrophic bacterium with a temperature range of growth between 4 and 30 °C, with a optimum growth at around 25 °C [2], suggesting that is a psychrotolerant bacterium. After several day of cultivation on solid nutrient medium, most of the strains form rubbery colonies that are violet colored due to the production of violacein. The MTR strain of Janthinobacterium lividum forms violet-colored colonies with a rubber-like aspect that cannot grow at temperatures above 30 °C, with an optimum growth temperature of 25 °C (Table 1). Under aerobic conditions, strain MTR grows as a planctonic culture without violacein production, while in static cultures it forms a thick dark violet biofilm in the air-broth interface. In static cultures, Janthinobacterium lividum MTR also shows capnophilic behavior that has not been described before among other members of the Janthinobacterium genus. Comparison of bacterium grown on Luria, Todd Hewitt, nutrient, Sabouraud, Mueller Hinton, and MacConkey solid media shows that colonies of Janthínobacterium lividum strain MTR grow best on Todd Hewitt, nutrient and MacConkey plates. No growth was observed on Sabouraud plates. Growth was more rapid on plates of nutrient medium, where violacein production was evident at only 24 h of culture at 25 °C, while violacein production was observed on plates of Luria and Mueller Hinton medium at 72 h of growth. In all tested culture media Janthinobacterium lividum MTR formed bright and convex colonies in the first 72 h. At 72 h, the colony morphology began to change, becoming dark violet, bright, rugous and raised by seven days of cultivation.
Table 1.
Classification and general features of Janthinobacterium lividum strain MTRT [40]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [41] | |
| Phylum Proteobacteria | TAS [42] | ||
| Class Betaproteobacteria | TAS [43, 44] | ||
| Order Burkholderiales | TAS [45] | ||
| Family Oxalobacteraceae | TAS [1, 43] | ||
| Genus Janthinobacterium | TAS [1] | ||
| Species Janthinobacterium lividum | TAS [1] | ||
| (Type) strain: Strain T MTR | |||
| Gram stain | Negative | TAS [30] | |
| Cell shape | Rod | TAS [1] | |
| Motility | Motile | TAS [30] | |
| Sporulation | Nonsporulated | TAS [1] | |
| Temperature range | 4–30 °C | TAS [46, 47] | |
| Optimum temperature | 25 °C | TAS [46, 47] | |
| pH range; Optimum | 5.0–8.0; 7.0 | IDA | |
| Carbon source | Heterotrophic | TAS [47] | |
| MIGS-6 | Habitat | Aquatic and soil | TAS [46] |
| MIGS-6.3 | Salinity | Non reported | NAS |
| MIGS-22 | Oxygen requirement | Aerobic | TAS [30] |
| MIGS-15 | Biotic relationship | Free-living | TAS [47] |
| MIGS-14 | Pathogenicity | Non-pathogen | NAS |
| MIGS-4 | Geographic location | Cajón del Maipo, Santiago Metropolitan Region, Chile | NAS |
| MIGS-5 | Sample collection | 2009 | IDA |
| MIGS-4.1 | Latitude | −33.56 | IDA |
| MIGS-4.2 | Longitude | −70.46 | IDA |
| MIGS-4.4 | Altitude | Non reported | NAS |
aEvidence codes - IDA Inferred from Direct Assay, TAS Traceable Author Statement (i.e. a direct report exists in the literature), NAS Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology Project [48]
Ultrastructural analysis by electron microscopy of ultrathin sections of Janthinobacterium lividum strain MTR confirmed the presence of an exopolymeric substance that would be involved in the formation of biofilms (see arrows in Fig. 1, c and d). High magnification showed the clearly layered structure of the Gram-negative cell envelope or outer membrane, the peptidoglycan cell wall, and the cytoplasmic or inner membrane (see arrows in Fig. 1a). It was also possible to observe high concentrations of ribosomes in the bacterial cytoplasm (see arrows in Fig. 1b), indicating a high level of protein biosynthesis. The bacterial cells are rod-shaped, approximately 2.5 μm long and 0.65 μm in diameter (Fig. 1).
Fig. 1.
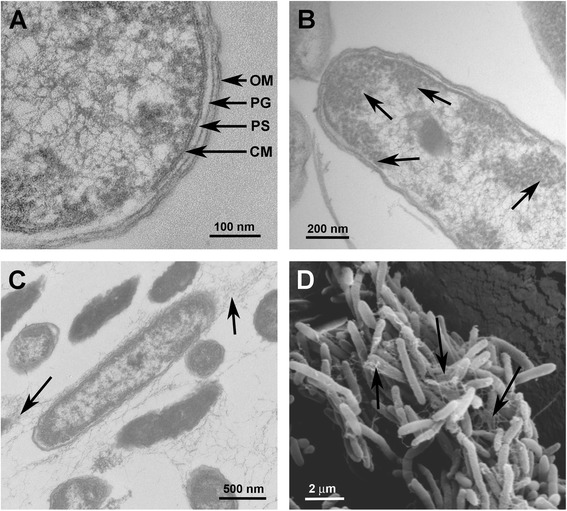
Electron micrographs of the Janthinobacterium lividum strain MTR. a, b and c, ultrathin sections visualized by transmission electron microscope. d, sample of bacterial cells shading with metallic gold and visualized by scanning electron microscope. The arrows in (a) indicate the outer membrane (OM), the peptidoglycan (PG), the periplasmic space (PS) and the cytoplasmic membrane (CM). The arrows in (b) show areas of high concentration of ribosomes. The arrows in (c) and (d) indicate regions where exopolymeric substances can be seen clearly (EPS)
Antibiogram profiles using the Kirby-Bauer method [13] showed that Janthinobacterium lividum MTR is resistant to clindamycin, tetracycline, ampicillin, penicillin, cefuroxime, cephaloridine, and piperacillin.
