Abstract
NADPH oxidases (NOXs) represent the only known dedicated source of reactive oxygen species (ROS) and thus a prime therapeutic target. Type 4 NOX is unique as it produces H2O2, is constitutively active, and has been suggested to localize to cardiac mitochondria, thus possibly linking mitochondrial and NOX-derived ROS formation. The aim of this study was to identify NOX4-binding proteins and examine the possible physiological localization of NOX4 to mitochondria and its impact on mitochondrial ROS formation. We here provide evidence that NOX4 can, in principle, enter protein–protein interactions with mitochondrial complex I NADH dehydrogenase subunits, 1 and 4L. However, under physiological conditions, NOX4 protein was neither detectable in the kidney nor in cardiomyocyte mitochondria. The NOX inhibitor, GKT136901, slightly reduced ROS formation in cardiomyocyte mitochondria, but this effect was observed in both wild-type and Nox4−/− mice. NOX4 may thus associate with mitochondrial complex I proteins, but in cardiac and renal mitochondria under basal conditions, expression is beyond our detection limits and does not contribute to ROS formation. Antioxid. Redox Signal. 23, 1106–1112.
Introduction
Reactive oxygen species (ROS) can originate from several sources, including the mitochondrial respiratory chain and monoamine oxidases (6, 7). NADPH oxidases (NOXs) represent the only known dedicated enzymatic source of ROS. Type 4 NOX stands out as it represents the quantitatively most abundant isoform, is constitutively active, and primarily regulated at the gene expression level. Furthermore, NOX4 is distinguished from the other isoforms by producing H2O2 in preference instead of O2− and by its independence of the interaction with classic NADPH oxidase cellular subunits, such as p47phox, p67phox, and p40phox. Upon cardiac stress, NOX4 has been described to be localized in mitochondria of cardiovascular tissues (1, 9). Moreover, the mitochondrial respiratory chain complex I was recently reported to be a functional target for NOX4 (8). NOX4 may thus mechanistically link NOX-dependent and mitochondrial respiratory chain-dependent ROS formation. Whether or not NOX4 is present and functional in mitochondria under physiological conditions and by which interactions is unclear. To this aim, NOX4 protein–protein interactions and its expression levels and possible role in mitochondrial ROS formation were investigated in the heart and kidney from wild-type (WT) and NOX4 knockout (KO) mice.
Innovation.
This is the first study to identify complex I NADH dehydrogenase subunits, 1 and 4L, as potential NOX4-binding proteins. Moreover, the localization and functional importance of NOX4 in the mouse heart and kidney mitochondria under basal conditions is investigated using two of the currently best available NOX4 antibodies and NOX inhibitors. Our data suggest that physiologically NOX4 is below the detection limit and functionally not relevant for reactive oxygen species formation in cardiac mitochondria. Under conditions of cardiac or other stress, this may change and NOX4-complex I interactions may become relevant.
NOX4 Protein Binds to Complex I Proteins
Using the DUAL membrane screening technology, 79 primary interactions were found, from which 67 plasmids were successfully rescued (Fig. 1). The BLAST output (nr database of GenBank) from prey clone sequencing, alignment, and grouping is shown in Table 1. None of the binding proteins common to NOX1 and NOX2 were found; instead, the majority of hits pointed surprisingly toward complex I NADH dehydrogenase subunits, 1 and 4L, as main NOX4 interacting proteins. These data suggested that NOX4 can indeed localize to mitochondria. Thus, NADH dehydrogenase subunits, 1 and 4L, are potentially interesting new NOX4-binding proteins, but require confirmation by immunoprecipitation or other suitable techniques in a tissue or cell model where NOX4 immunoreactive protein is detectable in mitochondria. Once confirmed, these protein–protein interactions would suggest inner mitochondrial membrane localization. Until then, mitochondrial NOX4 in complex I remains only a possibility.
FIG. 1.
