Abstract
Purpose
To investigate human brain metabolite discriminability and general measurement reproducibility of two-dimensional (2D) J-resolved 1H MRS and Prior Knowledge Fitting (ProFit).
Materials and Methods
2D J-resolved 1H MRS spectra were acquired from the anterior cingulate cortex (ACC) and the parietal-occipital cortex (POC) of 10 healthy subjects at a magnetic field strength of 2.9 Tesla. Amplitude correlation matrices were constructed for each subject and brain region to assess metabolite discriminability. ProFit-estimated metabolite peak areas were normalized to a water reference signal, and intra- and inter-subject reproducibility was evaluated.
Results
Favorable between-metabolite correlation coefficients (<20%) were observed for a range of metabolites. Lower correlation coefficients between a given pair of metabolite estimates were consistently observed for POC metabolites. The group mean correlation coefficient existing between glutamate and glutamine was calculated as −18% and −13% for ACC and POC, respectively. Most ACC and POC metabolites showed intra- and inter-subject CV values of <15% and <20%, respectively.
Conclusion
The observed Glu and Gln signal discrimination makes these techniques suitable for investigating a variety of psychiatric disorders. Intra- and inter-subject metabolite level reproducibility was comparable to the existing literature findings. These data serve as a valuable benchmark for assessing future modifications to 2D 1H MRS data acquisition and ProFit analysis.
Keywords: two-dimensional J-resolved proton magnetic resonance spectroscopy, metabolite peak discriminability, test–retest reliability
Low spectral resolution and severe metabolite peak overlap are associated with proton (1H) MR spectroscopy (MRS) data acquired from human brain using clinical MR imaging systems. Spatially localized variants of two-dimensional (2D) 1H MRS methods circumvent these issues by encoding a second frequency dimension and separating all uncoupled and scalar spin-spin (J)-coupled metabolite resonances across a 2D spectral surface. Localized 2D 1H MRS measurement sequences are analogous to high-resolution methods used extensively for elucidating the molecular connectivity of organic molecules in vitro, with localized 2D J-resolved spectroscopy (1–3) and 2D correlated spectroscopy (COSY) (4) being examples of two techniques adapted for human investigations. Applications for localized 2D J-resolved 1H MRS in human brain have included the resolution of γ-amino butyric acid (GABA) from creatine (Cre) resonances in substance abuse disorders (5,6), the investigation of amino acid neurotransmitter metabolism in bipolar disorder (7) and pain (8), and the separation of lactic acid (Lac) from lipids in brain tumors (9). Localized 2D COSY 1H MRS has been shown to enhance the available neurochemical information in healthy human brain tissue (4) and glioblastomas (10).
The implementation of localized 2D 1H MRS acquisition schemes onto clinical MRI/MRS systems is relatively straightforward, as is the reconstruction and visualization of the recorded 2D spectral data. However, quantification of 2D 1H MRS data remains particularly challenging, and several approaches having been adopted at different institutions. Earlier attempts for quantifying 2D J-resolved 1H MRS data were based on the extraction of a one-dimensional (1D) row where, for GABA, the extraction of a row positioned at 3.01 ppm / 7.45 Hz enables sufficient resolution from dominating Cre methyl resonance (3,11). The row extraction approach recently was improved to enable the automated 1D spectral fitting of all individual rows for multiple metabolites in a 2D J-resolved dataset (12). A popular method for quantifying 2D COSY 1H MRS data recorded in vivo involves the calculation of metabolite peak volume integrals and the reproducibility of that technique has been demonstrated at 1.5 Tesla (T) (13). A statistical peak-separation and object segmentation algorithm also has been proposed for the analysis of 2D COSY datasets (14).
Schulte and Boesiger recently introduced a novel algorithm termed Prior Knowledge Fitting (ProFit), which represents a notable advance in the automated analysis of 2D 1H MRS data (15). The ProFit algorithm iteratively fits in vivo 2D MRS data using a linear combination of suitable 2D model spectra, and can be adapted for fitting any given type of 2D spectral acquisition. The original report demonstrated the robust fitting of twenty individual metabolites to 2D J-resolved 1H MRS spectra acquired from healthy human volunteers including GABA, glutamate (Glu), glutamine (Gln), glutathione (GSH), and glycine (Gly). Data recorded from a single subject and from multiple subjects was used to calculate intra- and inter-subject metabolite fit standard deviations, respectively, which demonstrated a high degree of reproducibility (15). Spectral fit reliability also was evaluated using the Cramer-Rao lower bound (CRLB) values determined for each metabolite concentration. CRLB values are evaluated following inversion of the noise-normalized Fisher information matrix (16), and typically are outputted as the estimated standard deviation expressed as a percentage of the estimated metabolite concentration. Therefore, CRLB values represent the theoretical lower level of variability attainable for a metabolite at a given signal-to-noise (SNR). Compared to the fitting of conventional 1D 1H MRS data, 2D J-resolved 1H MRS data analyzed using ProFit resulted in significantly reduced CRLB values for the vast majority of metabolite species, including GABA, Gln, and Glu. The reproducibility of metabolite concentration, coupled with the low CRLB values detected for the amino neurotransmitter species, makes these approaches especially suitable for neuropsychiatric applications.
The off-diagonal elements of the inverted Fisher matrix also can be used to calculate the amplitude correlation coefficient for a pair of metabolite estimates, thus providing additional data reflecting the inter-dependence of the ProFit signal estimates (16). Note that as with CRLB values, the amplitude correlation coefficients are an estimate of the lowest amount of covariance between two fit parameters, thus providing a lower bound for the actual amount of observed covariance that cannot be readily extracted from in vivo data. Such measures could provide unique information regarding the relative discriminability of specific metabolite resonances such as Glu from Gln, or GABA from Cre, which are typical outcome measures in neuropsychiatric MRS studies. MRS discrimination of metabolite resonances is also expected to be region-specific, due to the differential static magnetic field homogeneity and signal linewidth that is realistically achievable across the human brain. The primary objective of this study was to interrogate the magnitude and variability of the between-metabolite correlation coefficients as calculated in human brain using 2D J-resolved 1H MRS and ProFit analysis. MRS data acquired from two distinct brain regions in healthy volunteers using a multi-scan protocol commonly used in serial studies focusing on metabolic changes underlying disease progression and/or the effects of exogenous agents. A second aim of this study was to use a ProFit-independent metabolite normalization scheme based on a CSF-corrected water reference signal to further assess the reproducibility of 2D 1H MRS and ProFit analysis, and those findings are briefly put into context with the precedent literature.
MATERIALS AND METHODS
Subject Selection
The local Institutional Review Board approved this investigation, which met the criteria for investigations in human subjects (see http://ohsr.od.nih.gov/guidelines/helsinki.html, last accessed on 01/26/2012). A total of 10 healthy volunteers (mean age ± standard deviation (SD) = 26 ± 3 years; 6 females, 4 males) were recruited from the general public for the study. One subject had undergone a single previous MRI scan unrelated to this study. MRI/MRS measurements were performed on 2 different days for each subject, with the second scan (S2) performed within 1 week of the baseline scan (S1).
