Abstract
Mitochondria cooperate with their host cells by contributing to bioenergetics, metabolism, biosynthesis, and cell death or survival functions. Reactive oxygen species (ROS) generated by mitochondria participate in stress signalling in normal cells but also contribute to the initiation of nuclear or mitochondrial DNA mutations that promote neoplastic transformation. In cancer cells, mitochondrial ROS amplify the tumorigenic phenotype and accelerate the accumulation of additional mutations that lead to metastatic behaviour. As mitochondria carry out important functions in normal cells, disabling their function is not a feasible therapy for cancer. However, ROS signalling contributes to proliferation and survival in many cancers, so the targeted disruption of mitochondria-to-cell redox communication represents a promising avenue for future therapy.
The relationship between mitochondria and their host cells began approximately 2 billion years ago, when an antecedent of modern-day mitochondria was engulfed by an archezoan cell, forming the first primitive eukaryote1,2. This relationship evolved over time, as gene transfer with other prokaryotes occurred or as genes were transferred from the endosymbiont to the nucleus3,4. The original symbiotic relationship probably succeeded because of the mutual benefits derived from the complementary roles in cellular energy production. For the host cell, oxidative phosphorylation, whereby ATP is generated from ADP and inorganic phosphate, is likely to have been the principal benefit. In exchange, the antecedent mitochondria enjoyed an intracellular environment that was rich in nutrients and protected from extremes of pH that could undermine their membrane transport functions. These symbiotic interactions persist in modern-day cells, but the relationship has grown more complex in terms of the number of shared responsibilities involved in a wide range of functions. Modern-day mitochondria now participate in the biosynthesis of haem and iron-sulphur centres, regulation of cytosolic calcium ion concentrations, regulation of cellular redox status, and the generation of substrates for protein and lipid biosynthesis. Mitochondria also facilitate cellular stress responses, including the response to hypoxia and the activation of programmed cell death via the release of pro-apoptotic molecules from the intermembrane space (IMS) to the cytosol. Under normal conditions, mitochondria trigger redox signalling in the cell through the release of reactive oxygen species (ROS) from the electron transport chain (ETC). Under pathophysiological conditions, ROS generation from mitochondria can also contribute to the initiation of cancer and to an amplification of the tumour cell phenotype. At the same time, mitochondrial ROS may render the tumour cell vulnerable to therapies that further stress their ability to regulate redox homeostasis, thereby opening opportunities for novel therapies.
This Review considers how mitochondria generate ROS, how these reactive molecules contribute to the transformation of healthy cells into tumours, and how redox signalling in established tumour cells can amplify the phenotypic behaviour in terms of proliferation, survival and migration. Although tumour cells rely on increased mitochondrial ROS signalling to regulate their phenotype, this characteristic puts them in dangerous territory, in terms of their vulnerability to therapeutic interventions that further stress their redox homeostasis. How this characteristic could be exploited represents both a major challenge and an important opportunity in the treatment of this disease.
Sources of mitochondrial ROS in cancer
Cancer cells are characterized by a need for ATP, which is required to support the anabolic processes involved in growth and proliferation. Mitochondria generate ATP by oxidizing lipids, amino acids and glucose, and by transferring the electrons derived from those reactions to the ETC, which ultimately delivers them to molecular O2. Free energy conserved in this process is then used to generate ATP. The oxidation and reduction steps in these reactions involve a diverse set of metalloproteins, quinones, flavin groups and haem moieties that function as electron ‘way-stations’, analogous to stepping-stones across a river. Collectively, these discrete sites constitute a discontinuous electrical conduction system, as electrons are routed from one site to the next. For the most part, this system is designed to limit the ability of electrons to engage in interactions that would divert them from the intended pathway. However, several factors undermine the ability of the system to prevent electron escape. First, the movement of electrons from one site to the next occurs sequentially, so a transient delay at one location generates a traffic backup of electrons at earlier sites. This delay creates opportunities for electrons that are stalled at a site to interact with O2, generating superoxide, a free radical. In addition, electrical charges moving within the mitochondrial inner membrane are subjected to a strong electrical field, arising from the potential difference between the matrix and intermembrane compartments. For example, a normal membrane potential of −180 millivolts across the inner membrane of 7-nanometre thickness produces an electrical field strength that is equivalent to 257,000 volts per centimetre. This considerable field strength exerts a powerful influence on the movement of ions in the inner membrane, potentially diverting electrons from their intended path. In addition, any superoxide anions formed within the lipid bilayer would be forcefully accelerated towards the IMS by this field, thus facilitating their access to the cytosol.
While several electron transfer sites in the mitochondria have been identified as potential sources of ROS generation, definitive measurements from every possible site have not been made and it is safe to assume that many sites can potentially generate ROS. The reactivity of reactive oxygen intermediates ranges from relatively low (superoxide) to moderate (hydrogen peroxide) to extremely high (hydroxyl radical). Excessive ROS generation, or failure of oxidant scavenging systems, can disrupt cellular function by causing oxidation of lipids, proteins and DNA5. Lower levels of oxidants act as signal transduction messengers in redox signalling pathways, which have important roles in the regulation of cell function, including proliferation. It has been estimated that about 2% of the O2 that is consumed by mitochondria is involved in ROS generation6, but the assumptions required for that calculation limit its usefulness. Cancer-inducing mutations in mitochondrial enzymes can augment the generation of ROS from multiple sites, as discussed later. The following sections summarize the known and potential sites of ROS generation within mitochondria.
The tricarboxylic acid cycle
The tricarboxylic acid cycle (TCA cycle) is comprised of a series of enzymes located in the mitochondrial matrix that remove electrons from intermediary metabolites and pass these to the ETC (FIG. 1). Several enzymes in the mitochondria use flavin-containing prosthetic groups — either flavin adenine dinucleotide (FAD) or flavin mononucleotide (FMN) — as electron way-stations. These enzymes include NADH dehydrogenase, pyruvate dehydrogenase, α-ketoglutarate and succinate dehydrogenases, acyl-CoA dehydrogenase involved in fatty acid oxidation and branched-chain 2-oxoacid dehydrogenase; all of which can potentially generate ROS7. Many other mitochondrial proteins that do not directly participate in the TCA cycle also incorporate flavin groups. Some flavoproteins undergo two-electron transfers, but others transfer only a single electron at a time, forming a semiquinone radical in the process. Electrons that become stranded on the flavin group have the potential to generate superoxide, making them important potential sites of ROS generation. Other enzymes incorporate metals, such as iron in the form of iron–sulphur clusters, which can also contribute to ROS generation. Mitochondrial aconitase, which contains a [4Fe–4S]2+ iron–sulphur cluster, is potentially capable of generating ROS but is better known for its inactivation by superoxide8. Inactivation of any site in the TCA cycle can augment ROS generation from earlier sites by stalling electrons on flavin-linked or iron–sulphur clusters.
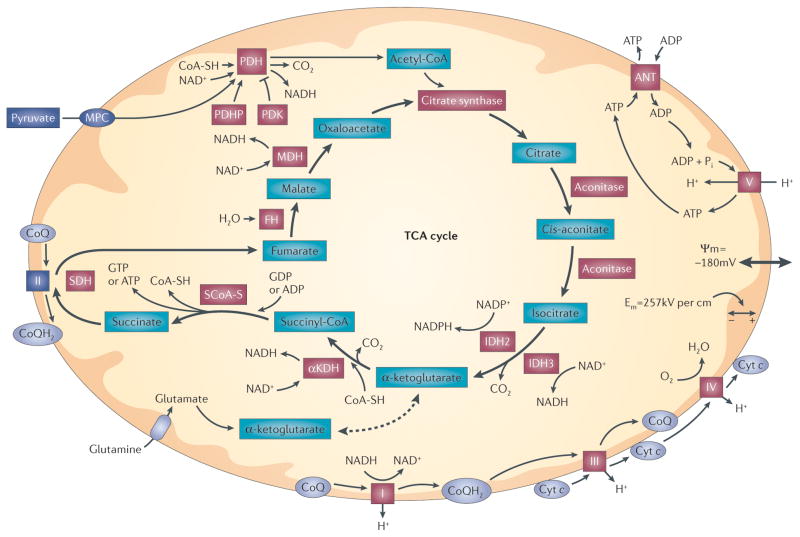
Figure 1. Mitochondrial bioenergetic function.
Pyruvate enters mitochondria via the mitochondrial pyruvate carrier (MPC), where it is decarboxylated and oxidized to acetyl-CoA by pyruvate dehydrogenase (PDH). Acetyl-CoA enters the tricarboxylic acid (TCA) cycle at citrate synthase. Subsequent steps in the cycle lead to the generation of reducing equivalents (NADH and NADPH) at dehydrogenase steps. Amino acids can also enter the TCA cycle by conversion to α-ketoglutarate, succinyl-CoA, fumarate, oxaloacetate or acetyl-CoA. For example, glutamine enters the TCA cycle after conversion to glutamate and subsequently to α-ketoglutarate. Reducing equivalents generated by the TCA cycle enter the electron transport chain and are eventually transferred to O2. Reducing equivalents generated at complex I (NADH–ubiquinone oxidoreductase) and complex II (succinate dehydrogenase (SDH)) are transferred to complex III by ubiquinol (CoQH2), a lipophilic quinone that carries a pair of electrons within the membrane. Electrons are transferred between complex III and complex IV by cytochrome c (cyt c), which carries a single electron coordinated to a haem group. Electrons that are transferred to complex IV (cyt c oxidase) are sequentially transferred to O2, generating H2O. Electron transfer steps at complexes I, III and IV are associated with proton translocation from the matrix to the intermembrane space, resulting in the generation of an electrochemical gradient across the inner membrane (ΔΨm). Complex V (ATP synthase) uses this gradient to catalyse the phosphorylation of ADP, yielding ATP. Exchange of ATP for ADP across the inner membrane is mediated by the adenine nucleotide transporter (ANT). Increases in ADP availability (as a result of cellular metabolic activity) tend to decrease ΔΨm slightly, which facilitates electron transfer at the steps involving proton extrusion. Hence, increases in cellular use of ATP cause an increase in mitochondrial respiration and ATP synthesis. By contrast, when ATP utilization falls, complex V activity decreases and the electron transport flux and oxygen consumption fall as a consequence of an increase in ΔΨm. Red highlighted proteins are known targets of reactive oxygen species (ROS). αKDH, α-ketoglutarate dehydrogenase; Em, electrical field within the membrane; FH, fumarate hydratase; IDH, isocitrate dehydrogenase; MDH, malate dehydrogenase; PDHP, pyruvate dehydrogenase phosphatase; PDK, pyruvate dehydrogenase kinase; Pi, inorganic phosphate; SCoA-S, succinyl-CoA synthase; SH, cysteine thiol.