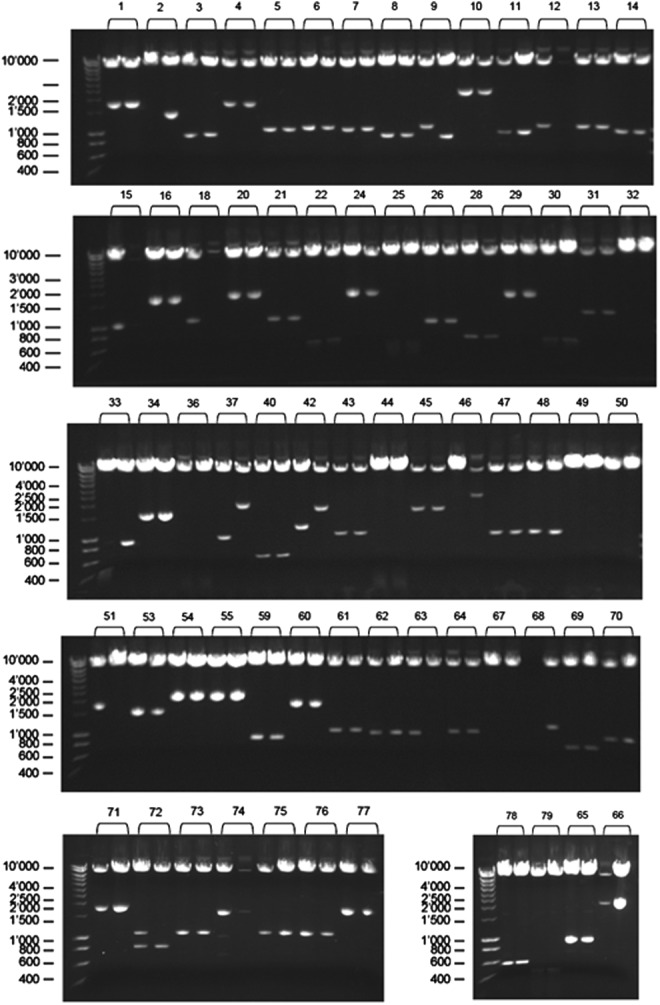
Plasmid rescue and retesting by insert analysis upon Sfi I digest. Of 79 primary NOX4 interactions from DUAL membrane screen, 67 plasmids were successfully rescued, digested with Sfi I, and analyzed by agarose gel electrophoresis. Marker size is given in bp.
Table 1.
Blast Results of NOX4 Bait-Dependent Prey Clones
| Protein | Number of picked clones | Translational table |
|---|---|---|
| cAMP-responsive element-binding protein 3 (Creb3) | 2 (group 5) | standard |
| LIM and senescent cell antigen-like domains 1 (Lims1) | 1 | standard |
| FUN14 domain containing 2 (Fundc2) | 1 | standard |
| Podocalyxin-like 2 (Podxl2) | 1 | standard |
| Ribosomal protein S28 homolog | 1 | standard |
| Glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (Grina) | 1 | standard |
| Surfeit gene 4 (Surf4) | 1 | standard |
| ORM1-like 3 (Ormdl3) | 1 | standard |
| Glycoprotein, synaptic 2 | 1 | standard |
| Solute carrier family 38, member 1 | 1 | standard |
| NADH dehydrogenase subunit 1 | 20 (group 2) | 2 |
| NADH dehydrogenase subunit 2 | 2 (group 4) | 2 |
| NADH dehydrogenase subunit 3 | 1 | 2 |
| NADH dehydrogenase subunit 4L | 12 (group 7) | 2 |
| NADH dehydrogenase subunit 5 | 2 (group 1) | 2 |
| Subunit I of cytochrome c oxidase | 2 (group 3) | 2 |
| COXIII | 1 | 2 |
Of 79 primary NOX4 interactions from DUAL membrane screen, 67 plasmids were successfully rescued (Fig. 1). Prey clones with insert were sequenced from the 5′ junction using the following primer: NXSEQfw 5′ GTCGAAAATTCAAGACAAGG 3′. All prey sequences were aligned and grouped. Preys belonging to the same group encode the same protein. Shown here is the BLAST output (nr database of GenBank) from prey clone sequencing, alignment, and grouping. The majority of hits point toward complex I NADH dehydrogenase subunits, 1 and 4L, as main NOX4 interacting proteins.