Phylogenetic reconstruction using the sequences of 16S rRNA obtained from the genome sequence of Janthinobacterium lividum MTR and EZtaxon database [14] confirmed that our strain belongs to the domain Bacteria, the phylum Proteobacteria, the class Betaproteobacteria, the order Burkholderiales, the family Oxalobacteraceae, the genus Janthinobacterium, and the species Janthinobacterium lividum, which is closely related to Janthinobacterium lividum BP01 isolated in Alaskan soil (Fig. 2) [15]. The 16S rRNA sequence of the MTR and BP01 strains of Janthinobacterium lividum share 100 % identity, although the 16S rRNA sequence of BP01 is shorter than it counterpart in the MTR strain.
Fig. 2.
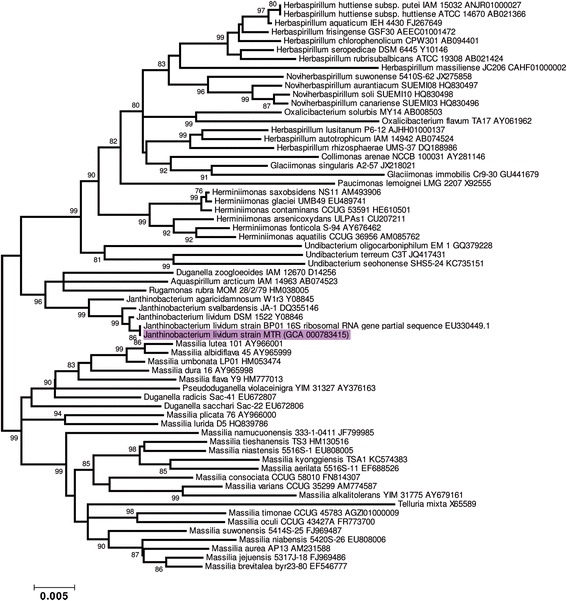
Phylogenetic relationship of the Janthinobacterium lividum strain MTR to other members of the Oxalobacteraceae family: The evolutionary history was inferred using the neighbor-joining method [49]. The optimal tree with a branch length sum = 0.15329841 is shown. The percentage of replicate trees (higher than 75) in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [50]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method [51] and are in the units of the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). The analysis involved 34 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1429 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 [52]. The Janthinobacterium lividum strain MTR is highlighted in violet
Janthinobacterium lividum strain MTR was isolated from soil samples (5 g) taken from a forest of native Drimys winteri var. chilensis A. Gray trees located in the Maipo Valley in central Chile. Samples were dissolved in 40 mL of sterile distilled water and incubated at 40 °C for 1 h to diminish the load of mesophile bacteria. This suspension (100 μL) was mixed with 3 mL of molten soft agar of salt medium (KNO3 2.0 g/L, MgSO4 ·7H2O 2.0 g/L, K2HPO4 2.0 g/L, MgSO4·7H2O 0.05 g/L, CaCO3 0.02 g/L, FeSO4·7H2O 0.01 g/L) and poured over salt medium plates supplemented with starch 1 %, casein 0.03 % and cycloheximide (10 μg/mL). After 10 days of culture at 25 °C, blue-violet colonies were streaked over plates of nutritive medium and grown at 25 °C. Each isolate was replated at least 4 times until an axenic culture was obtained. The initial identity of bacterial isolate was determined by 16S rRNA sequencing. Each isolate of Janthinobacterium lividum was grown in nutrient media in 2 mL static cultures at 25 °C, using carbon dioxide concentrations of 5 % and 0.03 %. After 48 h, growth was determined by CFU using the microdrop method. Janthinobacterium lividum strain MTR showed the best growth rate at 5 % of carbon dioxide and consequently this colony was selected for genomic sequencing.
Genome sequencing information
Genome project history
Janthinobacterium lividum strain MTR was selected for sequencing because of its capnophilic behavior, with optimum growth at carbon dioxide concentrations close to those estimated to be present on the skin of amphibian due to gas exchange. The genome project and draft genome sequence of the MTR strain of Janthinobacterium lividum were deposited at DDBJ/EMBL/GenBank under the accession code of the master record JRRH00000000, nucleotide sequences JRRH01000001-JRRH01000114. Sequencing was performed in March 2014 at the Genomic and Bioinformatic Center of the Universidad Mayor and released on November 14th 2014. Assembly and annotation were done by the bioinformatic support unit at the Universidad de Santiago and were completed in October 2014. The draft genome shows a mean coverage of 30x. Table 2 provides a summary of the project.