Data Acquisition
All subjects were scanned using a 2.9 Tesla (T) Siemens (Erlangen, Germany) TIM Trio™ whole-body MRI system running the “VB17” Siemens-supplied software. A circularly polarized body coil and a manufacturer-supplied 12-channel phased-array receive-only head coil were used for radiofrequency (RF) transmission and signal reception, respectively. Subjects were positioned supine and foam pads were used to fixate the subject’s head within the RF coil housing. Three orthogonal low-resolution proton-weighted gradient echo (repetition time/echo time [TR/TE] = 20/5 ms; field of view [FOV] = 280 × 280 mm; matrix size = 192 × 144; 8 mm slice thickness) localizer images initially were obtained to confirm optimal head positioning at the receive-only coil center. Subsequently, static magnetic field (B0) shimming was performed over the whole head FOV using a standard manufacturer-supplied phase map method. Three-dimensional (3D) high-contrast and high-resolution T1-weighted, magnetization-prepared, rapid gradient echo (MP-RAGE; TR/TE/TI = 2000/3.53/1100 ms; FOV = 256 × 256 × 224 mm; isotropic 1 mm in-plane resolution) MR images then were acquired and used to facilitate accurate MRS voxel positioning and for post hoc within-MRS voxel tissue-type segmentation.
For the S1 and S2 scan sessions, single-voxel MRS data were acquired from two distinct brain regions in all subjects: the anterior cingulate cortex (ACC) and the parietal occipital cortex (POC). The MRS voxel measured 25 × 25 × 30 mm3 for both regions, and was obliqued along the sagittal plane and positioned to cover predominantly gray matter within the ACC or POC. Voxel repositioning at S2 was performed manually, by visually inspecting the corresponding voxel placement at S1 on axial and sagittal image slices reconstructed from the S1 MP-RAGE dataset. Within-voxel B0 shimming was achieved using a manufacturer-supplied automated phase map procedure in combination with interactive manual shimming until a full-width at half-maximum (FHWM) of ≤11 Hz was observed for the real component of the ACC or POC unsuppressed water signal. A standard PRESS sequence was modified to enable 2D J-resolved 1H MRS measurements, where the first PRESS TE period (TE1) was fixed at 12 ms and the second TE period (TE2) was progressively incremented to sample the second (J) dimension. Spatial localization was achieved using a Hanning-filtered sinc RF pulse of 2.6 ms duration (bandwidth (BW) = 5 kHz) followed by two identical optimized sinc RF pulses of 7.0 ms duration (BW = 1 kHz) for slice-selective refocusing. Identical parameters were used to acquire 2D J-resolved 1H MRS data from both brain regions (TR/TE = 2400/31-229 ms; ΔTE = 2 ms; 4 signal averages per TE step with online averaging; 2D spectral width = 2000 × 500 Hz; 2D matrix size = 2048 × 100). The spectral data were obtained using a maximum-echo sampling scheme whereby the analogue-to-digital converter (ADC) ontime was fixed for all 100 TE steps (17). Outer-volume suppression (OVS) was achieved using six saturation bands positioned at least 1.5-cm away from the MRS voxel faces and band saturation was achieved using hyperbolic secant adiabatic full passage RF pulses. A three-pulse water elimination through T1-effects (WET; 18) scheme was interleaved with the OVS module for global water suppression. In addition, water unsuppressed 2D 1H MRS data were acquired from each voxel with 2 signal averages recorded for each TE step. The RF transmitter carrier frequency was set to 3.0 and 4.7 ppm for water suppressed and unsuppressed data, respectively.
Tissue Segmentation
Skull stripping and whole brain tissue-type segmentation was performed on MP-RAGE images using BET (19) and FAST (20), respectively, which are tools provided with the freely-available FMRIB software library (FSL; 21). Home-written MATLAB (version R2010b, The MathWorks, Natick, MA) functions then were used to extract the 3D volume corresponding to the positioned MRS voxel to obtain within-voxel gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF) tissue content for each subject. The GM percentage was calculated as the ratio to total brain matter, i.e., 100 × GM/(WM+GM).
Spectral Processing and Quantification
All 1H MRS data were stored as Siemens TWIX files and transferred to personal computer systems to be preprocessed by home-written functions written in MATLAB. The 12-channel receive-only head coil was operated in the 4-cluster mode with each cluster comprised of 3 coil elements. Before channel recombination, eddy current distortions initially were accounted for using a previously reported time-domain method (22), where each TE step from a given 2D 1H MRS dataset was corrected using the corresponding cluster-specific water unsuppressed recorded at the same TE. Coil cluster-specific signal weighting coefficients were determined on an individual subject basis using the real component of the phased unsuppressed water signal data. In brief, a weighting coefficient for a given cluster was calculated as the maximum amplitude of the corresponding water spectrum divided by the sum of the maximum amplitudes for all four coil clusters. The weighting coefficients determined for a given subject and voxel location subsequently were applied to all water suppressed and unsuppressed FID data before signal recombination. The eddy current corrected and signal weighted time-domain data from all four clusters were recombined on a TE-by-TE basis to afford a 2D matrix, characterized by 100 TE steps and 2048 complex points. The residual water signal was removed from each row of water suppressed 2D matrices using a Hankel singular value decomposition (HSVD) routine (23) written in MATLAB. Finally, the 2D matrix was reformatted to produce the individual file types required for ProFit read-in.
The ProFit algorithm was applied identically to all ACC and POC 2D 1H MRS data as detailed in (15) using the supplied 2D basis set generated without considering the effects of spatial localization. Before the 2D fast Fourier transformation (FFT), the raw 2D matrix was zero-filled to 200 points along the indirectly detected (J)-dimension. The basis set comprised of nineteen metabolites including Cre, N-acetyl aspartate (NAA), glycerophosphorylcholine (GPC), phosphorylcholine (PCh), alanine (Ala), aspartate (Asp), GABA, glucose (Glc), Gln, Glu, Gly, GSH, Lac, myo-inositol (Ins), N-acetyl aspartylglutamate (NAAG), phosphoethanolamine (PE), taurine (Tau), scyllo-inositol (sI), and ascorbic acid (Asc). The Cre methylene (CH2) and methyl (CH3) protons were fitted separately whereas the separate GPC and PCh peaks ultimately were considered as a composite resonance. For considering CRLB on the individual metabolite concentrations, the ProFit software first constructs the Fisher information matrix, F, as follows
| [1] |
where σ corresponds to the standard deviation of the real part of the noise, Re{Btotal} denotes the real part of the final basis matrix containing 2D spectral information for the included metabolites, and the superscript T is the transpose of the matrix (15). The CRLB on the concentration for a given metabolite m is then calculated following inversion of the Fisher matrix using
| [2] |
All metabolite Cramer-Rao Lower Bound (CRLB) values presented were calculated by the ProFit software with CRLB values <20% included in the final analysis. The Fisher matrix also can be used to construct the amplitude correlation matrix to describe the correlation ρl,m that exists between metabolite species l and m. Accordingly, the ProFit software was adapted to output a covariance matrix for each subject and brain region examined as follows
| [3] |
where ρl,m is a square matrix with diagonal elements of unity and off-diagonal elements corresponding to the amplitude correlation existing between metabolites l and m (16). Group averaged covariance matrices ultimately were constructed for both brain regions (using all S1 and S2 data) and were calculated after ensuring that equivalent signs existed for each matrix element across all subjects.