In Mitochondria Under Physiological Conditions, NOX4 is Below the Detection Limit
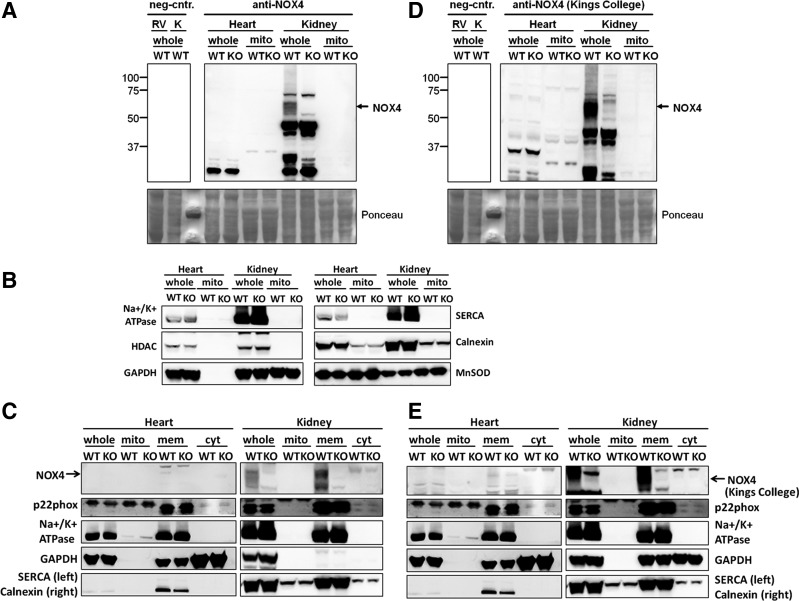
Since NOX4 is highly expressed in the kidney and a mitochondrial localization for NOX4 has been described in the heart (1, 5, 9), we next isolated mitochondria from these organs to validate this finding at an in vivo and physiological level. A 67-kDa band, which corresponded to the molecular weight of NOX4, was detected in kidneys (including membrane fractions) of WT, but not of Nox4−/− mice, confirming the specificity of both polyclonal antibodies used (Fig. 2A, C, D, E). However, in cardiac or kidney mitochondria, no NOX4 immunoreactive protein band was detected (Fig. 2A, D).
FIG. 2.
NOX4 is not detectable in cardiac and renal mitochondria under physiological conditions. (A) NOX4 Western blot from mitochondria (mito) isolated from WT and Nox4−/− mouse ventricles and kidneys. NOX4 protein was detected by Western blotting analysis using antibodies against NOX4 obtained from Abcam. A band at 67-kDa, which corresponds to the molecular weight of NOX4, was detected in kidneys of WT, but not in kidneys of Nox4−/− mice. In right ventricles (RVs), heart mitochondria, and kidney mitochondria no band for NOX4 protein was observed. For control, Western blot analysis with WT RVs and kidney lysates (whole) was performed using anti-rabbit IgG antibodies only. (B) Purity of the isolated mitochondria. Western blotting analysis with RVs and kidney lysates (whole) and with Percoll-purified mitochondria (mito) isolated from WT and Nox4−/− mouse left ventricles and kidneys was performed using antibodies against markers of the plasma membrane (Na+/K+-ATPase), the cytosol (GAPDH), the nucleus (HDAC), the sarcoplasmic reticulum (SERCA2 ATPase), the endoplasmic reticulum (calnexin), and mitochondria (MnSOD). (C) Analysis of subcellular NOX4 localization. NOX4 and p22phox protein were detected by Western blotting analysis using antibodies against Nox4 and p22phox. NOX4 protein band was detected in membrane fractions (mem) of WT kidneys, but not in WT ventricles (whole heart) and in cytosolic fractions (cyt). A band at 22-kDa, which corresponds to the molecular weight of p22phox, was detected in kidney lysates (whole kidney) and in the kidney and heart membrane of WT mice and Nox4−/− mice. Representative Western blot obtained from five experiments (n=5). (D) Stripped and reprobed NOX4 Western blot shown in (A) using antibodies against NOX4 obtained from Kings College. (E) Stripped and reprobed Western blot for subcellular NOX4 localization analysis using antibodies against NOX4 obtained from Kings College. KO, knockout; RV, right ventricle; WT, wild-type.
The purity of the mitochondrial preparation was confirmed by the absence of immunoreactivity for antibodies against markers of the plasma membrane (Na+/K+ ATPase), the nucleus (HDAC2), the cytosol (GAPDH), as well as the sarcoplasmic reticulum (SERCA2 ATPase) or endoplasmic reticulum (calnexin), and the presence of the mitochondrial marker, manganese superoxide dismutase (MnSOD) (Fig. 2B). The enrichment of membrane and cytosolic proteins was confirmed by the immunoreactivity of antibodies against markers of the plasma membrane (Na+/K+ ATPase), the endoplasmic reticulum (calnexin), and the cytosol (GAPDH). Moreover, p22phox is considered to be an obligatory NOX4-binding protein and was detected in kidney lysates and in kidney and heart membrane fractions of WT and Nox4−/− mice, but not in kidney mitochondria. Collectively, these data suggest that in mitochondria under physiological conditions, NOX4 is below the detection limit and—in the kidney—would lack its canonical binding partner, p22phox.