Most dehydrogenases in the TCA cycle use nicotinamide adenine dinucleotide (NAD) or NADP as electron carriers. These cofactors bind to the dehydrogenase and are then released to carry the reducing equivalents to the ETC. Although they function as electron carriers, NADH and NADPH do not directly participate in ROS generation, because the reducing equivalents are bound covalently. This covalent bond prevents the direct oxidation of the reduced carrier by O2.
The ETC
In mitochondria, electrons that are derived from the TCA cycle are passed down the ETC and transferred to O2 at complex IV. The electron transfer steps at complexes I, III and IV are coupled with proton translocation across the inner mitochondrial membrane, which drives ATP synthesis at complex V. As with TCA cycle enzymes, electrons can escape from flavin groups or iron–sulphur clusters in the ETC and be captured by O2, forming ROS (FIG. 2). Ubiquinone (also known as coenzyme Q), is a quinone with an isoprenoid side chain. It resides in the inner membrane and delivers pairs of electrons that are derived from multiple sources — including complexes I and II — to complex III. When the first electron is donated to complex III, a free radical, ubisemiquinone, is transiently created. Any delay in the release of its second electron increases the probability that the electron will instead be donated to O2, generating superoxide9. ETC sites that are implicated in ROS generation include complexes I, II, III and ubisemiquinone. Complex IV transfers electrons to O2, so one might expect that it could generate ROS. However, upon transfer of the first electron, the binding affinity for O2 increases markedly, assuring that release cannot occur until all four electrons have reduced the O2 to H2O (REF. 10). Complex V does not participate in electron transport, and there is no evidence that it directly generates ROS. However, changes in complex V activity can produce reciprocal changes in the membrane potential, which in turn can markedly affect ROS generation from the ETC.
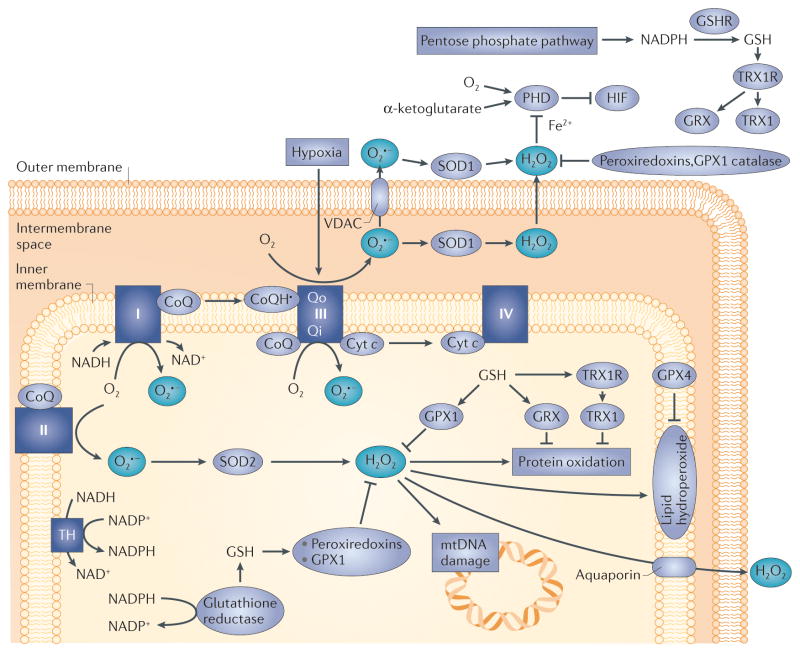
Figure 2. Mitochondrial reactive oxygen species (ROS) generation.
Electrons derived from the oxidation of metabolic intermediates can lead to the generation of ROS at specific sites in mitochondria. Transfer of a single electron to O2 yields superoxide (O2•−) which is converted to hydrogen peroxide (H2O2) by superoxide dismutase in the matrix (SOD2; also known as MnSOD), or in the intermembrane space (SOD1; also known as CuZn–SOD). The H2O2 is degraded in the matrix by glutathione peroxidase 1 (GPX1) or peroxiredoxins (PRDX3 or PRDX5) using reducing equivalents obtained from the oxidation of reduced glutathione (GSH)22–24. Oxidized glutathione (GSSG) is reduced by glutathione reductase, which obtains its equivalents from NADPH oxidation. H2O2 generated in the matrix can oxidize proteins, lipids or mitochondrial DNA (mtDNA). Oxidized proteins are repaired by thioredoxin 2 (TRX2) or glutaredoxin (GRX). TRX2 and GRX are subsequently reduced by thioredoxin reductase 2 or by glutathione. Lipid hydroperoxides are reduced by GPX4. Ultimately, all ROS removal depends on the availability of GSH, which is maintained by the availability of NADPH in the respective compartments. H2O2 can potentially leak to the intermembrane space and the cytosol when excessive ROS generation occurs or antioxidant mechanisms fail. Complexes I, II and III can potentially generate ROS in the matrix compartment. Superoxide can be released to the intermembrane space from complex III, owing to generation from ubisemiquinone at the outer ubiquinone binding site (Qo) of complex III. The electrical gradient across the inner membrane (−180 mV) creates a strong electrical field within the membrane (257 kV per cm) that accelerates superoxide anions from the membrane into the intermembrane space. Paradoxically, cellular hypoxia augments the rate of ROS generation at that site, leading to the production of H2O2 in the intermembrane space13,14,33,36,37. Subsequent diffusion to the cytosol triggers redox-dependent inhibition of prolyl hydroxylases (PHDs), negative regulators of hypoxia-inducible factor-α (HIFα) stabilization. Thus, mitochondria-derived ROS can promote cancer initiation through oxidative stress, and cancer cell progression through the activation of transcription by HIF. CoQ, ubiquinone; cyt c, cytochrome c; GSHR, glutathione reductase; Qi, inner ubiquinone binding site on complex III; TH, mitochondrial trans-hydrogenase; TRX1R, TRX1 receptor; VDAC, voltage-dependent anion channel.
Different subcellular compartments can be affected by ETC-derived ROS, depending on where the superoxide is generated. When generated at sites within the inner mitochondrial membrane, superoxide is driven towards the IMS by the strong electric field within the membrane. ROS that are derived from prosthetic groups of matrix-oriented subunits are released into the matrix compartment. ROS that are released in the matrix are scavenged by the antioxidant systems in that compartment; although high levels of matrix ROS may overwhelm that capacity and escape to the IMS and the cytosol. ROS that reach the IMS or the cytosol can then participate in redox signalling or cause oxidative damage. ROS have some ability to cross membranes, as H2O2 can travel through aquaporins and superoxide can pass through anion channels11,12. However, the cytosol, IMS and matrix contain their own collections of antioxidant enzymes, which allow the independent control of redox homeostasis in these sub-compartments13,14.
Antioxidant machinery
Scavenging of ROS is mediated by a set of ‘antioxidant’ enzymes that are expressed in various subcellular compartments. By degrading ROS, these systems limit their signalling effects and prevent oxidative damage. Superoxide dismutases (in the cytosol, the IMS and the matrix) redistribute electrons between two superoxide molecules to form hydrogen peroxide (FIG. 2). The degradation of hydroperoxides is achieved primarily by enzymes that supply electrons to reduce them to water. These enzymes include glutathione peroxidase15, which acts in concert with glutathione, a tripeptide containing a reactive cysteine residue. Reduced glutathione (GSH) is present in millimolar concentrations in the cell and is used in the scavenging of ROS. Glutathione peroxidase reduces H2O2 to water while it oxidizes GSH to form a dithiol (GSSG). Oxidized glutathione that is generated by these reactions is sequestered within vacuoles to maintain a high GSH/GSSG ratio in the cytosol16. GSSG is subsequently reduced by glutathione reductase in the cytosol and the mitochondria, using electrons obtained from NADPH.
Peroxiredoxins17,18 are a family of H2O2-scavenging enzymes that incorporate reactive cysteines, which become oxidized during the scavenging of H2O2; peroxiredoxins are re-reduced primarily by thioredoxins19. Oxidized thioredoxins in turn are restored to a reduced state by thioredoxin reductases, which use NADPH as the source of reducing equivalents.
NADPH has an important role in cancer cells because it is required for maintaining cellular redox balance. In certain cancer cells, futile redox cycling results in excessive consumption of NADPH, resulting in a chronic state of oxidative stress caused by limitations in the ability to maintain a sufficient pool of NADPH. Maintenance of NADPH levels in subcellular compartments is therefore essential for the regulation of redox in those environments. An important source of NADPH in the cytosol is the pentose phosphate pathway, which generates NADPH from the oxidation of glucose-6-phosphate by glucose-6-phosphate dehydrogenase (FIG. 2). This pathway diverts the glycolytic flux towards the synthesis of purines and the generation of NADPH in the cytosol. NADPH is also derived from folate metabolism involving methylene tetrahydrofolate oxidation to 10-formyl-tetrahydrofolate, generating NADPH. Recent studies suggest that this is a major source of NADPH in the cell20. In a separate study, Lewis et al.21 compared the fate of serine conversion to glycine in mitochondria and cytosol, by following the fate of NADPH subsequently generated by methylene tetrahydrofolate oxidation. They found that this mechanism is primarily operative in the mitochondria. Other sources of mitochondrial NADPH include isocitrate dehydrogenase 2 (IDH2) and the transhydrogenase system that exchanges reducing equivalents between NADH and NADPH (FIG. 2).