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | 30 × |
| MIGS-28 | Libraries used | Paired-end |
| MIGS 29 | Sequencing platforms | Illumina MiSeq |
| MIGS 31.2 | Fold coverage | 144 × |
| MIGS 30 | Assemblers | Velvet v.1.2.10 |
| MIGS 32 | Gene calling method | RAST |
| Locus tag | NC77 | |
| Genbank ID | JRRH01000001-JRRH01000114 | |
| GenBank date of release | 11/14/2014 | |
| GOLD ID | Gp0112111 | |
| BIOPROJECT | PRJNA263254 | |
| MIGS 13 | Source material identifier | MTR1474 |
| Project relevance | Environment |
Growth conditions and genomic DNA preparation
Janthinobacterium lividum strain MTR was grown in nutritive broth at 25 °C by 48 h. Total DNA was isolated from 3 mL of liquid culture using the MasterPureTM DNA purification kit (Epicentre®). Cells were first collected by centrifugation at 13,000 g for 5 min, resuspended in 300 μL of tissue and cell lysis solution and treated with 100 μg of lyzozyme at 37 °C for an hour. After lyzozyme treatment, cell lysis was achieved by adding 50 μg of proteinase K and incubation at 37 °C for an hour with brief agitation every 5 min. After lysis, RNA was removed by treatment with 10 μg of RNAse A for 5 min at 37 °C. Protein and cell debris was removed by adding 175 μL of MPC solution, and further centrifugation at 13,000 g for 10 min at 4 °C. The DNA was precipitated by adding 300 μL of isopropanol per each 400 μL of supernatant and was collected by centrifugation at 13000 g for 10 min at 4 °C. The pellet containing DNA was washed with ethanol 70 %, dried using a SpeedVac and resuspended in 50 μL of miliQ water. The DNA was stored at −20 °C until its subsequent use.
Genome sequencing and assembly
The genome of the Janthinobacterium lividum strain MTR was sequenced using Illumina MiSeq, a sequencing-by-synthesis technology, which generates 2 × 250 reads (paired-ends). The reads were analyzed by FastQC v.0.10.1 [16] and low-quality sequences were removed by Trimmomatic v.0.32 before assembly. The trimmed sequences were assembled de novo using an assembling coverage of 120x, with the Velvet Assembler program (v1.2.10 p).
Genome annotation
Annotations was performed using RAST [17], identifying a total of 5,774 protein encoding genes. The tRNAs were identified by tRNAscan-SE v.1.23 [18] and the rRNAs by RNAmmerv.1.2Server [19]. The proteins identified by RAST were used to determine clusters of orthologous groups with the WebMGA server [20], CRISPRs were evaluated by CRISPRfinder [21], transmembrane helix domains were determined by TMHMM 2.0c [22] and signal peptides were estimated by SignalP v.4.0 [23].
Genome properties
The draft genome properties and statistics of Janthinobacterium lividum strain MTR (Accession CP006828) are shown in Tables 3 and 4. The assembly the MTR strain resulted in 114 contigs, with sizes ranging from 209 to 472,391 bp (N50, 154,343 bp). The total draft genome had a length of 6,535,606 bp, with a G + C content of 62.37 %. The draft genome was shown to encode 5,876 putative genes, 5,362 protein-coding genes, 408 pseudogenes, and 106 genes for RNAs (87 genes encoding for tRNA, two complete 5S-16S-23S operons) (Fig. 3). Approximately 67.69 % of the genes encode for putative proteins with known functions (Table 3). Table 4 presents the distribution of genes in functional COG categories.
Table 3.
Genome statistics
| Attribute | Value | % of total |
|---|---|---|
| Genome size (bp) | 6,535,606 | 100.00 |
| DNA coding (bp) | 5,330,283 | 81.56 |
| DNA G + C (bp) | 4,076,257 | 62.37 |
| DNA scaffolds | 114 | 100.00 |
| Total genes | 5,876 | 100.00 |
| Protein coding genes | 5,362 | 91.25 |
| RNA genes | 106 | 1.80 |
| Pseudo-genes | 408 | 6.94 |
| Genes in internal clusters | 2,639 | 44.91 |
| Genes with predicted functions | 3,978 | 67.69 |
| Genes assigned to COGs | 4,181 | 79.94 |
| Genes with Pfam domains | 4,268 | 79.59 |
| Genes with signal peptides | 1,250 | 21.27 |
| Genes with transmembrane helices | 1,065 | 18.12 |
| CRISPR repeats | 0 | 0.00 |
Table 4.