On the other hand, a rather tight engulfment of NOX4 in complex I and thus hidden antibody epitope may explain our difficulties in detecting NOX4 protein. Alternatively, NOX4 protein may have been below the detection limit since previous studies support a localization of NOX4 in the heart and in cardiac and renal mitochondria (1, 3, 9). The differences between these findings and our results could also be due to the use of different anti-NOX4 antibodies. However, the two antibodies used in this study are highly validated and detected a protein band at the predicted molecular weight of NOX4 in the positive (WT), but not the negative (Nox4−/−) kidney lysates controls.
NOX Inhibitors Have No Specific Effect on Mitochondrial ROS
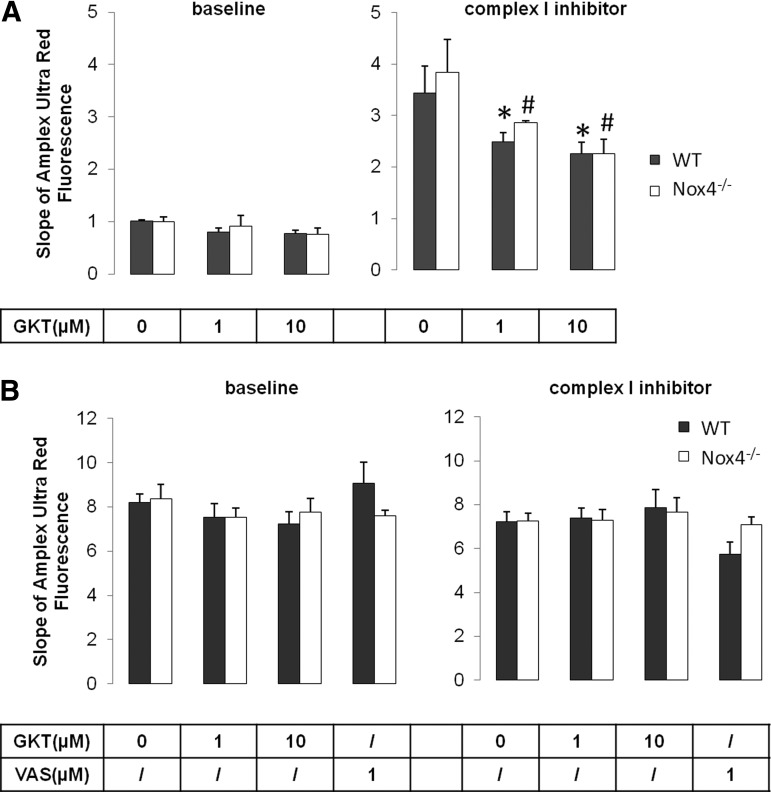
To verify the lack of functional NOX4 activity in cardiomyocyte mitochondria, ROS formation in mitochondria isolated from WT and Nox4−/− mouse left ventricles and kidneys were treated with the NOX inhibitors, GKT136901 (1 and 10 μM) and VAS2870 (1 μM, kidney only), or with 2 μl of the vehicle, 1% DMSO. ROS production was measured at baseline or in combination with the complex I inhibitor, rotenone. In heart mitochondria at baseline and after inhibition of complex I, ROS formation was decreased in a dose-dependent manner after treatment with GKT136901, in both WT and Nox4−/− mitochondria. This suggested NOX4-independent inhibitory effects on ROS formation (Fig. 3A).
FIG. 3.
The NOX inhibitors, GKT136901 and VAS2870, had no effect on ROS production in mitochondria isolated from ventricles and kidneys of WT and Nox4−/− mice. Mitochondria isolated from ventricles (A) and kidneys (B) in WT and Nox4−/− mice were treated with DMSO or with 1 and 10 μM of the NOX inhibitor, GKT136901 (GKT), or 1 μM of the NOX inhibitor, VAS2870 (VAS). Via Amplex Ultra Red method, ROS detection was performed at baseline (left) and in the presence of the complex I inhibitor, rotenone (right), using complex I substrates. Bar graphs indicate mean slope of the first three minutes±SEM obtained from 5 to 10 experiments (n=5–10). *p<0.05 vs. DMSO, #p<0.05 vs. rotenone. ROS, reactive oxygen species. Respiration control ratios (RCR) for glutamate/malate were within a range of 2; for succinate, 3 to 4.
Similar results were observed after treating heart mitochondria with the complex III inhibitor, antimycin A, in combination with the NOX inhibitor, GKT136901 (data not shown). In renal mitochondria, the NOX inhibitor, GKT136901, but also a different NOX inhibitor, VAS2870, had no effect on ROS formation, neither under basal conditions nor after complex I inhibition (Fig. 3B). Similarly to cardiac mitochondria, no differences between WT and Nox4−/− mitochondria were observed. Our data with two independent pharmacological NOX inhibitors therefore argue against a functional relevance of mitochondrial NOX4 in cardiac and renal mitochondria under physiological conditions.