The software was further modified to calculate signal-to-noise (SNR) ratios based on the NAA CH3 resonance at 2.0 ppm. Briefly, 2D spectral regions-of-interest (ROI) were defined between 9.0 and 10.0 ppm and 1.75 and 2.25 ppm, which corresponded to noise and NAA ROIs, respectively. All points along the J-dimension were used for reconstructing the 2D ROIs. The SNR was defined as the maximum absolute peak height calculated for the NAA ROI divided by the standard deviation of the real part of the noise ROI. The estimated metabolite 2D peak areas were normalized to the short TE = 31 ms unsuppressed water signal, which was calculated after fitting a Voigt line-shape to the real component of the phased frequency-domain unsuppressed water data (24). The nonlinear least-squares “lsqnonlin” function provided with the MATLAB Optimization Toolbox™ was used to fit the water data, with the initial estimate for signal amplitude being subject-specific and based on the maximum peak amplitude. An initial estimate of 8 Hz was used for signal linewidth (LW) with the lower and upper bounds set to 1 and 20 Hz, respectively. The resulting metabolite/water ratios were corrected for within-voxel CSF-fraction determined using the relevant segmented MRI data.
Statistical Analysis
Statistical analysis was performed using OriginPro 8 (OriginLab Corporation, Northampton, MA) with the coefficient of variation defined as the standard deviation divided by the mean. Both intra- and inter-subject CVs were calculated for all metabolites, subjects and brain regions. The inter-subject metabolite CV values were calculated using two methods as follows: the first approach used the averaged S1 and S2 values before calculating inter-subject CVs, whereas the second method considered only the S1 data.
Finally, to test for significant differences in covariance between the ACC and POC matrices, two-sample t-tests (equal variance assumed, two-tailed) were performed on an element-wise basis with the significance level (α) set to 0.05.
RESULTS
Figure 1a presents tissue-segmented axial and sagittal T1-weighted MR images, which also display the typical ACC and POC voxel locations used for the present study. The mean within-MRS voxel GM and CSF content together with their corresponding intra- and inter-subject CV values are presented in Table 1.
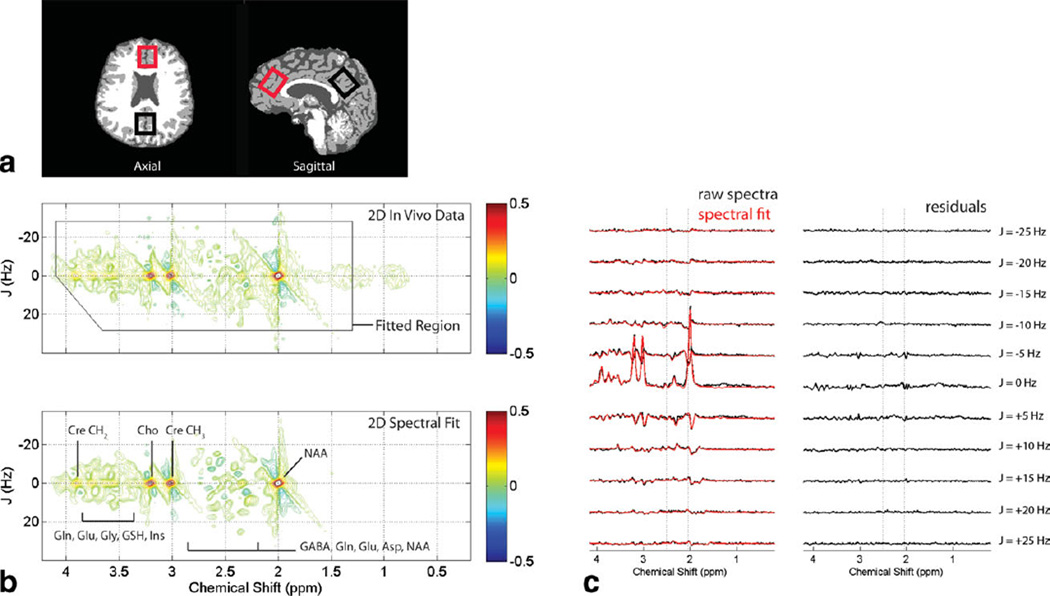
Figure 1.
a: Axial and mid-sagittal slices extracted from a tissue-segmented 3D MP-RAGE MRI dataset recorded from a single subject. The red and black rectangles depict the typical voxel positioning used for ACC and POC measurements, respectively. b: Phase-sensitive (real component) 2D J-resolved 1H MRS data recorded from the ACC voxel shown in (a) and fitted using the ProFit algorithm. The 2D data are characterized by a J-resolved (Hz) axis plotted against the chemical shift (ppm) dimension, with the raw 2D spectral data (top panel) and 2D spectral fit (bottom panel) displayed as expanded regions (first dimension: 0.2–4.2 ppm; second dimension: ± 40 Hz) of their respective full 2D matrices. Both 2D spectra are presented using identical scaling and the color bars show the minimum and maximum contour levels used (arbitrary units). The black box within the top panel represents the spectral region analyzed for 2D fitting, and tentative signal assignments are provided on the bottom panel. c: Eleven individual 1D spectral rows (J = ± 25 Hz) extracted from the 2D 1H MRS data shown in (b), presented together with their respective fits (red) and fit residuals. The dotted lines delineate the 2.05–2.50 ppm spectral region that is comprised primarily of Gln and Glu resonances.
Table 1.
Group Mean Within MRS-Voxel Tissue Content for the ACC and POC Brain Regions, Presented Together With the Corresponding Intra- and Inter-subject CV Values*
| Brain Region |
Tissue type |
Mean percentage (± SD) |
Intra-subject CV |
Inter-subject CV |
|---|---|---|---|---|
| ACC | GM | 72 ± 3 % | 1 % | 5 % |
| CSF | 14 ± 5 % | 4 % | 33 % | |
| POC | GM | 68 ± 4 % | 2 % | 6 % |
| CSF | 13 ± 6 % | 10 % | 46 % |
See text for details regarding within-voxel segmentation procedures.
Figure 1b displays an expanded region of the raw 2D J-resolved 1H MRS data recorded from the ACC voxel in Figure 1, together with the 2D spectral fit achieved using the ProFit algorithm. Tentative signal assignments for major proton resonances also are provided with the 2D spectral fit. Figure 1c displays a series of individual 1D spectral rows extracted from the raw 2D 1H MRS data presented in Figure 1b, shown together with the corresponding fits (overlaid in red) and fit residuals (i.e., fit subtracted from the corresponding raw data). To provide some insight into the overall consistency of spectral acquisitions, the group averaged SNR and solvent water-based LW measures are presented in Table 2 along with the calculated intra- and inter-subject measures.