Number of genes associated with general functional COG categories
| Code | Value | % age | Description |
|---|---|---|---|
| J | 183 | 3.41 | Translation, ribosomal structure and biogenesis |
| A | 3 | 0.06 | RNA processing and modification |
| K | 474 | 8.84 | Transcription |
| L | 148 | 2.76 | Replication, recombination and repair |
| B | 3 | 0.06 | Chromatin structure and dynamics |
| D | 39 | 0.73 | Cell cycle control, Cell division, chromosome partitioning |
| V | 82 | 1.53 | Defense mechanisms |
| T | 483 | 9.01 | Signal transduction mechanisms |
| M | 272 | 5.07 | Cell wall/membrane biogenesis |
| N | 190 | 3.54 | Cell motility |
| U | 178 | 3.32 | Intracellular trafficking and secretion |
| O | 181 | 3.38 | Posttranslational modification, protein turnover, chaperones |
| C | 273 | 5.09 | Energy production and conversion |
| G | 302 | 5.63 | Carbohydrate transport and metabolism |
| E | 374 | 6.98 | Amino acid transport and metabolism |
| F | 85 | 1.59 | Nucleotide transport and metabolism |
| H | 179 | 3.34 | Coenzyme transport and metabolism |
| I | 172 | 3.21 | Lipid transport and metabolism |
| P | 291 | 5.43 | Inorganic ion transport and metabolism |
| Q | 98 | 1.83 | Secondary metabolite biosynthesis, transport and catabolism |
| R | 534 | 9.96 | General function prediction only |
| S | 441 | 8.22 | Function unknown |
| - | 1,181 | 22.03 | Not in COGs |
The total is based on the total number of protein coding genes in the genome
Fig. 3.
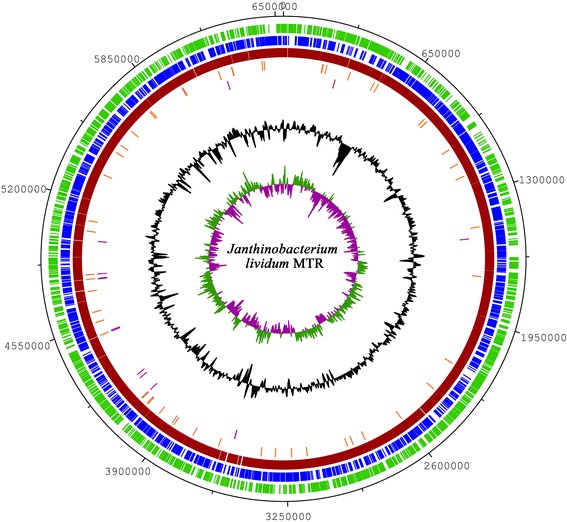
Circular representation of Janthinobacterium lividum strain MTR genome: The figure shows the forward CDS (green), reverse CDS (blue), total CDS, coding genome (red), tRNAs (orange), rRNAs (violet), GC content (black), and GC skew (purple/olive)
Insights from the genome sequence
Currently, the genomes of eight strains of Janthinobacterium spp. have been deposited in the GenBank database. The strains Janthinobacterium sp. CG3, Janthinobacterium sp. HH01, Janthinobacterium sp. Marseille, Janthinobacterium sp. RA13, Janthinobacterium agaricidamnosumNBRC102515, Janthinobacterium lividum PAMC25724, Janthinobacterium lividum RIT308 and Janthinobacterium lividum MTR have been sequenced to different levels. A complete genome has only been reported for Janthinobacterium sp. Marseille and Janthinobacterium agaricidamnosumNBRC102515. Although all these bacteria have been classified as belonging to the Janthinobacterium genus, detailed analysis of the 16S rRNA sequence using EZTaxon showed that Janthinobacterium sp. HH01 and Janthinobacterium sp. Marseille are closer to others members of the Oxalobactericeae family, Duganella phyllosphaerae and Herminiimonas glaciei, respectively. The 16S rRNA of Janthinobacterium sp. CG3 showed 98.43 % identity with 16S rRNA of Janthinobacteriumsvalbardensis, while the 16S rRNA of Janthinobacterium sp. RA13 had 100 % identity with the 16S rRNA of Janthinobacterium lividumDSM1522. This analysis also indicated that Janthinobacterium lividum strain MTR has a 99.86 % of identity with the 16S rRNA of Janthinobacterium lividumDSM1522. This suggest that there are only four genomes of the bonafide species Janthinobacterium in Genbank. Comparisons of these genomes shows that Janthinobacterium lividum strain MTR has a similar GC percentage (62.4 %) to those of the strains RIT308 and RA13, and higher than that of the strain PAMC25724. Janthinobacterium lividum strain MTR has the largest genome, with 6.5 Mb, followed by the strains RA13 (6.4 Mb), RIT308 (6.2 Mb) and PAMC25724 (4.98 Mb). Although Janthinobacterium lividum strain MTR has the largest genome among lividum strains, it encodes for only 5362 proteins, fewer than the 5712 and 5431 putative proteins encoded in RA13 and RIT308 strains, respectively. Comparison of the proteins encoded in Janthinobacterium lividum strain MTR shows that our strain is most closely related to Janthinobacterium lividum RIT308. Approximately 45.76 % of the putative proteins encoded in Janthinobacterium lividum MTR have their closest homologues in Janthinobacterium lividum RIT308, while 42.9 % and 4.70 % of genes have their closest homologues in Janthinobacterium sp RA13 and Janthinobacterium lividum PAMC25724, respectively. In relation to strain specific genes, 12 % of putative protein encoded by our strain share less than 80 % of identity with proteins encoded in the genome of others members of the lividum species. Whole genome comparison with blastN showed similar results. Janthinobacterium agaricidamnosum, Janthinobacterium sp. GC3, Janthinobacterium lividum RIT308, Janthinobacterium sp. RA13, and Janthinobacterium lividum PAMC25724 form a clade in which Janthinobacterium lividum MTR is closer to the bacterial strains Janthinobacterium