These findings are in contrast to previous studies suggesting a functional significance of NOX4 for mitochondria ROS formation. However, differences in species, animal model, underlying pathophysiology, isolation method, and ROS measurement assay could explain some differences in the previous and the present results. As a matter of concern, many of the previous studies mainly measured superoxide production instead of H2O2 (1, 3, 9), which is the main product of NOX4, or used diphenylene iodonium as an inhibitor (3), which is clearly not specific for NOX, but inhibits flavoproteins in general. Probably, a majority of articles may currently argue against a mitochondrial localization.
Interestingly, our protein–protein interaction data would fit to a report that NOX4 can decrease the activity of the mitochondrial respiratory chain complex I in human endothelial cells (8). In this previous publication, reduced mitochondrial respiratory capacity was observed in shRNA vector-treated cells stimulated with ADP or rotenone, whereas the respiratory activity was preserved in NOX4 knockdown cells. No differences in the rate of respiration were determined after addition of malate and glutamate only. In accordance with this, we also did not find any differences in ROS production between WT and Nox4−/− mitochondria under basal conditions (glutamate/malate only). Moreover, complex I inhibition with rotenone resulted in a slightly higher ROS signal in Nox4−/− cardiac mitochondria compared with WT mitochondria, but did not reach significance (Fig. 3A). The finding of significantly higher protein levels for complex I subunits in NOX4 knockdown HUVECs (8) could explain the trend toward higher ROS signals in Nox4−/− mitochondria.
In summary, our data point toward new NOX4-binding proteins in mitochondrial complex I. This appears to be of little physiological relevance. Under situations of increased NOX4 expression or activity (e.g., in hypoxia), NOX4 might (potentially) translocate to mitochondria (1, 3), and then contribute to mitochondrial ROS formation or regulate complex I activity. NOX4 has been suggested to contain a putative mitochondrial localization signal (3) and to transition easily from one intracellular compartment to another (10, 11). Additional studies, however, are required to show if, under conditions of stress, NOX4-complex I interaction may gain relevance.
Notes
Animals
All animal experiments were approved by the animal care and use committee of the Justus-Liebig University of Giessen, Germany, and were in accordance with the NIH Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85-23, 1996).
WT C57BL/6 mice (Janvier SAS, CS 4105, Le Genest Saint Isle, F-53941 St Berthevin Cedex), Nox4−/− mice, and their WT littermates on a C57BL/6 background were used for the isolation of mitochondria from hearts and kidneys.
Antibodies
The following antibodies were used in the experiments: mouse monoclonal anti-rat sodium/potassium (Na+/K+)-ATPase (Upstate), mouse monoclonal anti-dog sarcoplasmic calcium (SERCA2)-ATPase (Sigma-Aldrich), rabbit monoclonal anti-human histone deacetylase 2 (HDAC2; Abcam), mouse monoclonal anti-rabbit glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Hytest), rabbit polyclonal anti-human translocase of the outer membrane 20 (Tom20; Santa Cruz), rabbit polyclonal anti-human voltage-dependent anion channel (Abcam), rabbit polyclonal anti-human manganese superoxide dismutase (MnSOD; Upstate), rabbit polyclonal anti-mouse NADPH oxidase 4 (Nox4; Abcam), rabbit polyclonal anti-NADPH oxidase 4 (2), rabbit polyclonal anti-equine cytochrome c (Cyt c; Santa Cruz), rabbit polyclonal anti-human p22phox (p22phox; Santa Cruz), and rabbit polyclonal anti-human calnexin (calnexin; Abcam).
Primer/vectors/yeast reporter strain
The following primer was used in the experiments: NXSEQfw 5′ GTCGAAAATTCAAGACAAGG 3′, and pBT3-N bait and pNubG-x cDNA library vectors and yeast reporter strain, NMY32, in selection medium (SD-trp-leu and SD-trp-leu-his-ade) were from DualSystems Biotech.
