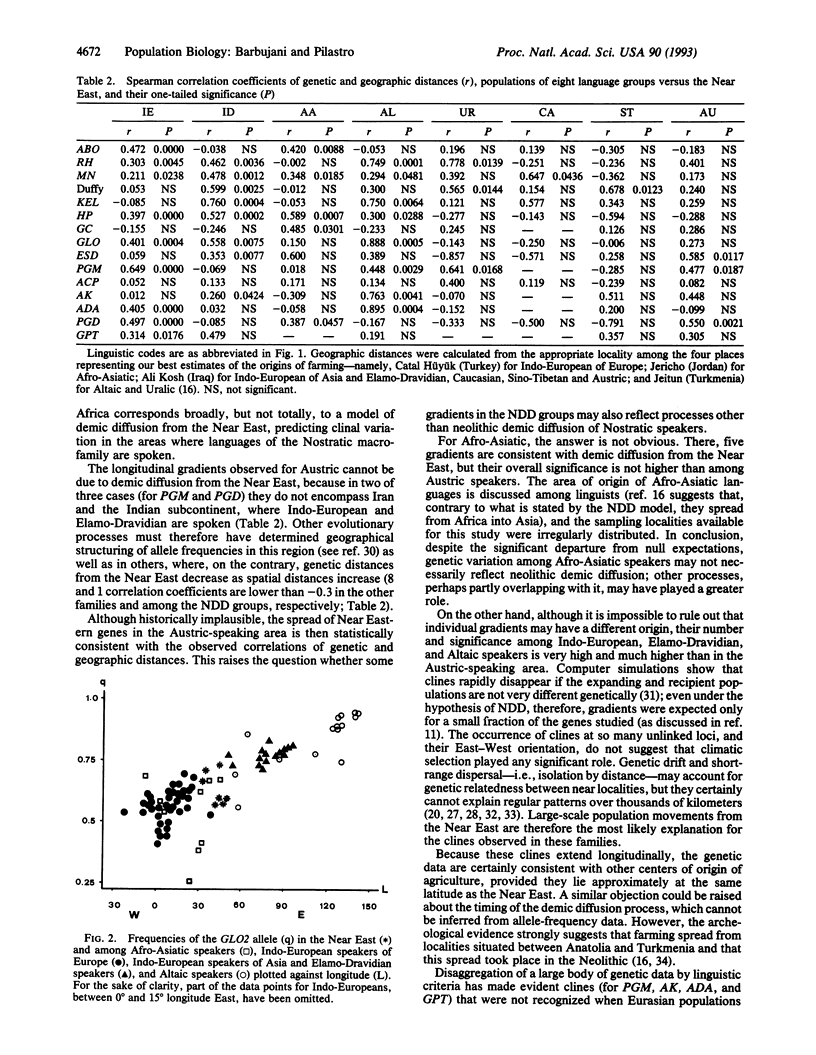
Abstract
Contemporary patterns of allele frequencies allow inferences on past evolutionary processes. L.L. CavalliSforza [(1988) Munibe 6, 129-137] and C. Renfrew [(1991) Cambridge Archaeol. J. 1, 3-23] proposed that neolithic farmers from the Near East propagated a group of related ancestral languages, from which three or four linguistic families developed. Here we show that genetic variation among Indo-European, Elamo-Dravidian, and Altaic speakers (grouped by some linguists in the Nostratic macrofamily) supports this hypothesis, whereas the evidence on Afro-Asiatic speakers is ambiguous. Gene-frequency clines within these linguistic families suggest that language diffusion was largely associated with population movements rather than with purely cultural transmission. Archeological, linguistic, and genetic evidence can be reconciled by envisaging a process of population growth and multidirectional dispersal from the Near East as the main factor shaping genetic and linguistic diversity in Eurasia and perhaps in North Africa.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbujani G. Autocorrelation of gene frequencies under isolation by distance. Genetics. 1987 Dec;117(4):777–782. doi: 10.1093/genetics/117.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbujani G. Diversity of some gene frequencies in European and Asian populations. III. Spatial correlogram analysis. Ann Hum Genet. 1987 Oct;51(Pt 4):345–353. doi: 10.1111/j.1469-1809.1987.tb01069.x. [DOI] [PubMed] [Google Scholar]
- Barbujani G., Jacquez G. M., Ligi L. Diversity of some gene frequencies in European and Asian populations. V. Steep multilocus clines. Am J Hum Genet. 1990 Nov;47(5):867–875. [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Edwards A. W. Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet. 1967 May;19(3 Pt 1):233–257. [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Minch E., Mountain J. L. Coevolution of genes and languages revisited. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5620–5624. doi: 10.1073/pnas.89.12.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Piazza A., Menozzi P., Mountain J. Reconstruction of human evolution: bringing together genetic, archaeological, and linguistic data. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6002–6006. doi: 10.1073/pnas.85.16.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L. Population structure and human evolution. Proc R Soc Lond B Biol Sci. 1966 Mar 22;164(995):362–379. doi: 10.1098/rspb.1966.0038. [DOI] [PubMed] [Google Scholar]
- Farabegoli A., Barbujani G. Diversity of some gene frequencies in European and Asian populations. VI. Geographic patterns of PGM and ACP. Hum Hered. 1990;40(6):313–321. doi: 10.1159/000153954. [DOI] [PubMed] [Google Scholar]
- Kimura M, Weiss G H. The Stepping Stone Model of Population Structure and the Decrease of Genetic Correlation with Distance. Genetics. 1964 Apr;49(4):561–576. doi: 10.1093/genetics/49.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menozzi P., Piazza A., Cavalli-Sforza L. Synthetic maps of human gene frequencies in Europeans. Science. 1978 Sep 1;201(4358):786–792. doi: 10.1126/science.356262. [DOI] [PubMed] [Google Scholar]
- Morton N. E. Kinship, information and biological distance. Theor Popul Biol. 1975 Apr;7(2):246–255. doi: 10.1016/0040-5809(75)90019-2. [DOI] [PubMed] [Google Scholar]
- Sokal R. R. Ancient movement patterns determine modern genetic variances in Europe. Hum Biol. 1991 Oct;63(5):589–606. [PubMed] [Google Scholar]
- Sokal R. R., Harding R. M., Oden N. L. Spatial patterns of human gene frequencies in Europe. Am J Phys Anthropol. 1989 Nov;80(3):267–294. doi: 10.1002/ajpa.1330800302. [DOI] [PubMed] [Google Scholar]
- Sokal R. R., Oden N. L., Wilson C. Genetic evidence for the spread of agriculture in Europe by demic diffusion. Nature. 1991 May 9;351(6322):143–145. doi: 10.1038/351143a0. [DOI] [PubMed] [Google Scholar]
- Stringer C. B., Andrews P. Genetic and fossil evidence for the origin of modern humans. Science. 1988 Mar 11;239(4845):1263–1268. doi: 10.1126/science.3125610. [DOI] [PubMed] [Google Scholar]
- Takahata N. Genealogy of neutral genes and spreading of selected mutations in a geographically structured population. Genetics. 1991 Oct;129(2):585–595. doi: 10.1093/genetics/129.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]