sp. RA13 and Janthinobacterium lividum RIT308. As was predicted by the 16S rRNA sequence analysis, whole genome comparison revealed that Janthinobacterium HH01 is closer to a member of Duganella while Janthinobacterium sp. Marseille is closer to a member of the Herminiimonas group (Additional file 1: Figure S1). Comparison using ANI (Average Nucleotide Index) indicated that Janthinobacterium lividum strain MTR share a 92.68 % and 92.38 % with RA13 and RIT308 strains, respectively. These values are below of the threshold percentage (95 %) to be considered members of the same species (Additional file 2: Table S2). Interesting, among the genomes analyzed, ANI values higher than 94 % were not observed. These results suggest that classification inside of Janthinobacterium genus must be carefully analyzed and reconsidered in future works.
Resistance to penicillin, first- and second-generation carbapenems and cephalosporins, may be due to the presence of a gene homologous to THIN-B (KHA75569.1), which is a broad spectrum β-lactamase [24]. Resistance to tetracycline may be due to the high number of efflux pumps identified in the genome (KHA80475.1, KHA80070.1, KHA80071.1, KHA79872.1, KHA78654.1, KHA78419.1, KHA78456.1, KHA77902.1, KHA76948.1, KHA76949.1, EZP37289.1, KHA75757.1) [25].
Extended insights
Comparison of metabolic characteristics deduced from COGs to those of other sequenced species of Janthinobacterium showed that Janthinobacterium lividum strain MTR has more genes in the categories C (energy production and conversion) and P (inorganic ion transport and metabolisms). With 279 genes in category C, our strain has the largest number of genes related to electron transport (citocromes, plastocianin, etc.) and enzymes with putative coenzyme F420-dependent N5,N10-methylene tetrahydromethanopterin reductase activity. These enzymes are present in methanogenic microorganisms and reduce CO2 to methane [26]. The large number of genes related to this function may explain the high growth rate of this strain observed in CO2 rich atmospheres, in which CO2 could serve as an alternative electron acceptor. In agreement with this observation, our strain has a large number of genes in category P encoding for ABC-type nitrate/sulfonate/bicarbonate transport systems and ferric uptake, suggesting that this strain has high requirements of CO2 and electron transport.
The reconstruction of metabolic pathways using KEGG showed the presence of enzymes that fix CO2 in C4 plants in dark CAM (Crassulacean acid metabolism). This pathway fixes CO2 in two steps, the first of which is catalyzed by phosphoenolpyruvate carboxylase and fixes CO2 through the formation of oxaloacetate from PEP and CO2. The second step is catalyzed by malate dehydrogenase and transforms oxaloacetate into malate [27]. In Janthinobacterium lividum strain MTR, this pathway may be used as an anaplerotic reaction to restore or increase the molecules that make up the TCA cycle, which improves metabolism under aerobic condition. Another possible role of the CAM pathway is that excess malate is used to produce piruvate, the molecule that initiates gluconeogenesis. Janthinobacterium lividum strain MTR encodes for a decarboxylating malate dehydrogenase (E1.1.1.39) that converts malate into pyruvate [28]. However, in this step, one carbon atom is lost as CO2 so that a positive carbon balance is not expected. Interestingly, our strain also encodes for the enzymes that make up the glyoxylate cycle, which facilitates the conversion of acetyl-CoA into malate and allows for avoidance of the decarboxylation steps of the TCA cycle. Malate in turn converts into pyruvate, which initiates gluconeogenesis [29]. These steps allow for retaining three quarters of the carbon atoms. Because the CAM pathway fixes one carbon atom, its coupling with the glyoxylate cycle should increase the fraction of carbon retained in gluconeogenesis. In the stationary phase, Janthinobacterium lividum forms a strong biofilm that is rich in exopolysaccharides [30]. The increase in TCA and gluconeogenesis metabolism by the combination of the CAM pathway, and glyoxylate cycle may improve the production of exopolysaccharide in the stationary phase in which complex carbon sources are not available. In fact, our strain contains all the enzymes required for glycolysis and gluconeogenesis so that it is highly plausible that our bacterium combines these pathways. Interestingly, the glyoxylate cycle is a virulence factor in some pathogenic microorganisms. Inhibition or deletion of isocitrate lyase, the key enzyme of glyoxylate cycle, reduces the survival of fungi and mycobacteria in hosts, and has been the target for designing novel antimicrobial agents [31–34].