Hydrogen peroxide that escapes clearance by antioxidant systems can affect cellular signalling, principally by causing protein thiol oxidation. Protein oxidation is normally reversed by thioredoxin, glutaredoxin22 and sulphiredoxin23. These enzymes rely on thioredoxin reductase and GSH for reduction, both of which are in turn reduced by NADPH. The glutaredoxin and thioredoxin systems are expressed independently in the cytosol (GRX, TRX1 and TRX1 reductase) and the mitochondria (GRX2, TRX2 and TRX2 reductase), allowing independent regulation of redox status in these subcellular compartments22. Some tumours exhibit increased expression of TRX and TRX reductase, which may contribute to resistance to chemotherapies24.
Some cancer cells upregulate their ability to deal with oxidative stress by augmenting their expression of enzymes responsible for degrading ROS. Nuclear respiratory factor 2 (NRF2) is a transcriptional activator of genes that mitigate oxidant stress; its activity is suppressed by kelch-like ECH-associated protein 1 (KEAP1). Some tumours exhibit constitutive activation of NRF2 (REF. 25), either through gain-of-function mutations or through inactivation of KEAP1. NRF2 gain-of-function confers resistance to therapies that act by augmenting oxidant stress26,27. In some tumours, inactivation of KEAP1 by post-translational modifications leads to the activation of NRF2 (REF. 28), which may help to enhance their resistance to chemotherapeutic agents that augment oxidant stress.
Although increases and decreases in antioxidant systems have been found in various tumour cell lines, a comprehensive study of redox status across a wide range of tumour cell types has not been carried out. Hence, our understanding of how redox shifts contribute to phenotype across a broad spectrum of tumour cell types is incomplete. Conceivably, selective pressures have led to the emergence of redox alterations that promote growth and survival in each cell line. When a better understanding of redox status is achieved across a wide range of tumour cell types, it may be possible to link redox disruptions with phenotypic behaviour, resistance to therapy, originating tissue, or metastatic potential.
Hypoxia and ROS generation in cancer
Although many tumours undergo a Warburg-type shift towards glycolytic metabolism29, many still maintain substantial mitochondrial oxygen consumption. This oxygen consumption seems to represent a metabolic shift in glucose utilization towards biosynthetic and redox regulatory functions, rather than a compensation for dysfunctional oxidative phosphorylation. For example, using a novel fluorescent protein dual reporter to detect the activation of hypoxia-inducible factors (HIFs) and proliferation, Le et al.30 detected a sub-population of non-cycling cells lacking HIF activation that had high expression levels of genes involved in mitochondrial function. These cells exhibited relatively high oxygen consumptions and mitochondrial capacity while they maintained the ability to form new tumours30. In general, many tumours develop profound hypoxia when growth outpaces their vascular supply. In the mitochondrial matrix, non-specific ROS generation decreases during hypoxia and increases when cells are made hyperoxic31,32. However, release of ROS from complex III to the IMS increases paradoxically during hypoxia13,33. Evidence for this response comes from a large number of studies from multiple laboratories using diverse methods to assess oxidant signalling13,34–38, targeted antioxidant studies39,40, genetic suppression or deletion models31,36–38 and studies of in vivo tumour growth39,41,42. Although the specific mechanism has not been described, a likely source is the outer ubiquinone binding site (Qo) of complex III, in which lowered oxygen concentrations may prolong the lifetime of the semiquinone radical. This could enhance superoxide generation by augmenting the conditions favouring its generation, even though the oxygen concentration is lower9. Targeting a hydrogen peroxide scavenger specifically to the mitochondrial IMS abolishes hypoxia-induced increases in oxidant stress in that compartment and the cytosol, and abrogates the downstream stabilization of HIF1α in the cytosol14. The enhanced generation of mitochondrial ROS in the hypoxic tumour microenvironment represents an important mechanism promoting growth, metabolic reprogramming and survival, as these oxidants can act both as signalling molecules and by damaging DNA.
Mitochondrial DNA and ROS in cancer
Mitochondrial DNA as a target of ROS
In the context of cancer initiation, an important target of mitochondrial ROS is mitochondrial DNA (mtDNA), as mutations in mtDNA seem to be capable of promoting tumorigenesis43,44. Human mitochondria contain redundant copies of a circular loop of DNA comprised of ~16.6 kilobases, encoding 13 polypeptide components of the oxidative phosphorylation system (OXPHOS system), 12S and 16S ribosomal RNAs, and 22 transfer RNAs. The polypeptides include seven of the nearly 45 subunits of complex I, one subunit of complex III, three subunits of complex IV and two subunits of complex V, the ATP synthase. The remaining ~1,500 proteins in the mitochondria are encoded by nuclear DNA. Each mitochondrion potentially carries dozens of mtDNA copies and each cell contains scores of mitochondria, so each cell can potentially contain many hundreds of mtDNA copies. mtDNA is maternally inherited, and mutations in mtDNA that contribute to cancer can either be inherited as germline mutations or appear as somatic mutations in specific tissues. Functionally, mutations can either be neutral, pathogenic or beneficial.
Unlike nuclear DNA, mtDNA is not protected by histones; therefore, the proofreading capacity is limited and ROS that is generated in the matrix can attack it45. Accordingly, the rate of mitochondrial mutation is much greater than for nuclear DNA46. In general, mutations can include deletions, insertions, point mutations and changes in mtDNA copy number. When a mutation arises, it can be passed to the daughter cells along with the normal mtDNA, resulting in a state of heteroplasmy. For reasons that are not fully understood, non-dichotomous segregation of mutant and wild-type mitochondria can occur during cell division, causing a subset of the tumour cell population to drift towards a state of homoplasmy, in which all mtDNA copies are of the mutant form. Both normal and cancer cell mitochondria seem to contain considerable amounts of homoplasmic and heteroplasmic mutations in mtDNA in different tissues of the same individual47, although the importance of this in cancer is not fully clear.
mtDNA mutations and the generation of ROS in cancer
Alterations in mtDNA were described in leukaemic cells even before the age of DNA sequencing48,49. Germline and somatic mtDNA mutations have been linked to colorectal cancer50, renal, lung, bladder, head and neck, gastric, thyroid, prostate and ovarian cancers, glioblastomas and hepatocellular carcinomas51 and others, as reviewed previously43,52. There is good evidence suggesting that at least some mtDNA mutations are tumour-specific43. In some cases the somatic mtDNA mutations identified in tumours represent a natural progression that is synonymous with the neutral mutations that developed in the general population during human evolution53. It is also possible that these changes represent selection against detrimental mutations54. In support of this idea, a progressive shift in haplotype has been tracked in a patient with myelodysplastic syndrome, during progression to acute leukaemia. The slow progress of this transition made it possible to document a shift from the original germline haplotype to a heteroplasmic state, and finally to a homoplasmic state55. Similar shifts over time have also been noted in other studies56. Somatic mutations that are rarely seen in the general population are sometimes seen in tumours, suggesting that the functional changes they introduce may confer some beneficial effect on the tumour growth and survival43.
More compelling evidence for the role of mtDNA mutations in cancer comes from the observation that the introduction of mutant mtDNA into a tumour cell line can alter its phenotype. Petros et al.57 compared the in vivo growth of PC3 prostate cancer cells that were homoplasmic for the mtDNA mutation T8993G to those with wild-type mtDNA (T8993T). The T8993G mutation was previously shown to result in an amino acid substitution in complex V that significantly decreased its activity58 and increased mitochondrial ROS generation59. The mutant mtDNA-containing cells grew faster than wild-type cells in immunocompromised mice, suggesting that mutations in mtDNA have the potential to alter the phenotype in an existing tumour cell line. Additional insight comes from the study of cybrids. These are cells generated by fusing enucleated cytoplasts containing mutant mitochondria with tumour cells that have been depleted of their endogenous mtDNA (ρ0 cells). The effects of different mitochondrial haplotypes on the tumorigenic behaviour of the cybrids can then be compared in the same nuclear genetic background. Shidara et al.60 studied cybrid lines containing mutations in the mitochondrial ATP synthase subunit VI, T8993G and T9176C, and found that these mutations conferred a growth advantage and apoptotic resistance when grown in nude mice. Collectively, these findings indicate that at least some mitochondrial mutations can markedly increase the tumour phenotype, potentially through the generation of ROS.
How do mtDNA mutations contribute to tumorigenicity? Missense and nonsense mtDNA mutations can potentially alter the function of the ETC or the ATP synthase. One conceivable mechanism is that the mutation causes a decrease in mitochondrial function that promotes a Warburg-like shift towards glycolysis. However, there are clear counter-examples of that explanation61. More likely, the decrease in activity caused by the mutation produces a shift in mitochondrial membrane potential or the redox status of electron carriers upstream or downstream from the altered site. Changes such as these do not necessarily inactivate oxidative phosphorylation, but they may introduce shifts in function that alter ETC redox status and thus ROS generation. For example, mutations that decrease but do not abolish activity of complex IV can shift the reduction state of electron carriers upstream in the chain, thereby augmenting the probability of superoxide generation from complex I, II or III60. To the extent that the ROS generated by this change affects cytosolic redox signalling involved in the regulation of proliferation, this could amplify the growth rate. The magnitude of these effects in cells with heteroplasmic versus homoplasmic mtDNA is not known, but it seems likely that the answer would vary depending on the specific mutation involved.
A clear illustration of how alterations in mtDNA can drive tumorigenesis comes from the study of Woo et al.42, who noted that mitochondrial transcription factor A (Tfam)+/− mice develop a modest decrease in mtDNA copy number and an increase in mtDNA oxidative damage, suggestive of increased basal mitochondrial ROS generation. When bred with the adenomatous polyposis coli (Apc)Min/+ mouse tumour model, the mice developed increased numbers of intestinal tumours that grew faster. The tumour-promoting effect was linked to an increase in mitochondrial ROS by breeding the ApcMin/+ mice with transgenic mice that overexpress catalase targeted to the mitochondrial matrix. Those mice showed fewer polyps in the intestine in association with a decrease in mitochondrial oxidant stress. These results provide an important link between mtDNA stability and tumorigenesis that is mediated by increases in mitochondrial ROS generation.