Table 2.
Group Mean SNR and LW Values for the ACC and POC Brain Regions, Presented Together With the Corresponding Intra- and Inter-subject CV Values*
| Brain region |
Parameter | Mean (± SD) |
Intra-subject CV |
Inter-subject CV |
|---|---|---|---|---|
| ACC | SNR | 437 ± 74 | 8 % | 17 % |
| LW | 8 ± 1 Hz | 7 % | 12 % | |
| POC | SNR | 441 ± 75 | 11 % | 17 % |
| LW | 6 ± 1 Hz | 5 % | 6 % |
See text for details regarding SNR and LW calculation.
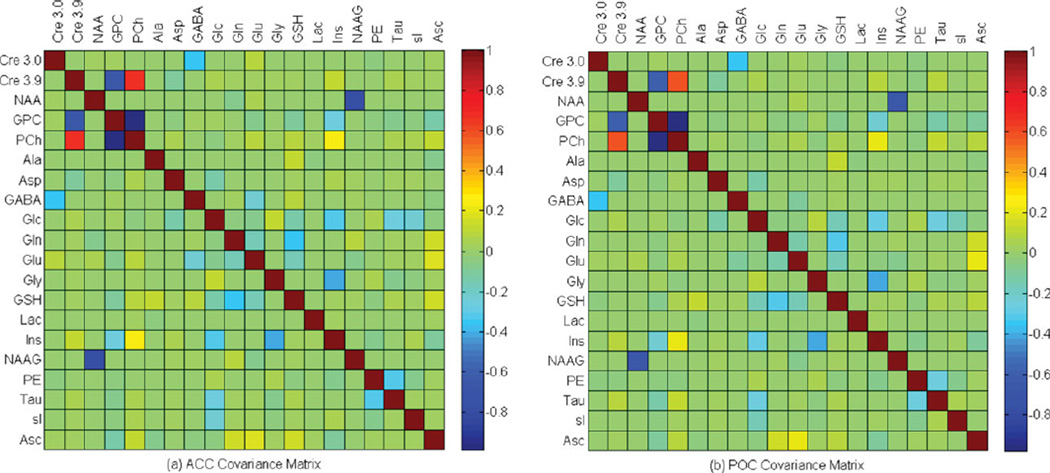
Figure 2 displays the ACC and POC metabolite covariance matrices calculated for the current study, which allow for a post hoc assessment of the metabolite discriminability achievable using 2D J-resolved 1H MRS and ProFit. Correlation coefficients (group mean ± SD) for pairwise interactions between ACC metabolites of interest were: Glu and Gln = −17.5 ± 4.1%, Glu and GABA = −22.3 ± 6.9%, Glu and NAA = 5.0 ± 0.9%, Gln and GABA = 1.45 ± 1.0%, Gln and NAA = −6.5 ± 2.9%, GABA and Cre = −36.4 ± 1.0%, Gly and mI = −42.2 ± 1.2%, GPC and PCh = −98.8 ± 0.4%, and NAA and NAAG = −78 ± 5.0%. Similarly, for the POC metabolites these values were: Glu and Gln = −12.7 ± 0.6%, Glu and GABA = −9.3 ± 3.9%, Glu and NAA = 4.1 ± 0.11%, Gln and GABA = 0.2 ± 0.1%, Gln and NAA = −1.2 ± 1.2%, GABA and Cre = −35.7 ± 0.4%, Gly and mI = −40.7 ± 0.9%, GPC and PCh = −98.1 ± 0.4%, and NAA and NAAG = −64.6 ± 5.3%. When considering one half of the symmetrical group-averaged ACC and POC covariance matrices, statistical analysis showed that a total of 168 of the 190 off-diagonal elements were significantly different between the two ROIs. For example, between-ROI differences were significant (P < 0.01) for the Glu ↔ Gln, Glu ↔ GABA and GABA ↔ Cre interactions, but not for sI ↔ Gly (P = 0.4) or Ala ↔ Tau (P = 0.4).
Figure 2.
Group averaged covariance matrices of the ProFit analysis calculated for the ACC (a) and POC (b) brain regions. The magnitude and sign of the correlation coefficients can be appreciated using the color bar provided with each plot, although specific correlation coefficients are provided within the text for ACC and POC metabolites of interest.
Table 3 provides the group mean ACC and POC water-normalized peak areas for all metabolites together with the corresponding CV values and CRLB estimates. The “Inc” column defines the number of spectra used in the final analysis that showed CRLB <20% for a given metabolite. Table 4 shows precedent literature values and results from the present study, which enables the direct comparison of intra- and inter-subject CV values obtained for selected metabolites in several brain regions of healthy volunteers. Also provided in Table 4 are the specific B0 field strength, voxel size, and total measurement time for each specific study.
Table 3.
ProFit-Estimated Group Mean Water-Normalized Peak Areas Together With the Corresponding Intra- and Inter-subject CVs and CRLB Values*
| Brain region |
Metabolite | Inc | Metabolite/water (Mean ± SD; ×10−5) |
Intra-subject CV (%) |
Inter-subject CV (%) |
S1 Inter-subject CV (%) |
CRLB (% ± SD) |
|---|---|---|---|---|---|---|---|
| ACC | Cre | 20 | 7.3 ± 1.0 | 4.5 | 15 | 16 | 0.5 ± 0.1 |
| NAA | 20 | 10.3 ± 1.2 | 7.0 | 10 | 14 | 0.5 ± 0.1 | |
| PCh/GPC | 20 | 2.0 ± 0.2 | 4.4 | 9 | 8 | 3.5 ± 0.6 | |
| Ala | 20 | 0.3 ± 0.1 | 26 | 29 | 29 | 17 ± 10 | |
| Asc | 20 | 3.6 ± 0.6 | 6.0 | 16 | 15 | 2.3 ± 0.3 | |
| Asp | 20 | 2.0 ± 0.4 | 6.5 | 21 | 25 | 6.2 ± 1.3 | |
| GABA | 20 | 1.3 ± 0.4 | 15 | 24 | 32 | 5.7 ± 1.3 | |
| Glc | 20 | 3.2 ± 0.6 | 8.1 | 17 | 16 | 3.9 ± 0.7 | |
| Gln | 20 | 1.8 ± 0.3 | 9.9 | 16 | 16 | 5.9 ± 1.3 | |
| Glu | 20 | 8.6 ± 1.2 | 4.5 | 14 | 14 | 1.4 ± 0.2 | |
| Gly | 20 | 0.8 ± 0.2 | 13 | 19 | 22 | 6.2 ± 1.8 | |
| GSH | 20 | 1.8 ± 0.3 | 7.5 | 17 | 16 | 2.2 ± 0.3 | |
| Ins | 20 | 5.4 ± 0.6 | 3.6 | 11 | 13 | 1.8 ± 0.2 | |
| Lac | 20 | 0.6 ± 0.2 | 12 | 30 | 31 | 7.4 ± 1.3 | |
| Tau | 20 | 1.2 ± 0.3 | 26 | 22 | 24 | 8.3 ± 3.4 | |
| sI | 20 | 0.4 ± 0.1 | 11 | 37 | 37 | 4.7 ± 1.4 | |
| NAAG | 4 | 1.0 ± 0.8 | 48 | 82 | 12 | 7.5 ± 6.5 | |
| PE | 20 | 2.6 ± 0.4 | 8.1 | 16 | 18 | 4 ± 0.5 | |
| POC | Cre | 20 | 8.4 ± 1.1 | 3.0 | 13 | 15 | 0.4 ± 0.1 |
| NAA | 20 | 13.0 ± 1.4 | 3.2 | 11 | 13 | 0.3 ± 0.1 | |