Although capnophilic behavior is present in several bacteria, among them human pathogens such as Helicobacterpilory, Campylobacter jejuni subsp jejuni and Aggregatibacter spp, the exact role of CO2 requirement is not well understand [35–37]. Several capnophilic bacteria are microaerophilic and do not grow well in high concentrations of O2, so that O2 displacement by CO2 may explain this characteristic. Interestingly Mannheimiasucciniciproducens, a heterotrophic bacterium incapable of aerobic respiration, requires CO2 to produce fumarate from PEP. Mannheimiasucciniciproducens lacks of some enzymes required for oxidative phosphorylation so that electron flux is maintained using fumarate as electron acceptor [38, 39]. In Campylobacterjujuni and Helicobacterpilori, the presence of PFOR:FldA:FqrB complexes generates pyruvate via CO2 fixation, which may explain the capnophilic behavior of the two bacteria. Since Janthinobacterium lividum strain MTR has the entire set of enzymes required for phosphorylative oxidation using O2 as an electron acceptor and does not encode a pyruvate ferredoxin oxidoreductase, our data suggest that J. lividum strain MTR and others species of Janthinobacterium are capnophilic due to different mechanisms, in which CO2 improves the metabolic rate of TCA and gluconeogenesis.
Conclusions
The genome sequence of Janthinobacterium lividum strain MTR revealed the presence of enzymes that allow carbon fixation (dark CAM pathway). These enzymes, in combination with the glyoxylate cycle, may increase the efficiency of gluconeogenesis by using intermediaries from the TCA cycle, which would explain capnophilic behavior. To our knowledge, this mechanism has not been proposed before and could be a strategy to improve growth in regions with high amounts of O2 and CO2, such as the skin of amphibians. Detailed comparison should be made of the Janthinobacterium lividum strain MTR genome to those of other sequenced strains of Janthinobacterium lividum or other capnophilic bacteria to determine if this proposed mechanism is common among these bacteria.
Acknowledgements
This work was supported by projects 020943TR (DICYT-USACH) and PBCT PDA20 (CONICYT) to MT, DIULA 09/2014 to AG, Project DGT-2014 AC to AC, Grants USA1298 to NV, and Fund Research Support 30/2014 DPI Universidad Autónoma de Chile to GC.
Abbreviations
- TCA
Tricarboxylic Acids
- PEP
Phosphoenolpiruvate
- CAM
Crassulacean acid metabolism
Additional files
Whole genome comparison among bacteria from Janthinobacterium genus. The figure shows the whole genome comparison of bacteria belonging to the genuses Janthinobacterium, Duganella and Herminiimonas. Comparison was performed with Gegenees software (PLoS One. 2012;7(6):e39107. doi:10.1371/journal.pone.0039107). Parameters were fixed as follow: fragments = 200 bp, overlapping regions = 100 pb. Genome comparisons were done with blastN algorithm. Panel A shows the average normalized BLAST scores for all fragments in each genome (compared against other genomes). Panel B shows a phylogenomic reconstruction inferred with the Neighbor-joining algorithm (by considering the distance matrix generated by Gegenees software). Reconstruction was done with Mega 6.0 software. Janthinobacterium lividum MTR is highlighted with a solid red rhomboid. (DOC 590 kb)
The table shows the ANI values among the strain analyzed. ANI values were calculated according to Goris et al. 2007 (Goris J., Konstantinidis K., Klappenbach J., Coenye T., Vandamme P., Tiedje J. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007 57(Pt 1):81–91), using web application (http://enve-omics.ce.gatech.edu/ani/) (Figueras M., Beaz-Hidalgo R., Hossain M., Liles M. Taxonomic affiliation of new genomes should be verified using average nucleotide identity and multilocus phylogenetic analysis. Genome Announc. 2014 2(6). pii: e00927-14. doi:10.1128/genomeA.00927-14). (DOC 235 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
NV participated in the genome assembly and sequencing analysis and drafted the original manuscript. PS and PA participated in the bacterial isolation and capnophilic characterization. LC participated in the electron microscopy characterization. AC conducted the electron microcopy characterization and revising the manuscript. AG participated in the sampling, bacterial isolation, and manuscript revision. GC participated in sequencing, providing the funds and drafting the original manuscript. MT originated the study, participated in the analysis of the genome sequence, and drafted and reviewed the original manuscript. All authors read and approved the final manuscript.