The understanding of how specific mtDNA alterations regulate the tumorigenic phenotype is still incomplete. Difficulty in addressing this question arises because it is often difficult to judge how a specific alteration affects mitochondrial function. One approach to solving this has been to introduce similar changes into simpler model systems that express orthologous complexes, and to use these to investigate function. For example, introduction of a G171D mutation in subunit I of the cytochrome oxidase in Rhodobacter sphaeroides produced a structural shift resulting in a decrease in activity arising from slowed intramolecular electron transfer, along with a specific proton leak across the inner membrane62. Another approach has been to replace the mtDNA in a cancer cell with a mutant form to determine the consequences in terms of growth, survival or metastatic behaviour. For example, Ishikawa et al.44 replaced the endogenous mtDNA in a poorly metastatic mouse tumour cell line (P29) with mtDNA carrying a loss-of-function mutation in complex I subunit 6 (NOD6) from a highly metastatic line (A11). This transfer conferred the highly meta-static phenotype to the recipient cells, in association with an increase in cellular ROS generation that was required for the altered metastatic behaviour. Examples such as this provide a proof-of-principle that some mtDNA mutations have the potential to shape tumour cell behaviour and survival, potentially through the generation of ROS. However, it seems likely that many other mutations, although correlated with oncogenic transformation, are merely innocent events that do not drive behaviour.
Mitochondrial ROS and genomic instability
Mitochondrial oxidants also contribute to genomic instability. In fibroblasts that are deficient in mitochondrial superoxide dismutase (SOD2; also known as MnSOD), Samper and colleagues63 detected increases in double-strand breaks and chromosomal translocation, along with loss of cell viability and proliferative capacity. Mice lacking Sod2 die shortly after birth, whereas heterozygotes (Sod2+/−) survive but develop mammary tumours as they age64. In single-cell zygotes, Liu and colleagues65 introduced mitochondrial dysfunction using the uncoupling agent carbonylcyanide-4-trifluorometh-oxyphenylhydrazone (FCCP) and observed increases in ROS, as well as telomere loss and chromosome damage that was prevented by antioxidants. Loss of caveolin 1, a structural component of membrane caveolae, occurs in response to transformation in cancer-associated fibroblasts, and its downregulation in breast cancer is a negative prognostic indicator. Loss of caveolin 1 in cancer-associated fibroblasts leads to mitochondrial dysfunction that is mediated by increased nitric oxide generation, leading to increased mitochondrial ROS generation, which in turn contributes to genomic instability in adjacent cancer cells66. In patients with Fanconi anaemia, a genetic disorder that is associated with bone marrow failure and increased risk of cancer, increased levels of DNA oxidative damage correlate with increased generation of ROS, apparently from mitochondria67. In lymphocytes from patients with Fanconi anaemia, Ponte et al.68 found that antioxidants significantly improved genomic stability. Finally, upregulation of matrix metalloproteinase 3 (MMP3) is frequently observed in cancer. It induces epithelial–mesenchymal transition (EMT), and its expression in transgenic mice leads to the formation of mammary tumours with genomic instability. Interestingly, MMP3 induces the expression of an alternatively spliced form of the small GTPase RAC1, which stimulated ROS generation from the mitochondria69. The resulting oxidant stress then contributes both to signalling by induction of the transcription factor SNAIL (also known as SNAI1), and to genomic instability. Collectively, these studies reveal that mitochondrial oxidants in cancer cells have the capacity to enhance the tumour phenotype by inducing genomic damage, and that mitochondrial oxidants in other disorders can promote oncogenic transformation, EMT and other tumour-inducing behaviours through the induction of genomic instability.
ROS as death inducers in tumour cells
Excessive mitochondrial oxidant stress can induce cell death, both in tumours and in healthy cells. However, the mechanism underlying this process has been controversial, with some studies implicating apoptosis and others involving the mitochondrial permeability transition pore (MPTP). Evidence for the involvement of apoptosis comes from studies showing that cytochrome c is released from the mitochondria to the cytosol after inducing oxidant stress, and that caspase activation ensues70. Other studies implicate the MPTP, a small yet incompletely defined channel that permits the transit of small solutes (<1.5 kDa) from the mitochondrial matrix to the cytosol71. Its molecular characterization has been controversial but, minimally, it seems to include a dimer of complex V and cyclophilin D72. Opening of the MPTP is crucial for cell death in ischaemia–reperfusion injury in the heart, as genetic deletion of cyclophilin D confers significant protection73. In the heart, reperfusion after an ischaemic insult causes the generation of excessive mitochondrial ROS74. These oxidants synergize with Ca2+ to cause opening of the MPTP, which depolarizes the mitochondria and permits redistribution of solutes such as NAD+ and NADH to escape to the cytosol75. ROS reaching the nucleus cause DNA damage, leading to poly(ADP-ribose) polymerase (PARP) activation and DNA repair. If left unchecked, however, PARP activity consumes cytosolic and/or mitochondrial NAD+, which halts glycolysis and causes a lethal bioenergetic crisis75. Many hours later, mitochondrial swelling in the injured cells triggers mitochondrial outer membrane rupture and cytochrome c release, but that process occurs in a cell that is already committed to death by a different pathway75. Indeed, excessive oxidant stress in cultured cells also causes MPTP opening as well as cytochrome c release, and significant protection is conferred by PARP inhibition. However, loss of either BAX or cytochrome c expression confers no protection in that model74. Collectively, these findings suggest that excessive mitochondrial oxidant stress triggers a death pathway involving MPTP activation. Although later release of pro-apoptotic factors from the mitochondria may occur, this represents a late event whose inhibition is not required for the death response. In tumours, a similar cell death process may occur when intermittent blood flow causes transient ischaemia in the interior core, followed by reperfusion and an associated burst of mitochondrial ROS generation76. This occurrence may help to explain why some tumours demonstrate necrotic cores even though genetic mutations may have inactivated their ability to undergo apoptosis.
ROS as signalling agents in cancer
ROS and signalling
Oncogenic transformation is frequently associated with a shift in cytosolic thiol redox balance to a more oxidized state, which may enhance the proliferative phenotype77. This shift may also contribute to genomic instability, as ROS in the cytosol can enter the nucleus during DNA replication to cause additional mutations. Although excessive oxidant stress is lethal, low levels of mitochondrial ROS have crucial roles in signalling and the regulation of protein function. Important targets of ROS signalling include reactive cysteine groups on proteins, which can be oxidized by H2O2 but not by superoxide. Protein thiol oxidation can cause conformational and functional changes and can also result in intra- or inter-molecular dithiol linkages. An important group of proteins subject to ROS attack are the lipid and protein phosphatases, which are inactivated by oxidation of the reactive cysteine thiol at their catalytic site78,79. Oxidative inactivation of phosphatases can cause major changes in the phosphorylation of protein targets of the MAPK–ERK and AKT kinase pathways that contribute to the proliferation and survival of cancer cells. In a broader sense, it is possible that the effects of genetic mutations in certain genes could be mimicked in tumour cells that develop continuous increases in basal ROS generation. For example, the continuous oxidation-mediated inactivation of PTEN by constitutive increases in ROS generation could potentially mimic the phenotypical changes induced by genetic inactivation of that gene. In that regard, oxidant stress-induced enzyme dysfunction could synergize with genetic mutations to amplify tumorigenic behaviour in some cancer cells. This might explain why growth inhibition in response to antioxidant treatment has been observed in some studies80.
ROS and hypoxia
The hypoxic environment within tumours promotes the stabilization of HIFs81–83. Destabilization of the HIFα subunits of HIF1 and HIF2 is mediated by prolyl hydroxylases (PHDs), which hydroxylate conserved proline residues located within an oxygen-dependent degradation domain84 in a 2-oxoglutarate- and oxygen-dependent reaction85,86. That modification then facilitates interaction with the E3 ubiquitin ligase, von Hippel–Lindau tumour suppressor protein (VHL), targeting the HIFα subunit for proteasomal degradation87. Whereas PHD activity is abolished in the absence of oxygen (anoxia), regulation of PHD activity under more physiological hypoxia is regulated by a paradoxical increase in ROS generation by mitochondria33,88. During physiological hypoxia, ROS originate from complex III (FIG. 2), as knockdown of the Rieske iron–sulphur protein (UQCRFS1), a subunit required for electron flux in that complex, attenuates the hypoxia-induced ROS signalling and the inhibition of PHD activity and HIF1α stabilization in hypoxia36,37,89. However, exogenous ROS can rescue HIF1α stabilization in the absence of the complex. Hypoxia-induced mitochondrial ROS signals are released from the Qo site of complex III to the mitochondrial IMS, and they subsequently enter the cytosol. Accordingly, fluorescent redox reporter sensors targeted to the mitochondrial matrix, the IMS or the cytosol detect increases in protein thiol oxidation in the IMS and the cytosol during hypoxia, while oxidant stress in the matrix simultaneously decreases14. Expression of a peroxide scavenger, peroxiredoxin 5, within the IMS attenuates hypoxia-induced thiol oxidation in that compartment and in the cytosol, while it also attenuates hypoxia-induced HIF1α stabilization14. These observations demonstrate that during hypoxia, ROS that are released from the inner mitochondrial membrane cross the IMS to reach the cytosol, where they inhibit PHDs and thereby contribute to the activation of HIF transcription factors. Although the mechanism of PHD inhibition is not known, it may involve oxidation of the Fe2+ within PHDs. Importantly, hypoxia-induced ROS signalling by mitochondria is a characteristic of both primary and malignant cells14,31 and seems to function as a normal oxygen-sensing mechanism9. Hence, the enhancement of HIF activation by ROS signalling within the tumour is an example of how mitochondria carrying out their normal functions are co-opted by the tumour cells to promote growth and survival through the induction of HIF target genes.