| PCh/GPC | 20 | 1.9 ± 0.3 | 4.0 | 15 | 16 | 3.5 ± 0.7 | |
| Ala | 20 | 0.4 ± 0.1 | 11 | 26 | 27 | 14 ± 3.3 | |
| Asc | 20 | 3.7 ± 0.6 | 5.5 | 16 | 18 | 2.6 ± 0.5 | |
| Asp | 20 | 2. 5 ± 0.4 | 6.4 | 14 | 15 | 5.6 ± 1.5 | |
| GABA | 20 | 1.4 ± 0.2 | 8.3 | 13 | 15 | 5.7 ± 1.5 | |
| Glc | 20 | 3.5 ± 0.8 | 8.3 | 23 | 26 | 4.1 ± 0.9 | |
| Gln | 20 | 2.2 ± 0.3 | 12.5 | 8.6 | 10 | 5.4 ± 1.6 | |
| Glu | 20 | 9.3 ± 0.9 | 2.5 | 10 | 10 | 1.4 ± 0.3 | |
| Gly | 20 | 1.3 ± 0.3 | 12 | 21 | 28 | 4.5 ± 1.3 | |
| GSH | 20 | 1.8 ± 0.4 | 5.3 | 20 | 24 | 2.4 ± 0.4 | |
| Ins | 20 | 6.0 ± 0.7 | 4.3 | 11 | 13 | 1.8 ± 0.4 | |
| Lac | 20 | 0.5 ± 0.1 | 11 | 20 | 23 | 12 ± 2.8 | |
| Tau | 20 | 1.1 ± 0.3 | 22 | 15 | 23 | 10.9 ± 8.4 | |
| sI | 20 | 0.5 ± 0.1 | 5.9 | 32 | 33 | 4.1 ± 1.3 | |
| NAAG | 5 | 0.6 ± 0.4 | ** | ** | 57 | 7.0 ± 3.4 | |
| PE | 20 | 2.3 ± 0.6 | 14 | 23 | 32 | 5.1 ± 1.7 |
The ‘Inc’ column denotes the total number of spectra used in the final analysis that showed CRLB <20% for a given metabolite.
None of the ten subjects showed NAAG CRLB values <20% at both S1 and S2, thus preventing the calculation of these CV values.
Table 4.
Precedent Literature Values and Results From the Present Study Comparing Intra- and Inter-Subject CV Values Obtained for Selected Metabolites in Frontal, Parietal or Occipital Lobe Regions in Healthy Adult Subjects*
|
1H MRS Method Brain region |
Reference | B0 Voxel size scan time |
Metabolite | Intra-subject CV (%) |
Inter-subject CV (%) |
|---|---|---|---|---|---|
| 2D J-resolved Frontal Lobe | Present Study | 3.0 T 18.75 cc 16 min | Cre | 7 | 10 |
| NAA | 4 | 9 | |||
| PCh/GPC | 5 | 15 | |||
| Glu | 5 | 14 | |||
| Gln | 10 | 16 | |||
| GABA | 15 | 24 | |||
| Ins | 4 | 11 | |||
| 2D J-resolved Frontal and Parietal Lobe | 15 | 3.0 T 15 cc 7 min | Cre | ||
| NAA | 6 | 6 | |||
| PCh/GPC | 5 | 5 | |||
| Glu | 6 | 6 | |||
| Gln | 19 | 19 | |||
| GABA | 17 | 17 | |||
| Ins | 10 | 10 | |||
| 1D PRESS Frontal Lobe | 37 | 3.0 T 12 cc 4 min | Cre | 3 | - |
| NAA | 3 | - | |||
| PCh/GPC | 3 | - | |||
| Glu | 5 | - | |||
| Gln | 37 | - | |||
| GABA | 13 | - | |||
| Ins | 15 | - | |||
| 2D J-resolved Frontal Lobe | 7 | 4.0 T 8 cc 26 min | Cre | 9 | - |
| NAA | 11 | - | |||
| PCh/GPC | 8 | - | |||
| Glu | 13 | - | |||
| Gln | 23 | - | |||
| GABA | - | - | |||
| Ins | 9 | - | |||
| 2D J-resolved Occipital Lobe | 11 | 1.5 T 27 cc 35 min | Cre | 8 | - |
| NAA | 4 | - | |||
| PCh/GPC | 8 | - | |||
| GABA | 26 | - | |||
| 2D COSY Frontal Lobe | 13 | 1.5 T 27 cc 34 min | Cre | 16+ | 14+ |
| NAA | 4 | 5 | |||
| PCh/GPC | 15++ | 10++ | |||
| Glu/Gln | 6 | 7 | |||
| GABA | 22 | 17 | |||
| Ins | 10 | 10 |
The intra- and inter-subject measures given by Schulte et al (15) were calculated using Cre-normalized data acquired from frontal and parietal lobes, respectively.
Based on the raw volume calculated for the diagonal Cre resonance.
Based on the mean of the Cre-normalized PCh/GPC diagonal and cross peak volumes.
T = Tesla; cc = centimeter cubed; min = minutes.
DISCUSSION
Assessment of metabolite discriminability and the general measurement reproducibility of 2D 1H MRS data analyzed using ProFit is of critical importance for establishing the clinical utility of these techniques, especially when considering their usage in longitudinal studies focusing on disease progression and/or the pharmacodynamic effects of psychotropic drugs. This report set out to evaluate both measures in a setting that is representative of MRS studies performed in psychiatric populations and, to achieve that goal, we recorded 2D J-resolved 1H MRS data from two brain regions in 10 healthy control subjects with each subject undergoing MRI/MRS scan procedures on two distinct scan days. Nine of the ten volunteers reported as MRI/MRS procedure-naive at the S1 scan time. The brain regions examined included the ACC, which is commonly investigated in a range of psychiatric disorders, and the POC, which typically serves as a control region for frontal lobe MRS measurements (7,25). The MRS data obtained from this study design enabled us to assess how metabolite discriminability varies between important brain regions and to further interrogate the general reproducibility of these emerging acquisition and processing MRS methods.