References
- 1.Garrity GM, Bell JA, Lilburn TE. Family II. Oxalobacteraceae fam. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. 2. New York: Springer; 2005. p. 623. [Google Scholar]
- 2.Gillis M, De Ley J. The Genera Chromobacterium and Janthinobacterium. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes: Handbook on the Biology of Bacteria. 3. New York: Springer; 2006. p. 737. [Google Scholar]
- 3.Duran N, Justo GZ, Ferreira CV, Melo PS, Cordi L, Martins D. Violacein: properties and biological activities. Biotechnol Appl Biochem. 2007;48(Pt 3):127–33. doi: 10.1042/BA20070115. [DOI] [PubMed] [Google Scholar]
- 4.Rodrigues AL, Gocke Y, Bolten C, Brock NL, Dickschat JS, Wittmann C. Microbial production of the drugs violacein and deoxyviolacein: analytical development and strain comparison. Biotechnol Lett. 2012;34(4):717–20. doi: 10.1007/s10529-011-0827-x. [DOI] [PubMed] [Google Scholar]
- 5.Matz C, Deines P, Boenigk J, Arndt H, Eberl L, Kjelleberg S, et al. Impact of violacein-producing bacteria on survival and feeding of bacterivorous nanoflagellates. Appl Environ Microbiol. 2004;70(3):1593–9. doi: 10.1128/AEM.70.3.1593-1599.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matz C, Webb JS, Schupp PJ, Phang SY, Penesyan A, Egan S, et al. Marine biofilm bacteria evade eukaryotic predation by targeted chemical defense. PLoS One. 2008;3(7):e2744. doi: 10.1371/journal.pone.0002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker MH, Brucker RM, Schwantes CR, Harris RN, Minbiole KP. The bacterially produced metabolite violacein is associated with survival of amphibians infected with a lethal fungus. Appl Environ Microbiol. 2009;75(21):6635–8. doi: 10.1128/AEM.01294-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris RN, Brucker RM, Walke JB, Becker MH, Schwantes CR, Flaherty DC, et al. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. Isme J. 2009;3(7):818–24. doi: 10.1038/ismej.2009.27. [DOI] [PubMed] [Google Scholar]
- 9.Becker MH, Harris RN, Minbiole KP, Schwantes CR, Rollins-Smith LA, Reinert LK, et al. Towards a better understanding of the use of probiotics for preventing chytridiomycosis in Panamanian golden frogs. Ecohealth. 2011;8(4):501–6. doi: 10.1007/s10393-012-0743-0. [DOI] [PubMed] [Google Scholar]
- 10.Loudon AH, Holland JA, Umile TP, Burzynski EA, Minbiole KP, Harris RN. Interactions between amphibians’ symbiotic bacteria cause the production of emergent anti-fungal metabolites. Front Microbiol. 2014;5:441. doi: 10.3389/fmicb.2014.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brucker RM, Harris RN, Schwantes CR, Gallaher TN, Flaherty DC, Lam BA, et al. Amphibian chemical defense: antifungal metabolites of the microsymbiont Janthinobacterium lividum on the salamander Plethodon cinereus. J Chem Ecol. 2008;34(11):1422–9. doi: 10.1007/s10886-008-9555-7. [DOI] [PubMed] [Google Scholar]
- 12.Emilio MG. Gas exchanges and blood concentrations in the frog Rana ridibunda. J Exp Biol. 1974;60(3):901–8. doi: 10.1242/jeb.60.3.901. [DOI] [PubMed] [Google Scholar]
- 13.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493–6. [PubMed] [Google Scholar]
- 14.Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007;57(Pt 10):2259–61. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- 15.Schloss PD, Allen HK, Klimowicz AK, Mlot C, Gross JA, Savengsuksa S, et al. Psychrotrophic strain of Janthinobacterium lividum from a cold Alaskan soil produces prodigiosin. DNA Cell Biol. 2010;29(9):533–41. doi: 10.1089/dna.2010.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrews S. FastQC a quality-control tool for high-throughput sequence data. 2014. [Google Scholar]
- 17.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–64. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–8. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu S, Zhu Z, Fu L, Niu B, Li W. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35(Web Server issue):W52–7. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–82. [PubMed] [Google Scholar]
- 23.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 24.Docquier JD, Lopizzo T, Liberatori S, Prenna M, Thaller MC, Frere JM, et al. Biochemical characterization of the THIN-B metallo-beta-lactamase of Janthinobacterium lividum. Antimicrob Agents Chemother. 2004;48(12):4778–83. doi: 10.1128/AAC.48.12.4778-4783.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178(1):306–8. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berg IA. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol. 2011;77(6):1925–36. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dodd AN, Borland AM, Haslam RP, Griffiths H, Maxwell K. Crassulacean acid metabolism: plastic, fantastic. J Exp Bot. 2002;53(369):569–80. doi: 10.1093/jexbot/53.369.569. [DOI] [PubMed] [Google Scholar]