Although oxidant stress in malignant cells probably contributes to the acquisition of additional mutations, some studies suggest that the amplifying effects on HIF regulation are also important. In a study assessing genomic instability in tumours, Gao et al.80 administered N-acetyl cysteine (NAC) in a mouse model of MYC-dependent human B cell lymphoma and found that growth was inhibited, whereas genomic instability was unaffected. The beneficial effect of NAC was traced to its inhibition of HIF activation in a PHD- and VHL-dependent manner. Moreover, expression of an oxygen-independent mutant HIF1α abolished the protective effect of NAC. These findings reveal the importance of hypoxia-induced ROS for the regulation of hypoxia-induced responses that determine the tumour phenotype.
Pseudohypoxic activation of HIF in cancer
HIFα stabilization occurs when PHD activity declines, as occurs during hypoxia. Pseudohypoxic stabilization of HIFα refers to conditions in which PHD is inhibited not by a lack of O2, but rather by a lack of 2-oxoglutarate, or chelation of its adducted iron, or by pharmacological inhibitors such as dimethyloxallyl-glycine (DMOG) (FIG. 3). Low levels of exogenous ROS can also inhibit PHD during normoxia, presumably by oxidizing its iron to the ferric state36. Succinate, a TCA cycle intermediate with structural similarities to 2-oxoglutarate, can also theoretically inhibit PHD90. Depending on their mechanism of action, pseudohypoxic activators can also trigger other responses that can alter the phenotype of tumour cells (FIG. 3).
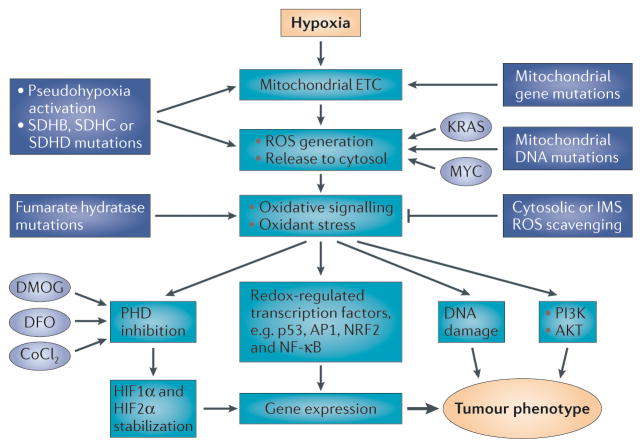
Figure 3. Hypoxia and pseudohypoxia activate mitochondrial reactive oxygen species (ROS) generation and oxidant signalling that drives the tumour cell phenotype.
Hypoxia triggers ROS generation by the mitochondrial electron transport chain (ETC), leading to activation of hypoxia-inducible factor 1 (HIF1) and HIF2 through prolyl hydroxylase (PHD) inhibition88. Hypoxia also activates other responses and transcription factors through ROS-dependent signalling. Pseudohypoxia mimetics trigger the effects of hypoxia by activating or inhibiting steps in the hypoxia response pathway. Oncogenes including KRAS and MYC can further drive ROS generation. Mutations in mitochondrial DNA (mtDNA)- or nuclear DNA-encoded proteins can augment mitochondrial ROS generation and mimic the effects of hypoxia57,59,63. Succinate dehydrogenase (SDH) mutations in subunits SDHB, SDHC or SDHD augment ROS generation from the ETC. Fumarate hydratase loss-of-function mutations lead to oxidative stress that activates redox signalling. Cobalt chloride (CoCl2) increases oxidant generation by redox cycling, thereby augmenting ROS generation33. Depending on the site of action, pseudohypoxic activators may only mimic some of the responses seen during authentic hypoxia. For example, chemical inhibitors of PHD (dimethyloxallyl glycine (DMOG) and desferrioxamine (DFO)) only trigger the activation of HIF1 and HIF2 without activating other pathways. AP1, activating protein 1; IMS, intermembrane space; NF-κB, nuclear factor-κB; NRF2, nuclear respiratory factor 2.
Succinate dehydrogenase (SDH) is a TCA cycle enzyme comprised of four nuclear-encoded subunits (A–D). This heterotetramer links the TCA cycle to the ETC by oxidizing succinate to fumarate and passing the electrons to ubiquinone, a mobile electron carrier that delivers them to complex III (FIG. 2). Mutations in the A subunit of SDH (SDHA) diminish enzymatic activity and thereby impair mitochondrial oxidative phosphorylation. The mutation-induced loss of function causes a bioenergetic deficiency that manifests as encephalopathy, myopathy and Leigh syndrome91. However, with one exception92, SDHA mutations have not been associated with cancer. By contrast, mutations in the SDHB, SDHC or SDHD subunits produce a tumorigenic phenotype that is associated with pheochromocytomas and paragangliomas in humans93–96. Paragangliomas are benign, highly vascular tumours the incidence of which is increased in populations living at high altitude97. It was suggested that succinate accumulation caused by decreased SDH activity inhibits PHD, thereby stabilizing HIFα90. But that cannot explain why HIF is activated by SDHB, SDHC or SDHD subunit mutations but not by those in SDHA. A more likely mechanism is that defects in the SDHB, SDHC or SDHD subunits trigger HIFα by augmenting ROS generation at the SDHA subunit98. SDHA catalyses the oxidation of succinate to fumarate, using an FAD group as an electron carrier. Other electron carriers within SDH include three iron–sulphur clusters and a haem moiety in the SDHB, SDHC and SDHD subunits. Mutations in the SDHA subunit interfere with catalytic activity and prevent oxidation of succinate. However, if SDHA is normal and there are defects in the SDHB, SDHC or SDHD subunits, then the oxidation of succinate proceeds but the shuttling of electrons within the complex to the electron carrier ubiquinone is impaired28,98. Hence, the electron obtained from succinate remains on the FAD group, where it can react with O2 to form superoxide. That event clears the FAD group and permits SDHA to oxidize another succinate, creating a futile redox cycle yielding superoxide. Consistently, Guzy and colleagues98 showed that genetic suppression of SDHB but not SDHA expression results in an increase in ROS signalling in tumour cells, leading to an increase in HIF1α stabilization, an increase in cell proliferation and increased tumour xenograft growth. Cells with stable suppression of SDHB exhibited HIF1α stabilization in normoxic conditions and this suppression was inhibited by antioxidants, whereas no increase in HIF1α was observed in cells with SDHA knockdown. Interestingly, mutations in SDHD have also been shown to contribute to genomic instability through the increased generation of superoxide and H2O2 (REF. 99). This provides an attractive mechanism to explain how a mutation in a single gene encoding SDHB, SDHC, or SDHD could, over time, lead to the ROS-mediated accumulation of additional genomic mutations resulting in the tumour phenotype. In a broader sense, it shows how a somatic or germline mutation could cause a small but continuous increase in ROS generation that undermines genomic stability progressively over time, thereby pushing the cell (or cells) towards neoplastic transformation.
A parallel story has emerged with respect to fumarate hydratase (FH), a TCA cycle enzyme that converts fumarate to malate (FIG. 4). Humans with mutations in FH are predisposed to hereditary leiomyomatosis and renal clear cell cancer (HLRCC)100. Individuals with a germline mutation in one allele of FH typically undergo somatic loss of heterozygosity, often in the kidney. Tissue analysis in HLRCC tumours revealed excessive HIF1α levels in the cytosol and nucleus101, consistent with the idea that biallelic loss of FH contributes to tumour formation through the activation of HIF1 (REF. 102). Increased oxidant stress seems to explain this phenotype. Loss of FH activity abolishes oxidative phosphorylation, forcing the cells to rely entirely on glycolysis for energy production103. Although the TCA cycle is blocked at FH, it can still operate in reverse to metabolize glutamate to citrate by reductive carboxylation104. This reaction yields citrate, which is exported to the cytosol for use in the biosynthesis of lipids105. TCA cycle inhibition in this manner can potentially increase the generation of ROS by halting the flux of electrons at iron–sulphur centres or flavin groups. Trapped in the ‘traffic gridlock’, these electrons may be captured by O2 to generate superoxide that is sub-sequently dismuted to H2O2. Indeed, Sudarshan et al.103 found that ROS levels were constitutively elevated in HLRCC-derived tumour cells and in renal epithelial cells in which FH had been genetically suppressed. Importantly, the increase in HIF1α was dependent on ROS generation, presumably because oxidants inhibit the activity of PHDs.
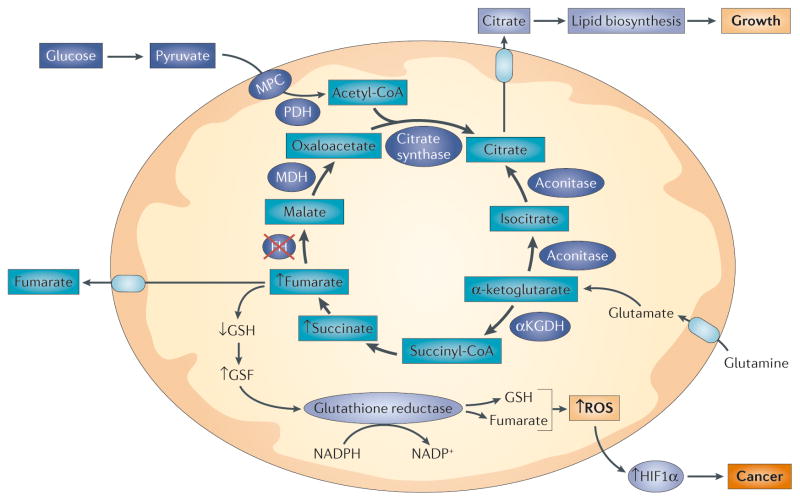
Figure 4. Mutations in tricarboxylic acid (TCA) cycle enzymes drive tumour cell progression through the generation of oxidant signalling.