Identification of NOX4-binding proteins
To identify NOX4-binding proteins, the DUAL membrane screen was performed (GATC). Unlike the yeast two-hybrid system, the DUAL membrane system is not dependent on the localization of bait and prey in the nucleus. Both, cytosolic interaction partners and membrane proteins can be identified in a screen. The NOX4 bait (PCR product 1783 bp) was cloned using Sfi I into pBT3-N and expressed as a fusion to the C-terminal half of ubiquitin (Cub) and an artificial transcription factor (LexA-VP16). Using the pNubG-x vector, prey from a mouse adult brain library (average insert size 1.3 kb, complexity 3×106) was expressed as a fusion to a mutated N-terminal half of ubiquitin (NubG). Verification of correct expression was done by means of Western blot analysis of total yeast extracts using an antibody directed against the lexA domain of the fusion protein (Santa Cruz Technologies). Correct expression of the bait was also assayed using several positive controls. In this step, the bait was coexpressed with a selection of yeast integral membrane proteins fused to NubI (WT Nub).
Before the actual library screen, several small-scale pilot screens were carried out to optimally adjust the screening conditions. This included monitoring of background as well as interaction with positive controls and also includes a step where the bait was assayed on selective plates containing varying amounts of the compound 3-amino-1,2,4-triazole (3-AT) to modulate the sensitivity of the HIS3 reporter gene. The optimal screening conditions determined in the pilot screens were then used for the actual library screen. In the library screen, the reporter strain expressing the bait was transformed with the cDNA library. Primary selection was carried out on selective medium lacking the amino acids, histidine and adenine, and secondary selection was carried out using a semiquantitative color assay for β-galactosidase. Several prey clones that are positive in the primary and secondary selection were picked up, restreaked, and retested for both activities. The clones yielding the highest levels of β-galactosidase activity were taken further for plasmid rescue.
Library plasmids were isolated from the primary yeast clones and amplified in Escherichia coli. Since yeast cells have the potential to take up several independent library plasmids during the transformation procedure, two independent E. coli transformants were picked per original yeast clone for plasmid isolation. Insert sizes of the isolated library plasmids were determined by restriction digest. If the two plasmids derived from the same original yeast clone were identical in size, only one plasmid was processed further, but if the insert sizes were different, both plasmids were processed. After isolation, the plasmids are reintroduced into the reporter strain together with the original bait and the resulting transformants were retested for growth on selective medium and β-galactosidase activity. Only clones that scored positive in both assays were processed further. Finally, clones were sequenced from the 5′end to determine the junction between the NubG sequence and the cDNA and to identify the reading frame and the encoded protein. Prey clones with insert were sequenced from the 5′ junction using the following primer: NXSEQfw 5′ GTCGAAAATTCAAGACAAGG 3′. All prey sequences were aligned and grouped. Preys belonging to the same group encoded the same protein. DNA sequences were used and the database number of GenBank was searched using the BLAST algorithm.
Mitochondria preparation and Western blot
Mice were sacrificed by cervical dislocation and hearts and kidneys were removed. All steps were performed at 4°C to maintain mitochondrial integrity. Left ventricles were used for the isolation of subsarcolemmal mitochondria (SSM), and whole kidneys were used for the isolation of renal mitochondria. For functional analysis, left ventricles and kidneys were separately washed in buffer A (100 mM KCl, 50 mM 3-[N-morpholino]-propane sulfonic acid (MOPS), 5 mM MgSO4, 1 mM ATP, 1 mM EGTA, pH7.4) and minced in 5 ml of buffer B, that is, buffer A plus 0.04% bovine serum albumin (BSA) with scissors, and then with two (kidney) or six (heart) strokes of a Teflon pistle in a glass potter. The homogenates were centrifuged at 1000 g for 10 min at 4°C and the supernatant centrifuged again at 8000 g for another 10 min. The mitochondria were washed in buffer A, centrifuged at 8000 g for 10 min, and the resulting pellet was resuspended in 250–300 μl (SSM) or 600–800 μl (renal mitochondria) of homogenization buffer (100 mM KCl, 50 mM MOPS, 5 mM MgSO4, 1 mM EGTA, pH 7.4).
For Western blot analysis, SSM were isolated as described previously (4). In brief, left ventricles were minced in isolation buffer containing 250 mM sucrose, 10 mM HEPES, 1 mM EDTA, and 0.5% BSA, adjusted to pH 7.4 with Tris base, homogenized with an Ultra Turrax, and centrifuged at 1000 g for 10 min. The supernatant was again homogenized with two strokes of a Teflon pistle, centrifuged at 1000 g for 10 min, and the resulting supernatant was centrifuged for 10 min at 8000 g. The supernatant was collected and the pellet was resuspended in BSA free isolation buffer and centrifuged thrice at 8000 g for 10 min. The previously collected supernatant was separated into cytosolic (supernatant) and membrane fraction (pellet) by ultracentrifugation at 252,000 g for 2 h at 4°C. The cytosolic fraction was further fourfold concentrated via Vivaspin concentrators at 5000 g for 1 h at 4°C.