- 28.Saz HJ, Hubbard JA. The oxidative decarboxylation of malate by Ascaris lumbricoides. J Biol Chem. 1957;225(2):921–33. [PubMed] [Google Scholar]
- 29.Ensign SA. Revisiting the glyoxylate cycle: alternate pathways for microbial acetate assimilation. Mol Microbiol. 2006;61(2):274–6. doi: 10.1111/j.1365-2958.2006.05247.x. [DOI] [PubMed] [Google Scholar]
- 30.Pantanella F, Berlutti F, Passariello C, Sarli S, Morea C, Schippa S. Violacein and biofilm production in Janthinobacterium lividum. J Appl Microbiol. 2007;102(4):992–9. doi: 10.1111/j.1365-2672.2006.03155.x. [DOI] [PubMed] [Google Scholar]
- 31.Dunn MF, Ramirez-Trujillo JA, Hernandez-Lucas I. Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology. 2009;155(Pt 10):3166–75. doi: 10.1099/mic.0.030858-0. [DOI] [PubMed] [Google Scholar]
- 32.Lorenz MC, Fink GR. The glyoxylate cycle is required for fungal virulence. Nature. 2001;412(6842):83–6. doi: 10.1038/35083594. [DOI] [PubMed] [Google Scholar]
- 33.Kratky M, Vinsova J. Advances in mycobacterial isocitrate lyase targeting and inhibitors. Curr Med Chem. 2012;19(36):6126–37. doi: 10.2174/092986712804485782. [DOI] [PubMed] [Google Scholar]
- 34.Cheah HL, Lim V, Sandai D. Inhibitors of the glyoxylate cycle enzyme ICL1 in Candida albicans for potential use as antifungal agents. PLoS One. 2014;9(4):e95951. doi: 10.1371/journal.pone.0095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cover TL. Perspectives on methodology for in vitro culture of Helicobacter pylori. Methods Mol Biol. 2012;921:11–5. doi: 10.1007/978-1-62703-005-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.St Maurice M, Cremades N, Croxen MA, Sisson G, Sancho J, Hoffman PS. Flavodoxin:quinone reductase (FqrB): a redox partner of pyruvate:ferredoxin oxidoreductase that reversibly couples pyruvate oxidation to NADPH production in Helicobacter pylori and Campylobacter jejuni. J Bacteriol. 2007;189(13):4764–73. doi: 10.1128/JB.00287-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nerad J, Seville M, Snydman D. Miscellaneous Gram-negative bacilli: Acinetobacter, Cardiobacterium, Actinobacillus, Chromobacterium, Capnocytopaga, and others. In: Gorbach S, Bartlett J, Blacklow N, editors. Infectious Diseases. 2. Philadelphia: WB Saunders Company; 1998. pp. 1871–87. [Google Scholar]
- 38.Song H, Lee JW, Choi S, You JK, Hong WH, Lee SY. Effects of dissolved CO2 levels on the growth of Mannheimia succiniciproducens and succinic acid production. Biotechnol Bioeng. 2007;98(6):1296–304. doi: 10.1002/bit.21530. [DOI] [PubMed] [Google Scholar]
- 39.Hong SH, Kim JS, Lee SY, In YH, Choi SS, Rih JK, et al. The genome sequence of the capnophilic rumen bacterium Mannheimia succiniciproducens. Nat Biotechnol. 2004;22(10):1275–81. doi: 10.1038/nbt1010. [DOI] [PubMed] [Google Scholar]
- 40.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26(5):541–7. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87(12):4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garrity GM, Bell JA, Lilburn TE. Phylum XIV. Proteobacteria phyl. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. 2. New York: Springer; 2005. p. 1. [Google Scholar]
- 43.Hornung C, Poehlein A, Haack FS, Schmidt M, Dierking K, Pohlen A, et al. The Janthinobacterium sp. HH01 genome encodes a homologue of the V. cholerae CqsA and L. pneumophila LqsA autoinducer synthases. PLoS One. 2013;8(2):e55045. doi: 10.1371/journal.pone.0055045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garrity GM, Bell JA, Lilburn TE. Class II. Betaproteobacteria. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. 2. New York: Springer; 2005. p. 575. [Google Scholar]
- 45.Garrity GM, Bell JA, Lilburn TE. Order 1. Burkholderiales. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. 2. New York: Springer; 2005. p. 575. [Google Scholar]
- 46.O’Sullivan J, McCullough J, Johnson JH, Bonner DP, Clark JC, Dean L, et al. Janthinocins A, B and C, novel peptide lactone antibiotics produced by Janthinobacterium lividum. I. Taxonomy, fermentation, isolation, physico-chemical and biological characterization. J Antibiot (Tokyo). 1990;43(8):913–9. doi: 10.7164/antibiotics.43.913. [DOI] [PubMed] [Google Scholar]
- 47.Kampfer P, Falsen E, Busse HJ. Reclassification of Pseudomonas mephitica Claydon and Hammer 1939 as a later heterotypic synonym of Janthinobacterium lividum (Eisenberg 1891) De Ley et al. 1978. Int J Syst Evol Microbiol. 2008;58(Pt 1):136–8. doi: 10.1099/ijs.0.65450-0. [DOI] [PubMed] [Google Scholar]
- 48.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 50.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 51.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101(30):11030–5. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]