Inactivating mutations in fumarate hydratase (FH) block TCA cycle function, causing accumulation of fumarate and succinate. Fumarate reacts with reduced glutathione (GSH) to produce succinated glutathione (GSF), an oncometabolite that is degraded by glutathione reductase at the expense of NADPH28. Released GSH is then available to recombine with fumarate in a futile cycle that consumes NADPH. This impairs the reactive oxygen species (ROS) detoxifying capacity of mitochondria, leading to an increased ROS release to the cytosol that inhibits prolyl hydroxylase and increases the stabilization of hypoxia inducible factor 1α (HIF1α). Mitochondria continue to metabolize α-ketoglutarate to produce citrate through reverse carboxylation104. The supply of α-ketoglutarate is maintained by glutamate derived from glutamine deamination. Citrate that is produced by this reverse operation is then exported to the cytosol, where it is used to generate acetyl-CoA that is needed for lipid biosynthesis. Thus, TCA cycle inhibition can produce diverse effects in a tumour cell by driving ROS-dependent and ROS-independent signalling. αKGDH, α-ketoglutarate dehydrogenase; Acon, aconitase; KEAP1, kelch-like ECH-associated protein 1; MDH, malate dehydrogenase; MPC, mitochondrial pyruvate carrier; NRF2, nuclear respiratory factor 2; PDH, pyruvate dehydrogenase.
Further insight into the mechanism by which FH loss augments ROS signalling and HIF activation comes from Sullivan et al.28, who observed that mitochondria lacking FH oxidize glutamine to fumarate in the TCA cycle. As fumarate accumulates, it reacts with GSH to yield succinated glutathione (GSF), a cancer-associated metabolite that is subsequently metabolized by glutathione reductase at the expense of NADPH. As NADPH levels become depleted by the recursive generation of GSF, the ability of glutathione reductase to reduce GSSG becomes compromised. This causes a redox shift in the cell towards a more oxidized state, leading to an increase in ROS-dependent signalling, PHD inhibition and HIF1α stabilization28. Thus, fumarate constitutes a proto-oncometabolite by undermining cellular antioxidant defences, thereby favouring oxidant-driven pathways that are involved in tumorigenic and metastatic behaviour. These findings reveal that disrupted mitochondrial function can promote tumour progression through the activation of HIF in a ROS-dependent manner.
Oncogenes and mitochondrial ROS
Oncogenic transformation promotes tumorigenesis by increasing mitochondrial ROS generation106. For example, KRAS activation increases oxidant stress in the mitochondrial matrix as detected by a fluorescent redox sensor39. Mitochondria-targeted nitroxide scavengers of superoxide attenuated the mitochondrial ROS levels and abolished anchorage-independent cell growth, compared to cells treated with chemically similar untargeted nitroxides. The mitochondria-targeted antioxidants also induced cell proliferation arrest. These findings link the matrix ROS generation with the ability to proliferate in soft agar39.
To determine the mitochondrial site that is responsible for ROS generation, 143B osteosarcoma ρ0 cells were reconstituted with either normal mitochondria or with others containing a mutation in the mtDNA-encoded cytochrome b gene39. The ρ0 cells lack mtDNA, their ETC is dysfunctional, they do not respire, and their mitochondrial ROS levels were undetectable. The cytochrome b mutants also lacked respiration but could still generate ROS because they sustain electron transport into complex III. Unlike ρ0 cells, the cytochrome b mutants demonstrated anchorage-independent cell growth, which was abolished by genetic suppression of complex III. To test this idea in vivo, these investigators used an inducible KRAS-driven mouse model of lung cancer (LSL-Kras-G12D+/fl mice) combined with genetic deletion of Tfam. The loss of TFAM leads to loss of mtDNA, mitochondrial transcription and inactivation of the ETC. In that model, loss of Tfam led to a decrease in the number and size of lung tumours and in tumour cell proliferation. These results underscore the importance of the ETC and mitochondrial ROS for the growth and proliferation in KRAS-G12D-induced tumorigenesis.
Other oncogenes also manipulate mitochondrial ROS generation to support proliferation and survival. The transcription factor MYC is activated by extra-cellular mitogenic signals that regulate the MAPK–ERK signalling and normal cell proliferation107. In lymphomas, oncogenic MYC activation causes an acceleration of mitochondrial glutaminolysis108, enhanced mitochondrial biogenesis109, and activation of lactate dehydrogenase A (LDHA) expression110 to drive glycolytic metabolism. These changes are associated with an increase in ROS signalling that renders the cells more susceptible to exogenous oxidant stress77. These ROS signals also contribute to proliferation by activating transcription factors such as HIF or nuclear factor-κB111, and by activating AKT, possibly through the oxidative inactivation of the phosphatase PTEN79.
Summary and therapeutic implications
Are mitochondrial ROS the initiators, amplifiers or the Achilles’ heel in cancer? The answer is all of the above. Mitochondrial ROS can alter cellular redox regulation, induce nuclear DNA and mtDNA damage and influence the activation of cancer-promoting transcription factors, thereby contributing in diverse ways to the initiation and progression of cancer. At least some cancer cells exhibit increased basal levels of oxidant stress. This oxidative shift may render the cells vulnerable to chemotherapeutic agents that act by augmenting oxidant generation112 or inhibiting antioxidant capacity113. Indeed, the selective toxicity of some chemotherapeutics in cancer cells relative to normal cells may arise because the former sit closer to the ‘precipice’, in terms of their ability to defend against excessive oxidant stress. Given the importance of mitochondrial ATP production in normal tissues, pharmacological inhibition of the ETC as a means of limiting ROS generation represents an unlikely therapeutic target for the treatment of cancer. However, many cancer cells benefit from mitochondrial ROS generation through its effects on redox signalling. Therefore, one possibility would be to develop small molecules that interfere with the ability of mitochondria to release oxidant signals, thereby preventing their activation of protective mechanisms such as HIF and AMP-activated protein kinase (AMPK). Clinical trials of ‘antioxidants’ have been uniformly unsuccessful in the prevention114 or the treatment of cancer115. This is perhaps not surprising, as the broad range of redox-dependent processes in cells make it just as likely that non-specific antioxidants will disrupt as protect a cell. However, it is conceivable that therapeutics aimed at intercepting the redox signals between the mitochondria and the host cell could be achieved without disrupting oxidative phosphorylation, thereby inactivating an important aspect of the communication between them116. Such an approach might not be sufficient as a single agent, but it could help to ‘tip the scale’ in the battle between the patient and the cancer cells by limiting the ability of mitochondria to function as initiators or amplifiers, thereby turning them into ‘innocent bystanders’.
Acknowledgments
The authors were supported by the US National Institutes of Health (NIH) Grants HL35440 and HL122062.
Glossary
- Mitochondria
Organelles within eukaryotic cells that participate in energy production, biosynthetic processes, redox regulation, cell survival, signalling and cell death pathways
- ATP
(Adenosine triphosphate). A high-energy molecule that is hydrolysed by enzymes to provide the exergonic free energy required to carry out endergonic reactions
- Hypoxia
A condition in which the molecular oxygen concentration is decreased relative to normal physiological levels
- Reactive oxygen species
(ROS). Reactive molecules generated by the reduction of O2 with a single electron (superoxide), two electrons (hydrogen peroxide) or three electrons (hydroxyl radical)
- ROS signalling
(Reactive oxygen species signalling). A cellular signal transduction mechanism involving oxidation–reduction reactions, usually resulting in a reversible alteration of protein structure and function that elicits a subsequent cellular response. ROS signalling frequently involves redox alterations of cysteine thiol (SH) groups in proteins
- Free radical
A molecule or atom containing an unpaired valence electron that renders it chemically reactive. Free radicals can potentially oxidize or reduce other molecules
- Tricarboxylic acid cycle
(TCA cycle). A system within mitochondria that participates in intermediary metabolism involved in energy production, inter-conversion of metabolites, and synthesis of small molecules needed for lipid or protein synthesis
- NADPH
A cofactor that is used by enzymes mediating electron transfer steps in energy production, lipid and nucleic acid synthesis, and the maintenance of intracellular oxidation–reduction status
- Superoxide dismutases
A family of enzymes that redistribute electrons between two superoxide anions to form a single molecule of hydrogen peroxide
- Hypoxia-inducible factors
(HIFs). A family of heterodimeric transcription factors that become activated during hypoxia or pseudohypoxia in a cell, and are responsible for potentially altering the expression of hundreds of genes involved in regulating cellular responses to hypoxia
- Mitochondrial DNA
(mtDNA). Circular loops of DNA containing ~16.6 kilobases, located in the matrix of mitochondria. This DNA encodes 13 proteins, ribosomal RNAs and transfer RNAs that are required for a functional oxidative phosphorylation system
- Cybrids
Experimental cells that are formed by fusing a cell lacking mitochondrial DNA with an enucleated cytoplast containing mutant mitochondria
- Pseudohypoxic activators
Stimuli that trigger activation of cellular responses to hypoxia, even though the O2 level in the cell is normal
- AMP-activated protein kinase
(AMPK). A complex consisting of three proteins that has a central role in the regulation of cellular energy production and energy utilization
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 2.Martin W, Hoffmeister M, Rotte C, Henze K. An overview of endosymbiotic models for the origins of eukaryotes, their ATP-producing organelles (mitochondria and hydrogenosomes), and their heterotrophic lifestyle. Biol Chem. 2001;382:1521–1539. doi: 10.1515/BC.2001.187. [DOI] [PubMed] [Google Scholar]