Kidneys were washed and minced with scissors in 5 ml of isolation buffer containing 250 mM sucrose, 10 mM HEPES, 1 mM EDTA, and 0.5% BSA (pH 7.4 adjusted with Tris base), homogenized with an Ultra Turrax, and centrifuged at 1000 g for 10 min. The resulting supernatant was further homogenized with six strokes of a Teflon pistle in a glass potter and centrifuged for 10 min at 1000 g. The supernatant was centrifuged for 10 min at 10780 g. The resulting supernatant was collected and the pellet was resuspended in BSA free isolation buffer and centrifuged thrice at 7650 g for 5 min. The previously collected supernatant was separated into cytosolic (supernatant) and membrane fraction (pellet) by ultracentrifugation at 252,000 g for 2 h at 4°C. The cytosolic fraction was further fourfold concentrated via Vivaspin concentrators at 5000 g for 1 h at 4°C.
For analysis on the purity of mitochondrial proteins, freshly isolated ventricular SSM or mitochondria from kidneys were bedded on 30% Percoll gradient, followed by ultracentrifugation at 35,000 g for 30 min at 4°C. This procedure resulted in the separation of mitochondria from other cellular constituents. The enriched mitochondrial fractions were collected, washed thrice in isolation buffer containing 250 mM sucrose, 10 mM HEPES, and 1 mM EDTA, and then centrifuged at 10,200 g for 5 min. Kidney mitochondria were further purified via anti-TOM22 MicroBeads (Mitochondria Isolation Kit; Miltenyi Biotec). As positive control, frozen mouse tissue samples of right ventricles and kidneys were homogenized with a glass pistle in a glass potter. The procedure was performed on ice to prevent heat generation.
Percoll-purified mitochondria and ∼30 mg tissue powder were denaturated in 300 μl 95°C bromphenolblue-free Laemmli buffer (62.5mM Tris-HCl, 2% SDS, 5% 2-mercaptoethanol, 10% glycerol) and subsequently incubated at 65°C for 10 min. The suspensions were sonicated (three cycles of 15 s, amplitude 30%) and centrifuged at 16,100 g for 10 min. The protein concentration in the supernatant was determined by a modified protocol described by Zaman and Verwilghen using BSA as a standard. In brief, 4 μl of the protein or standard samples was diluted to 10 μl with ddH2O, and then 90 μl of 100 mM potassium phosphate buffer, pH 7.4, was added. After mixing and incubation for 5–10 min, the samples were centrifuged (5.000 g) for 10 min at room temperature. Nineteen microliters of the clear supernatant was transferred into a well of a 96-well plate and 209 μl of the Coomassie brilliant blue G-250 solution (0.05% solution of Coomassie brilliant blue G-250 in 0.5 M perchloric acid) was added. After mixing and standing for 5 min, the absorbance at 620 nm was measured in a Tecan spectrophotometer (Infinity 200 PRO; Tecan). Forty micrograms of protein extract was electrophoretically separated on a 10% SDS polyacrylamide gel.
Detection of ROS
H2O2 was detected by the Amplex Ultra Red (A36006; Invitrogen) method. Fifty micrograms of freshly isolated cardiac mitochondria (SSM) and kidney mitochondria was treated with DMSO, with 1 or 10 μM of the NOX inhibitor, GKT136901 (Department of Synthetic Organic Chemistry, University Leipzig, Germany), and with 1 μM of the pan-NOX inhibitor, VAS2870 (Vasopharm GmbH). H2O2 production was measured in 96-well plates at baseline and in the presence of the complex I inhibitor rotenone (2 μM) using glutamate/malate incubation buffer (125 mM KCl, 10 mM Tris-MOPS, 1.2 mM MgCl2, 1.2 mM KH2PO4, 20 μM EGTA, 5 mM glutamate, 2.5 mM malate, pH 7.4). Fluorescence reading (maximum 8 wells/reading) was performed at 37°C with excitation and emission wavelengths of 565 and 581 nm, respectively, in a Cary Eclipse spectrophotometer (Agilent, Santa Clara, CA). Respiration of 0.1 mg/ml mitochondrial proteins isolated from left ventricles of WT mice was measured in a Clark-type oxygen electrode (Strathkelvin) at 25°C in the incubation buffer (containing in mM: 125 KCl, 10 Tris-MOPS, 1.2 Pi-Tris, 1.2 MgCl2, 0.02 EGTA-Tris, pH 7.4).