Metabolite discriminability was assessed for each subject and brain region through the use of covariance matrices, which were outputted following appropriate manipulation of the Fisher information matrix (16). The covariance matrix diagonal elements of unity indicate the complete indiscrimination of the equivalent metabolite, whereas off-diagonal elements can be used to assess the strength of pairwise correlations between two distinct metabolite species. We actually presented the group averaged ACC and POC covariance matrices, which for 168 of the total 190 possible metabolite interactions showed significantly lower values for the off-diagonal elements for the POC data. That observation is consistent with the improved B0 shimming achieved in the POC region giving rise to lower LW values and an improved metabolite discriminability (see Table 2). The majority of the metabolite pairwise correlations were <20% for both brain regions. Particularly noteworthy are the mean Gln ↔ Glu correlation coefficients of −17.5% (SD = ± 4.1%) and −12.7% (SD = ± 0.6%) calculated within the ACC and POC, respectively, indicating minimal interference of the measured Glu and Gln signals with low subject variability. 1H MRS performed in human subjects have implicated glutamatergic dysfunction in a range of psychiatric illnesses including schizophrenia (26,27), bipolar disorder (7,28–30), depression (31,32), anxiety disorders (33), and substance abuse (34,35). Therefore, the reliable separation of Glu from Gln is of vital importance for characterizing the precise nature of glutamatergic abnormalities. Our data suggest that 2D J-resolved 1H MRS data analyzed using ProFit provides a sufficiently robust Glu/Gln peak resolution within the ACC, which is deemed a major locus of pathology in psychiatric disorders. Moderately low correlation coefficients of −36.4% (ACC) and −35.7% (POC) were detected for GABA ↔ Cre signifying reasonable metabolite discriminability for these two metabolites. Note that these coefficients, depending on a preset covariance threshold, might warrant the use of GABA levels as a covariate if the Cre 3.0 ppm resonance is used as an internal normalization reference. Another commercially available MRS fitting package (36) specifies that if the correlation between two metabolites is consistently more negative than −50%, then their level should be expressed as the sum of the pair. However, future work should aim to use realistic physiological phantoms for investigating covariance thresholds specific to metabolite pairs measured using 2D J-resolved MRS and ProFit. Following on from this, the integrity of the covariance matrices is reinforced after considering the strong negative pairwise correlations between overlapping singlet species, namely NAA ↔ NAAG and PCh ↔ GPC. For both brain regions, we observed a fewer number of successful NAAG fits (i.e., CRLBNAAG <20%) compared to previously reported findings at 3.0T (15), which might stem from the slightly lower spectral resolution associated with a B0 of 2.89T. Given that 2D J-resolved 1H MRS is based on the acquisition of FIDs at multiple TE steps, we are investigating a separate postprocessing pipeline based on TE-averaged 1H MRS and a regularized lineshape deconvolution, which recently was reported as an alternative means for resolving NAA and NAAG (37).
Schulte and Boesiger originally evaluated the intra- and inter-subject reproducibility of 2D J-resolved 1H MRS and ProFit using Cre-normalized data acquired from a single subject and 27 subjects, respectively (15). The second objective of the present study was to more rigorously establish the reliability of 2D J-resolved 1H MRS and ProFit In contrast to the Cre-based normalization scheme used by Schulte and Boesiger (15), we normalized our ProFit-derived metabolite peak areas using the PRESS CSF-corrected tissue water signal before calculation of CV values. This normalization scheme provided a ProFit-independent approach that (i) eliminated the potential for Cre 2D fitting errors to propagate into the metabolite normalization, and (ii) allowed the determination of Cre reproducibility. Our within-voxel segmentation data demonstrate that the within-subject variation of tissue type was negligible, indicative of accurate voxel repositioning between S1 and S2 scan times. In addition, the within-subject SNR and signal LW also were highly reproducible in the ACC and POC regions. Both the ACC and POC intra- and inter-subject CV values calculated for the current study agree well with those previously reported using 2D J-resolved 1H MRS and ProFit (15). The metabolite inter-subject CVs are notably higher than the corresponding intra-subject CVs, reflecting the likely differences in metabolite concentrations between subjects. For most metabolites, inter-subject CVs calculated using only S1 values are typically higher than those calculated using averaged S1 and S2 values, as errors related to intra-subject variability are included the analysis. The majority of metabolites showed CRLB values <20% for both ACC and POC regions for all analyzed subject data with the exception of NAAG (see earlier discussion). For some metabolites including Cre, NAA, and Gln, lower CRLB values were observed for the POC region, which might be explained by the lower group mean LW recorded for this brain region and the theoretical dependency of CRLB on the observed T2* (23). Compared to previous findings (15), we also observed lower CRLB values for the majority of metabolites in both brain regions, an observation that is likely explained by the 25% increase in voxel used for the present study combined with the use of a 12-channel phased array receive-only coil as opposed to a single-channel transmit-receive coil. For both brain regions, it was crucial to verify that our measured metabolite peak areas relative to Cre were consistent with previous findings. Our calculated ACC metabolite/Cre ratios for NAA, Cho, Glu, Gln, and GABA were 1.41, 0.27, 1.18, 0.25, and 0.18, respectively, which can be compared to Schulte and Boesiger’s corresponding ratios of 1.38, 0.33, 1.18, 0.19, and 0.12 (15). The POC metabolite/Cre ratios for NAA, Cho, Glu, Gln, and GABA were calculated as 1.55, 0.23, 1.11, 0.26, and 0.17, respectively, which are closely similar to the previously reported values of 1.48, 0.28, 1.28, 0.21, and 0.17 (15).
This section briefly discusses the present reproducibility data compared with precedent literature 1D and 2D MRS findings. Note that the sensitivity of each MRS technique should be considered with respect to B0 field strength, MRS voxel size, and total measurement time (see Table 4). Mullins and colleagues reported the use of a PRESS sequence with an optimized TE (40 ms) that showed good intra-subject reliability for frontal lobe NAA, Cre, GPC/PCh, GABA, and Glu with poorer reliability recorded for Gln (38). We acknowledge that for a direct comparison our own CV values should be multiplied by √2 to be consistent with their method of CV calculation. However, the intra-subject CV values presented in that publication were based on same-session scanning thus removing potential for voxel repositioning errors. Ongur et al (7) and Lymer et al (11) reported intra-subject CV values for frontal lobe and occipital lobe 2D 1H MRS data, respectively, using 1D row extraction and fitting methods for spectral analysis. The corresponding intra-subject metabolite CV data from the present study is comparable or superior to those values, although neither report documented inter-subject CV data. The reproducibility of 2D COSY 1H MRS, as evaluated based on the measurement of metabolite volume integrals (13), generally showed higher intra-subject CV values compared to those presented in the current study. However, the inter-subject CV values presented in (13) were lower than those from the present study, which may represent natural between-subject differences in the two distinct cohorts of subjects examined. The 2D 1H MRS quantification schemes discusses (7,11,13) were unable to investigate metabolite signal interaction and discriminability as construction of the required Fisher information matrix was unfeasible.