- 3.Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nature Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 4.Adams KL, Palmer JD. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol. 2003;29:380–395. doi: 10.1016/s1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- 5.Murphy MP, et al. Unraveling the biological roles of reactive oxygen species. Cell Metab. 2011;13:361–366. doi: 10.1016/j.cmet.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 7.Quinlan CL, et al. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. J Biol Chem. 2014;289:8312–8325. doi: 10.1074/jbc.M113.545301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner PR. Superoxide-driven aconitase FE-S center cycling. Biosci Rep. 1997;17:33–42. doi: 10.1023/a:1027383100936. [DOI] [PubMed] [Google Scholar]
- 9.Guzy RD, Schumacker PT. Oxygen sensing by mitochondria at complex III: the paradox of increased reactive oxygen species during hypoxia. Exp Physiol. 2006;91:807–819. doi: 10.1113/expphysiol.2006.033506. [DOI] [PubMed] [Google Scholar]
- 10.Fabian M, Palmer G. Hydrogen peroxide is not released following reaction of cyanide with several catalytically important derivatives of cytochrome c oxidase. FEBS Lett. 1998;422:1–4. doi: 10.1016/s0014-5793(97)01561-5. [DOI] [PubMed] [Google Scholar]
- 11.Han D, Antunes F, Canali R, Rettori D, Cadenas E. Voltage-dependent anion channels control the release of the superoxide anion from mitochondria to cytosol. J Biol Chem. 2003;278:5557–5563. doi: 10.1074/jbc.M210269200. [DOI] [PubMed] [Google Scholar]
- 12.Bienert GP, et al. Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J Biol Chem. 2007;282:1183–1192. doi: 10.1074/jbc.M603761200. [DOI] [PubMed] [Google Scholar]
- 13.Waypa GB, et al. Hypoxia triggers subcellular compartmental redox signaling in vascular smooth muscle cells. Circ Res. 2010;106:526–535. doi: 10.1161/CIRCRESAHA.109.206334. This study demonstrates how hypoxia-induced changes in mitochondrial ROS generation affect redox status in subcellular compartments differently. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabharwal SS, Waypa GB, Marks JD, Schumacker PT. Peroxiredoxin-5 targeted to the mitochondrial intermembrane space attenuates hypoxia-induced reactive oxygen species signalling. Biochem J. 2013;456:337–346. doi: 10.1042/BJ20130740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mills GC. Hemoglobin catabolism. I Glutathione peroxidase, an erythrocyte enzyme which protects hemoglobin from oxidative breakdown. J Biol Chem. 1957;229:189–197. [PubMed] [Google Scholar]
- 16.Morgan B, et al. Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nature Chem Biol. 2013;9:119–125. doi: 10.1038/nchembio.1142. [DOI] [PubMed] [Google Scholar]
- 17.Chae HZ, Rhee SG. A thiol-specific antioxidant and sequence homology to various proteins of unknown function. Biofactors. 1994;4:177–180. [PubMed] [Google Scholar]
- 18.Kim K, Kim IH, Lee KY, Rhee SG, Stadtman ER. The isolation and purification of a specific “protector” protein which inhibits enzyme inactivation by a thiol/Fe(III)/O2 mixed-function oxidation system. J Biol Chem. 1988;263:4704–4711. [PubMed] [Google Scholar]
- 19.Rhee SG, Chae HZ, Kim K. Peroxiredoxins: A historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 20.Fan J, et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis CA, et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 23.Rhee SG, Jeong W, Chang TS, Woo HA. Sulfiredoxin, the cysteine sulfinic acid reductase specific to 2-Cys peroxiredoxin: its discovery, mechanism of action, and biological significance. Kidney Int Suppl. 2007:S3–S8. doi: 10.1038/sj.ki.5002380. [DOI] [PubMed] [Google Scholar]
- 24.Arner ES, Holmgren A. The thioredoxin system in cancer. Semin Cancer Biol. 2006;16:420–426. doi: 10.1016/j.semcancer.2006.10.009. This excellent review summarizes the role of thioredoxin and its contributions to the cellular phenotype of cancer cells. [DOI] [PubMed] [Google Scholar]
- 25.Padmanabhan B, et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Singh A, Bodas M, Wakabayashi N, Bunz F, Biswal S. Gain of Nrf2 function in non-small-cell lung cancer cells confers radioresistance. Antioxid Redox Signal. 2010;13:1627–1637. doi: 10.1089/ars.2010.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata T, et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci USA. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sullivan LB, et al. The proto-oncometabolite fumarate binds glutathione to amplify ROS-dependent signaling. Mol Cell. 2013;51:236–248. doi: 10.1016/j.molcel.2013.05.003. This study examines the mechanisms by which FH deficiency drives tumour cell behaviour in a redox-dependent manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. This excellent review summarizes the role of the Warburg effect on cell proliferation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le A, et al. Tumorigenicity of hypoxic respiring cancer cells revealed by a hypoxia-cell cycle dual reporter. Proc Natl Acad Sci USA. 2014;111:12486–12491. doi: 10.1073/pnas.1402012111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waypa GB, et al. Superoxide generated at mitochondrial complex III triggers acute responses to hypoxia in the pulmonary circulation. Am J Respir Crit Care Med. 2013;187:424–432. doi: 10.1164/rccm.201207-1294OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farrow KN, et al. Brief hyperoxia increases mitochondrial oxidation and increases phosphodiesterase 5 activity in fetal pulmonary artery smooth muscle cells. Antioxid Redox Signal. 2012;17:460–470. doi: 10.1089/ars.2011.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandel NS, et al. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci USA. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. This study was the first to demonstrate that mitochondria-derived ROS can regulate transcription through their control of HIF1α stability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waypa GB, et al. Increases in mitochondrial reactive oxygen species trigger hypoxia-induced calcium responses in pulmonary artery smooth muscle cells. Circ Res. 2006;99:970–978. doi: 10.1161/01.RES.0000247068.75808.3f. [DOI] [PubMed] [Google Scholar]
- 35.Chi AY, Waypa GB, Mungai PT, Schumacker PT. Prolonged hypoxia increases ROS signaling and RhoA activation in pulmonary artery smooth muscle and endothelial cells. Antioxid Redox Signal. 2010;12:603–610. doi: 10.1089/ars.2009.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guzy RD, et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. This study demonstrated the importance of mitochondrial complex III in hypoxia-induced ROS generation that controls HIF1α stability. [DOI] [PubMed] [Google Scholar]
- 37.Mansfield KD, et al. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-α activation. Cell Metab. 2005;1:393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell EL, Emerling BM, Chandel NS. Mitochondrial regulation of oxygen sensing. Mitochondrion. 2005;5:322–332. doi: 10.1016/j.mito.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci USA. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. This study examines the role of mitochondrial ROS signalling in the tumorigenic behaviour induced by KRAS activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanjuan-Pla A, et al. A targeted antioxidant reveals the importance of mitochondrial reactive oxygen species in the hypoxic signaling of HIF-1α. FEBS Lett. 2005;579:2669–2674. doi: 10.1016/j.febslet.2005.03.088. [DOI] [PubMed] [Google Scholar]
- 41.Hamanaka RB, et al. Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle development. Sci Signal. 2013;6:ra8. doi: 10.1126/scisignal.2003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woo DK, et al. Mitochondrial genome instability and ROS enhance intestinal tumorigenesis in APCMin/+ mice. Am J Pathol. 2012;180:24–31. doi: 10.1016/j.ajpath.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. This is an excellent review of the evidence linking mtDNA mutations and cancer. [DOI] [PubMed] [Google Scholar]
- 44.Ishikawa K, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. This study demonstrates that mtDNA mutations can amplify tumour progression by increasing cellular ROS generation. [DOI] [PubMed] [Google Scholar]
- 45.Alexeyev M, Shokolenko I, Wilson G, Ledoux S. The maintenance of mitochondrial DNA integrity—critical analysis and update. Cold Spring Harb Perspect Biol. 2013;5:a012641. doi: 10.1101/cshperspect.a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larsen NB, Rasmussen M, Rasmussen LJ. Nuclear and mitochondrial DNA repair: similar pathways? Mitochondrion. 2005;5:89–108. doi: 10.1016/j.mito.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 47.He Y, et al. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clayton DA, Vinograd J. Circular dimer and catenate forms of mitochondrial DNA in human leukaemic leucocytes. Nature. 1967;216:652–657. doi: 10.1038/216652a0. [DOI] [PubMed] [Google Scholar]
- 49.Clayton DA, Vinograd J. Complex mitochondrial DNA in leukemic and normal human myeloid cells. Proc Natl Acad Sci USA. 1969;62:1077–1084. doi: 10.1073/pnas.62.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polyak K, et al. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nature Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 51.Kulawiec M, Salk JJ, Ericson NG, Wanagat J, Bielas JH. Generation, function, and prognostic utility of somatic mitochondrial DNA mutations in cancer. Environ Mol Mutagen. 2010;51:427–439. doi: 10.1002/em.20582. [DOI] [PubMed] [Google Scholar]
- 52.Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 53.Coller HA, et al. High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nature Genet. 2001;28:147–150. doi: 10.1038/88859. [DOI] [PubMed] [Google Scholar]
- 54.Zhidkov I, Livneh EA, Rubin E, Mishmar D. MtDNA mutation pattern in tumors and human evolution are shaped by similar selective constraints. Genome Res. 2009;19:576–580. doi: 10.1101/gr.086462.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Linnartz B, Anglmayer R, Zanssen S. Comprehensive scanning of somatic mitochondrial DNA alterations in acute leukemia developing from myelodysplastic syndromes. Cancer Res. 2004;64:1966–1971. doi: 10.1158/0008-5472.can-03-2956. This study tracked the association between the increase in mtDNA mutations over time and the progression from myelodysplastic syndrome to acute myeloid leukaemia in patients. [DOI] [PubMed] [Google Scholar]
- 56.Kirches E, et al. High frequency of mitochondrial DNA mutations in glioblastoma multiforme identified by direct sequence comparison to blood samples. Int J Cancer. 2001;93:534–538. doi: 10.1002/ijc.1375. [DOI] [PubMed] [Google Scholar]
- 57.Petros JA, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci USA. 2005;102:719–724. doi: 10.1073/pnas.0408894102. This study used cellular cybrids to examine the role of mtDNA mutations on ROS generation and tumorigenicity in prostate cancer cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trounce I, Neill S, Wallace DC. Cytoplasmic transfer of the mtDNA nt 8993 T-->G (ATP6) point mutation associated with Leigh syndrome into mtDNA-less cells demonstrates cosegregation with a decrease in state III respiration and ADP/O ratio. Proc Natl Acad Sci USA. 1994;91:8334–8338. doi: 10.1073/pnas.91.18.8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mattiazzi M, et al. The mtDNA T8993G (NARP) mutation results in an impairment of oxidative phosphorylation that can be improved by antioxidants. Hum Mol Genet. 2004;13:869–879. doi: 10.1093/hmg/ddh103. [DOI] [PubMed] [Google Scholar]