After recording of basal oxygen consumption (5 mM glutamate and 2.5 mM malate as substrates for complex I or 2 μM rotenone [inhibits complex I] and 5 mM succinate as substrate for complex II), respiration was determined after the addition of 40 μM ADP. The mitochondrial respiratory control, defined as the respiration rate after ADP stimulation divided by the basal respiration rate, was calculated as quality control for the mitochondrial preparation.
Statistics
Data are presented as mean values±SEM. Statistical analysis between inhibitor-treated (1 μM and 10 μM GKT or 1 μM VAS) and control mitochondria (DMSO or rotenone only) was performed by one-way ANOVA (and post hoc Fisher's comparison test). Results were considered statistically significant with a p-value less than 0.05.
Abbreviations Used
- ROS
reactive oxygen species
- WT
wild-type
- KO
knockout
- SSM
subsarcolemmal mitochondria
- BSA
bovine serum albumin
Acknowledgments
This work was supported by the European Cooperation in Science and Technology (COST Action BM1203/EU-ROS), an international research training group, 1566 Giessen, Bad Nauheim, Barcelona “Protecting the Heart from Ischemia” (PROMISE), awarded to R.S. and an ERC Advanced Grant (294683-RADMED) awarded to H.H.H.W.S. We thank Pamela W. M. Kleikers for her support in transgenic mouse management and supply.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Ago T, Kuroda J, Pain J, Fu C, Li H, and Sadoshima J. Upregulation of Nox4 by hypertrophic stimuli promotes apoptosis and mitochondrial dysfunction in cardiac myocytes. Circ Res 106: 1253–1264, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anilkumar N, Weber R, Zhang M, Brewer A, and Shah AM. Nox4 and nox2 NADPH oxidases mediate distinct cellular redox signaling responses to agonist stimulation. Arterioscler Thromb Vasc Biol 28: 1347–1354, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Block K, Gorin Y, and Abboud HE. Subcellular localization of Nox4 and regulation in diabetes. Proc Natl Acad Sci U S A 106: 14385–14390, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boengler K, Dodoni G, Rodriguez-Sinovas A, Cabestrero A, Ruiz-Meana M, Gres P, Konietzka I, Lopez-Iglesias C, Garcia-Dorado D, Di Lisa F, Heusch G, and Schulz R. Connexin 43 in cardiomyocyte mitochondria and its increase by ischemic preconditioning. Cardiovasc Res 67: 234–244, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Dai DF, Chen T, Szeto H, Nieves-Cintron M, Kutyavin V, Santana LF, and Rabinovitch PS. Mitochondrial targeted antioxidant peptide ameliorates hypertensive cardiomyopathy. J Am Coll Cardiol 58: 73–82, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaludercic N, Carpi A, Menabo R, Di Lisa F, and Paolocci N. Monoamine oxidases (MAO) in the pathogenesis of heart failure and ischemia/reperfusion injury. Biochim Biophys Acta 1813: 1323–1332, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaludercic N, Carpi A, Nagayama T, Sivakumaran V, Zhu G, Lai EW, Bedja D, De Mario A, Chen K, Gabrielson KL, Lindsey ML, Pacak K, Takimoto E, Shih JC, Kass DA, Di Lisa F, and Paolocci N. Monoamine oxidase B prompts mitochondrial and cardiac dysfunction in pressure overloaded hearts. Antioxid Redox Signal 20: 267–280, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koziel R, Pircher H, Kratochwil M, Lener B, Hermann M, Dencher NA, and Jansen-Durr P. Mitochondrial respiratory chain complex I is inactivated by NADPH oxidase Nox4. Biochem J 452: 231–239, 2013 [DOI] [PubMed] [Google Scholar]
- 9.Kuroda J, Ago T, Matsushima S, Zhai P, Schneider MD, and Sadoshima J. NADPH oxidase 4 (Nox4) is a major source of oxidative stress in the failing heart. Proc Natl Acad Sci U S A 107: 15565–15570, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyle AN, Deshpande NN, Taniyama Y, Seidel-Rogol B, Pounkova L, Du P, Papaharalambus C, Lassegue B, and Griendling KK. Poldip2, a novel regulator of Nox4 and cytoskeletal integrity in vascular smooth muscle cells. Circ Res 105: 249–259, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Lohneysen K, Noack D, Wood MR, Friedman JS, and Knaus UG. Structural insights into Nox4 and Nox2: motifs involved in function and cellular localization. Mol Cell Biol 30: 961–975, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]