The current implementation of 2D J-resolved 1H MRS and ProFit has several methodological limitations. The first of these concerns is the lack of inclusion of a macromolecule (MM) spectrum in the 2D basis set. Jensen et al (39) demonstrated an approximate 15% contribution of MM resonances to the GABA peak detected at and around 3.0 ppm in 2D J-resolved 1H MRS data, and MM resonances may also contribute to the resonances of Lac, Glu, Gln, and NAA (40). The MM basis function could be created empirically using metabolite-nulling based on inversion recovery methods (41) and later incorporated into the final ProFit analysis. The use of a simulated basis set generated without considering the effects of spatial localization is a further limitation of the present study. Edden et al (42) investigated J-coupling evolution in localized 2D J-resolved 1H MRS demonstrating that, within a given PRESS-localized volume, J-coupling evolves as expected or can be partially or fully refocused due to an insufficient refocusing RF pulse bandwidth. Additional metabolite signals termed “J-refocused peaks” ultimately can be detected in the final 2D spectrum, which might be more accurately accounted for by incorporating the specific shaped RF waveforms used and localization gradients into the basis set simulation. Finally, given the single-voxel nature of the present methods and the relatively long measurement time required for two discrete regions, the recent advances in rapid acquisition of 2D spatial/2D spectral 1H MRS techniques should be investigated to enable increased brain coverage within a single scan (43).
In conclusion, the data from the present study assess metabolite peak discrimination and general technique reproducibility that is achievable when using ProFit to analyze 2D J-resolved 1H MRS data recorded from the ACC and POC of human subjects. A high degree of Glu and Gln signal discrimination was detected across all subjects for both brain regions making these techniques particularly suitable for investigating a variety of psychiatric disorders. Using a CSF-corrected tissue water signal normalization scheme we observed excellent intra- and inter-subject reproducibility for a wide range of metabolites, which was at least comparable or superior to the existing literature findings. The metabolite peak correlation matrix data together with the test–retest reliability findings presented, serve as a valuable benchmark for assessing future modifications to 2D 1H MRS data acquisition and ProFit processing strategies.
ACKNOWLEDGMENT
The authors thank Drs. Rolf Schulte, Anke Henning, and Alexander Fuchs for their guidance in implementing the ProFit methodology. The authors also thank the Rebekah Hubah and Kristen DelMastro for study coordination. Dr. Renshaw is a consultant to Kyowa Hakko, Novartis and Roche and receives research support from Roche and GlaxoSmithKline. P.F.R. was supported by NIH and additional funding provided by a University of Utah Brain Institute and Utah Center for Advanced Imaging Research Pilot Program in Imaging Research grant (APP).
Contract grant sponsor: NIH; Contract grant number: K24DA015116.
REFERENCES
- 1.Ryner LN, Sorenson JA, Thomas MA. 3D localized 2D NMR spectroscopy on an MRI scanner. J Magn Reson Series B. 1995;107:126–137. doi: 10.1006/jmrb.1995.1068. [DOI] [PubMed] [Google Scholar]
- 2.Ryner LN, Sorenson JA, Thomas MA. Localized 2D J-resolved 1H MR spectroscopy: strong coupling effects in vitro and in vivo. Magn Reson imaging. 1995;13:853–869. doi: 10.1016/0730-725x(95)00031-b. [DOI] [PubMed] [Google Scholar]
- 3.Ke Y, Cohen BM, Bang JY, Yang M, Renshaw PF. Assessment of GABA concentration in human brain using two-dimensional proton magnetic resonance spectroscopy. Psychiatry Res. 2000;100:169–178. doi: 10.1016/s0925-4927(00)00075-5. [DOI] [PubMed] [Google Scholar]
- 4.Thomas MA, Yue K, Binesh N, et al. Localized two-dimensional shift correlated MR spectroscopy of human brain. Magn Reson Med. 2001;46:58–67. doi: 10.1002/mrm.1160. [DOI] [PubMed] [Google Scholar]
- 5.Ke Y, Streeter CC, Nassar LE, et al. Frontal lobe GABA levels in cocaine dependence: a two-dimensional, J-resolved magnetic resonance spectroscopy study. Psychiatry Res. 2004;130:283–293. doi: 10.1016/j.pscychresns.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Streeter CC, Hennen J, Ke Y, et al. Prefrontal GABA levels in cocaine-dependent subjects increase with pramipexole and venlafaxine treatment. Psychopharmacology. 2005;182:516–526. doi: 10.1007/s00213-005-0121-5. [DOI] [PubMed] [Google Scholar]
- 7.Ongur D, Jensen JE, Prescot AP, et al. Abnormal glutamatergic neurotransmission and neuronal-glial interactions in acute mania. Biol Psychiatry. 2008;64:718–726. doi: 10.1016/j.biopsych.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prescot A, Becerra L, Pendse G, et al. Excitatory neurotransmitters in brain regions in interictal migraine patients. Mol Pain. 2009;5:34. doi: 10.1186/1744-8069-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas MA, Ryner LN, Mehta MP, Turski PA, Sorenson JA. Localized 2D J-resolved 1H MR spectroscopy of human brain tumors in vivo. J Magn Reson Imaging. 1996;6:453–459. doi: 10.1002/jmri.1880060307. [DOI] [PubMed] [Google Scholar]
- 10.Ramadan S, Andronesi OC, Stanwell P, Lin AP, Sorensen AG, Mountford CE. Use of in vivo two-dimensional MR spectroscopy to compare the biochemistry of the human brain to that of glioblastoma. Radiology. 2011;259:540–549. doi: 10.1148/radiol.11101123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lymer K, Haga K, Marshall I, Sailasuta N, Wardlaw J. Reproducibility of GABA measurements using 2D J-resolved magnetic resonance spectroscopy. Magn Reson Imaging. 2007;25:634–640. doi: 10.1016/j.mri.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 12.Jensen JE, Licata SC, Ongur D, et al. Quantification of J-resolved proton spectra in two-dimensions with LCModel using GAMMA-simulated basis sets at 4 Tesla. NMR Biomed. 2009;22:762–769. doi: 10.1002/nbm.1390. [DOI] [PubMed] [Google Scholar]
- 13.Binesh N, Yue K, Fairbanks L, Thomas MA. Reproducibility of localized 2D correlated MR spectroscopy. Magn Reson Med. 2002;48:942–948. doi: 10.1002/mrm.10307. [DOI] [PubMed] [Google Scholar]
- 14.Cocuzzo D, Lin A, Ramadan S, Mountford C, Keshava N. Algorithms for characterizing brain metabolites in two-dimensional in vivo MR correlation spectroscopy. Conf IEEE Eng Med Biol Soc. 2011;2011:4929–4934. doi: 10.1109/IEMBS.2011.6091222. [DOI] [PubMed] [Google Scholar]