- 60.Shidara Y, et al. Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res. 2005;65:1655–1663. doi: 10.1158/0008-5472.CAN-04-2012. [DOI] [PubMed] [Google Scholar]
- 61.Vazquez F, et al. PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Namslauer I, Brzezinski P. A mitochondrial DNA mutation linked to colon cancer results in proton leaks in cytochrome c oxidase. Proc Natl Acad Sci USA. 2009;106:3402–3407. doi: 10.1073/pnas.0811450106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Samper E, Nicholls DG, Melov S. Mitochondrial oxidative stress causes chromosomal instability of mouse embryonic fibroblasts. Aging Cell. 2003;2:277–285. doi: 10.1046/j.1474-9728.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- 64.VanRemmen H, et al. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genom. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- 65.Liu L, Trimarchi JR, Smith PJ, Keefe DL. Mitochondrial dysfunction leads to telomere attrition and genomic instability. Aging Cell. 2002;1:40–46. doi: 10.1046/j.1474-9728.2002.00004.x. [DOI] [PubMed] [Google Scholar]
- 66.Martinez-Outschoorn UE, et al. Oxidative stress in cancer associated fibroblasts drives tumor-stroma co-evolution: A new paradigm for understanding tumor metabolism, the field effect and genomic instability in cancer cells. Cell Cycle. 2010;9:3256–3276. doi: 10.4161/cc.9.16.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Degan P, et al. In vivo accumulation of 8-hydroxy-2′-deoxyguanosine in DNA correlates with release of reactive oxygen species in Fanconi’s anaemia families. Carcinogenesis. 1995;16:735–741. doi: 10.1093/carcin/16.4.735. [DOI] [PubMed] [Google Scholar]
- 68.Ponte F, et al. Improvement of genetic stability in lymphocytes from Fanconi anemia patients through the combined effect of α-lipoic acid and N-acetylcysteine. Orphanet J Rare Dis. 2012;7:28. doi: 10.1186/1750-1172-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Radisky DC, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436:123–127. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gerasimenko JV, et al. Menadione-induced apoptosis: roles of cytosolic Ca2+ elevations and the mitochondrial permeability transition pore. J Cell Sci. 2002;115:485–497. doi: 10.1242/jcs.115.3.485. [DOI] [PubMed] [Google Scholar]
- 71.Bernardi P. Mitochondrial transport of cations: Channels, exchangers, and permeability transition. Physiol Rev. 1999;79:1127–1155. doi: 10.1152/physrev.1999.79.4.1127. [DOI] [PubMed] [Google Scholar]
- 72.Giorgio V, et al. Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc Natl Acad Sci USA. 2013;110:5887–5892. doi: 10.1073/pnas.1217823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baines CP, et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 74.Loor G, et al. Mitochondrial oxidant stress triggers cell death in simulated ischemia-reperfusion. Biochim Biophys Acta. 2011;1813:1382–1394. doi: 10.1016/j.bbamcr.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schriewer JM, Peek CB, Bass J, Schumacker PT. ROS-mediated, PARP activity undermines mitochondrial function after permeability transition pore opening during myocardial ischemia-reperfusion. J Am Heart Assoc. 2013;2:e000159. doi: 10.1161/JAHA.113.000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dewhirst MW, Cao Y, Moeller B. Cycling hypoxia and free radicals regulate angiogenesis and radiotherapy response. Nature Rev Cancer. 2008;8:425–437. doi: 10.1038/nrc2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trachootham D, et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by β-phenylethyl isothiocyanate. Cancer Cell. 2006;10:241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 78.Lee SR, et al. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. This study demonstrated how H2O2 generation can lead to the reversible inactivation of the lipid phosphatase PTEN, with important implications for cancer cell growth driven by excessive ROS signalling. [DOI] [PubMed] [Google Scholar]
- 79.Kwon J, et al. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc Natl Acad Sci USA. 2004;101:16419–16424. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao P, et al. HIF-dependent antitumorigenic effect of antioxidants in vivo. Cancer Cell. 2007;12:230–238. doi: 10.1016/j.ccr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. This is an excellent review of the role of HIF1 in cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Semenza GL. Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin Cancer Biol. 2009;19:12–16. doi: 10.1016/j.semcancer.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 84.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. This is a classic paper describing the regions of the HIF1α protein that confer sensitivity to hypoxia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Epstein AC, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. This study demonstrates the importance of SDH mutations in the formation of hereditary paragangliomas, thereby linking mitochondria to tumorigenic behaviour. [DOI] [PubMed] [Google Scholar]
- 86.Ivan M, et al. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 87.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 88.Chandel NS, et al. Reactive oxygen species generated at mitochondrial Complex III stabilize HIF-1-α during hypoxia: A mechanism of O2 sensing. J Biol Chem. 2000;275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 89.Brunelle JK, et al. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1:409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 90.Selak MA, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 91.Rustin P, Roetig A. Inborn errors of complex II - Unusual human mitochondrial diseases. Biochim Biophys Acta. 2002;1553:117–122. doi: 10.1016/s0005-2728(01)00228-6. [DOI] [PubMed] [Google Scholar]
- 92.Burnichon N, et al. SDHA is a tumor suppressor gene causing paraganglioma. Hum Mol Genet. 2010;19:3011–3020. doi: 10.1093/hmg/ddq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ackrell BAC. Progress in understanding structure-function relationships in respiratory chain complex II. FEBS Lett. 2000;466:1–5. doi: 10.1016/s0014-5793(99)01749-4. [DOI] [PubMed] [Google Scholar]
- 94.Baysal BE, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 95.Gimenez-Roqueplo AP, et al. The R22X mutation of the SDHD gene in hereditary paraganglioma abolishes the enzymatic activity of complex II in the mitochondrial respiratory chain and activates the hypoxia pathway. Am J Hum Genet. 2001;69:1186–1197. doi: 10.1086/324413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dekker PBD, et al. SDHD mutations in head and neck paragangliomas result in destabilization of complex II in the mitochondrial respiratory chain with loss of enzymatic activity and abnormal mitochondrial morphology. J Pathol. 2003;201:480–486. doi: 10.1002/path.1461. [DOI] [PubMed] [Google Scholar]
- 97.Astrom K, Cohen JE, Willett-Brozick JE, Aston CE, Baysal BE. Altitude is a phenotypic modifier in hereditary paraganglioma type 1: evidence for an oxygen-sensing defect. Hum Genet. 2003;113:228–237. doi: 10.1007/s00439-003-0969-6. [DOI] [PubMed] [Google Scholar]
- 98.Guzy RD, Sharma B, Bell E, Chandel NS, Schumacker PT. Loss of SdhB, but not SdhA, subunit of Complex. II triggers ROS-dependent HIF activation and tumorigenesis. Mol Cell Biol. 2007;28:718–731. doi: 10.1128/MCB.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Owens KM, et al. Genomic instability induced by mutant succinate dehydrogenase subunit D (SDHD) is mediated by O2−• and H2O2. Free Radic Biol Med. 2012;52:160–166. doi: 10.1016/j.freeradbiomed.2011.10.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tomlinson IP, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nature Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 101.Pollard PJ, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1α in tumours which result from germline FH and SDH mutations. Hum Mol Genet. 2005;14:2231–2239. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- 102.Isaacs JS, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–153. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 103.Sudarshan S, et al. Fumarate hydratase deficiency in renal cancer induces glycolytic addiction and hypoxia-inducible transcription factor 1α stabilization by glucose-dependent generation of reactive oxygen species. Mol Cell Biol. 2009;29:4080–4090. doi: 10.1128/MCB.00483-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123:3664–3671. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hu Y, et al. K-rasG12V transformation leads to mitochondrial dysfunction and a metabolic switch from oxidative phosphorylation to glycolysis. Cell Res. 2012;22:399–412. doi: 10.1038/cr.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dang CV, et al. Function of the c-Myc oncogenic transcription factor. Exp Cell Res. 1999;253:63–77. doi: 10.1006/excr.1999.4686. [DOI] [PubMed] [Google Scholar]
- 108.Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li F, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shim H, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci USA. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chandel NS, Trzyna WC, McClintock DS, Schumacker PT. Role of oxidants in NF-κB activation and TNF-α gene transcription induced by hypoxia and endotoxin. J Immunol. 2000;165:1013–1021. doi: 10.4049/jimmunol.165.2.1013. [DOI] [PubMed] [Google Scholar]
- 112.Schumacker PT. Reactive oxygen species in cancer cells: live by the sword, die by the sword. Cancer Cell. 2006;10:175–176. doi: 10.1016/j.ccr.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 113.Hashemy SI, Ungerstedt JS, Avval FZ, Holmgren A. Motexafin gadolinium, a tumor-selective drug targeting thioredoxin reductase and ribonucleotide reductase. J Biol Chem. 2006;281:10691–10697. doi: 10.1074/jbc.M511373200. [DOI] [PubMed] [Google Scholar]
- 114.Goodman M, Bostick RM, Kucuk O, Jones DP. Clinical trials of antioxidants as cancer prevention agents: past, present, and future. Free Radic Biol Med. 2011;51:1068–1084. doi: 10.1016/j.freeradbiomed.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 115.Creagan ET, et al. Failure of high-dose vitamin C (ascorbic acid) therapy to benefit patients with advanced cancer. A controlled trial. N Engl J Med. 1979;301:687–690. doi: 10.1056/NEJM197909273011303. [DOI] [PubMed] [Google Scholar]
- 116.Jung HJ, et al. Terpestacin inhibits tumor angiogenesis by targeting UQCRB of mitochondrial complex III and suppressing hypoxia-induced reactive oxygen species production and cellular oxygen sensing. J Biol Chem. 2010;285:11584–11595. doi: 10.1074/jbc.M109.087809. [DOI] [PMC free article] [PubMed] [Google Scholar]