- 15.Schulte RF, Boesiger P. ProFit: two-dimensional prior-knowledge fitting of J-resolved spectra. NMR Biomed. 2006;19:255–263. doi: 10.1002/nbm.1026. [DOI] [PubMed] [Google Scholar]
- 16.Cavassila S, Deval S, Huegen C, van Ormondt D, Graveron-Demilly D. Cramer-Rao bound expressions for parametric estimation of overlapping peaks: influence of prior knowledge. J Magn Reson. 2000;143:311–320. doi: 10.1006/jmre.1999.2002. [DOI] [PubMed] [Google Scholar]
- 17.Schulte RF, Lange T, Beck J, Meier D, Boesiger P. Improved two-dimensional J-resolved spectroscopy. NMR Biomed. 2006;19:264–270. doi: 10.1002/nbm.1027. [DOI] [PubMed] [Google Scholar]
- 18.Ogg RJ, Kingsley PB, Taylor JS. WET, a T1- and B1-insensitive water-suppression method for in vivo localized 1H NMR spectroscopy. J Magn Reson Series B. 1994;104:1–10. doi: 10.1006/jmrb.1994.1048. [DOI] [PubMed] [Google Scholar]
- 19.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 21.Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 22.Klose U. In vivo proton spectroscopy in presence of eddy currents. Magn Reson Med. 1990;14:26–30. doi: 10.1002/mrm.1910140104. [DOI] [PubMed] [Google Scholar]
- 23.Cabanes E, Confort-Gouny S, Le Fur Y, Simond G, Cozzone PJ. Optimization of residual water signal removal by HLSVD on simulated short echo time proton MR spectra of the human brain. J Magn Reson. 2001;150:116–125. doi: 10.1006/jmre.2001.2318. [DOI] [PubMed] [Google Scholar]
- 24.Marshall I, Higinbotham J, Bruce S, Freise A. Use of Voigt line-shape for quantification of in vivo 1H spectra. Magn Reson Med. 1997;37:651–657. doi: 10.1002/mrm.1910370504. [DOI] [PubMed] [Google Scholar]
- 25.Brennan BP, Hudson JI, Jensen JE, et al. Rapid enhancement of glutamatergic neurotransmission in bipolar depression following treatment with riluzole. Neuropsychopharmacology. 2010;35:834–846. doi: 10.1038/npp.2009.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartha R, Williamson PC, Drost DJ, et al. Measurement of glutamate and glutamine in the medial prefrontal cortex of never-treated schizophrenic patients and healthy controls by proton magnetic resonance spectroscopy. Arch Gen Psychiatry. 1997;54:959–965. doi: 10.1001/archpsyc.1997.01830220085012. [DOI] [PubMed] [Google Scholar]
- 27.Theberge J, Bartha R, Drost DJ, et al. Glutamate and glutamine measured with 4.0 T proton MRS in never-treated patients with schizophrenia and healthy volunteers. Am J Psychiatry. 2002;159:1944–1946. doi: 10.1176/appi.ajp.159.11.1944. [DOI] [PubMed] [Google Scholar]
- 28.Castillo M, Kwock L, Courvoisie H, Hooper SR. Proton MR spectroscopy in children with bipolar affective disorder: preliminary observations. AJNR Am J Neuroradiol. 2000;21:832–838. [PMC free article] [PubMed] [Google Scholar]
- 29.Michael N, Erfurth A, Ohrmann P, et al. Acute mania is accompanied by elevated glutamate/glutamine levels within the left dorsolateral prefrontal cortex. Psychopharmacology. 2003;168:344–346. doi: 10.1007/s00213-003-1440-z. [DOI] [PubMed] [Google Scholar]
- 30.Dager SR, Friedman SD, Parow A, et al. Brain metabolic alterations in medication-free patients with bipolar disorder. Arch Gen Psychiatry. 2004;61:450–458. doi: 10.1001/archpsyc.61.5.450. [DOI] [PubMed] [Google Scholar]
- 31.Auer DP, Putz B, Kraft E, Lipinski B, Schill J, Holsboer F. Reduced glutamate in the anterior cingulate cortex in depression: an in vivo proton magnetic resonance spectroscopy study. Biol Psychiatry. 2000;47:305–313. doi: 10.1016/s0006-3223(99)00159-6. [DOI] [PubMed] [Google Scholar]
- 32.Hasler G, van der Veen JW, Tumonis T, Meyers N, Shen J, Drevets WC. Reduced prefrontal glutamate/glutamine and gamma-aminobutyric acid levels in major depression determined using proton magnetic resonance spectroscopy. Arch Gen Psychiatry. 2007;64:193–200. doi: 10.1001/archpsyc.64.2.193. [DOI] [PubMed] [Google Scholar]
- 33.Phan KL, Fitzgerald DA, Cortese BM, Seraji-Bozorgzad N, Tancer ME, Moore GJ. Anterior cingulate neurochemistry in social anxiety disorder: 1H-MRS at 4 Tesla. Neuroreport. 2005;16:183–186. doi: 10.1097/00001756-200502080-00024. [DOI] [PubMed] [Google Scholar]
- 34.Yang S, Salmeron BJ, Ross TJ, Xi ZX, Stein EA, Yang Y. Lower glutamate levels in rostral anterior cingulate of chronic cocaine users - A (1)H-MRS study using TE-averaged PRESS at 3 T with an optimized quantification strategy. Psychiatry Res. 2009;174:171–176. doi: 10.1016/j.pscychresns.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prescot AP, Locatelli AE, Renshaw PF, Yurgelun-Todd DA. Neurochemical alterations in adolescent chronic marijuana smokers: a proton MRS study. Neuroimage. 2011;57:69–75. doi: 10.1016/j.neuroimage.2011.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Li S, Marenco S, Shen J. Quantitative measurement of N-acetyl-aspartyl-glutamate at 3 T using TE-averaged PRESS spectroscopy and regularized lineshape deconvolution. Magn Reson Med. 2011;66:307–313. doi: 10.1002/mrm.23029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mullins PG, Chen H, Xu J, Caprihan A, Gasparovic C. Comparative reliability of proton spectroscopy techniques designed to improve detection of J-coupled metabolites. Magn Reson Med. 2008;60:964–969. doi: 10.1002/mrm.21696. [DOI] [PubMed] [Google Scholar]
- 39.Jensen JE, Frederick BD, Wang L, Brown J, Renshaw PF. Two-dimensional, J-resolved spectroscopic imaging of GABA at 4 Tesla in the human brain. Magn Reson Med. 2005;54:783–788. doi: 10.1002/mrm.20644. [DOI] [PubMed] [Google Scholar]
- 40.Behar KL, Ogino T. Characterization of macromolecule resonances in the 1H NMR spectrum of rat brain. Magn Reson Med. 1993;30:38–44. doi: 10.1002/mrm.1910300107. [DOI] [PubMed] [Google Scholar]
- 41.Behar KL, Rothman DL, Spencer DD, Petroff OA. Analysis of macromolecule resonances in 1H NMR spectra of human brain. Magn Reson Med. 1994;32:294–302. doi: 10.1002/mrm.1910320304. [DOI] [PubMed] [Google Scholar]
- 42.Edden RA, Barker PB. If J doesn’t evolve, it won’t J-resolve: J-PRESS with bandwidth-limited refocusing pulses. Magn Reson Med. 2011;65:1509–1514. doi: 10.1002/mrm.22747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andronesi OC, Gagoski BA, Adalsteinsson E, Sorensen AG. Correlation chemical shift imaging with low-power adiabatic pulses and constant-density spiral trajectories. NMR Biomed. 2012;25:195–209. doi: 10.1002/nbm.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]