Abstract
Clinically relevant weight loss is achievable through lifestyle modification, but unintentional weight regain is common. We investigated whether recently discovered genetic variants affect weight loss and/or weight regain during behavioral intervention. Participants at high-risk of type 2 diabetes (Diabetes Prevention Program [DPP]; N = 917/907 intervention/comparison) or with type 2 diabetes (Look AHEAD [Action for Health in Diabetes]; N = 2,014/1,892 intervention/comparison) were from two parallel arm (lifestyle vs. comparison) randomized controlled trials. The associations of 91 established obesity-predisposing loci with weight loss across 4 years and with weight regain across years 2–4 after a minimum of 3% weight loss were tested. Each copy of the minor G allele of MTIF3 rs1885988 was consistently associated with greater weight loss following lifestyle intervention over 4 years across the DPP and Look AHEAD. No such effect was observed across comparison arms, leading to a nominally significant single nucleotide polymorphism×treatment interaction (P = 4.3 × 10−3). However, this effect was not significant at a study-wise significance level (Bonferroni threshold P < 5.8 × 10−4). Most obesity-predisposing gene variants were not associated with weight loss or regain within the DPP and Look AHEAD trials, directly or via interactions with lifestyle.
Introduction
Adipose tissue is essential for fecundity and survival (1,2). Chronic excess adiposity (obesity) was probably uncommon throughout the majority of human evolution because foods were hunted or gathered by hand and those of very high–energy density may have been scarce. Hence, the human genome probably evolved to protect against leanness by promoting efficient energy expenditure and utilization. The heritability of BMI is ∼50–90% (3), underscoring the strong, biologically encoded nature of obesity.
Recently published (4,5) large-scale meta-analyses have identified ∼100 common obesogenic loci, but the clinical relevance of most of these is undetermined. Genetic data might be clinically valuable if it helped identify patients who are likely to respond well or poorly to clinical interventions, thereby facilitating targeted treatment. Testing if specific genotypes modify treatment effects in randomized controlled trials (RCTs) could help determine this.
The objective of these analyses was to test the genotype associations and treatment interactions for a comprehensive set of BMI-associated loci with intentional weight loss and weight regain in the Diabetes Prevention Program (DPP) and the Look AHEAD (Action for Health in Diabetes) studies, two RCTs of intensive lifestyle intervention conducted in multiethnic cohorts of overweight or obese adults with prediabetes or type 2 diabetes at enrollment.
Research Design and Methods
DPP
The DPP was a 27-site multicenter parallel arm RCT that assessed the effects of metformin treatment or an intensive lifestyle intervention on type 2 diabetes incidence in persons with prediabetes at baseline. The full details of the study are published elsewhere (6,7). Briefly, a total of 3,234 overweight or obese adults with elevated fasting and postchallenge glucoses were randomized to placebo control, metformin treatment (850 mg twice daily), or intensive lifestyle intervention (primarily fat gram, calorie, and physical activity goals) aimed at ∼7% weight loss. The intensive lifestyle intervention included 16 individual sessions within the first 6 months with in-person or phone follow-up at least monthly thereafter. In years 2 and beyond, group classes and campaigns were offered to reinforce lifestyle changes. In the placebo arm, standard lifestyle recommendations (annually) and inactive tablets were given. The trial’s primary outcome was the development of diabetes. Numerous other phenotypes, including weight, waist circumference, and abdominal adipose tissue distribution (from CT scans), were measured at baseline and intermittently as the trial progressed; this report focuses on weight change and weight regain in the placebo control and lifestyle arms only. Analyses for a small subset of single nucleotide polymorphisms (SNPs) examined here have been published previously in the DPP (8).
DPP Participants
Prior to initiating the study protocol, each participant provided written informed consent (88% consented to genetic analyses), and each study center obtained approval from its respective institutional review board. The analytic sample for the weight-change analyses consists of 1,824 comparison and lifestyle arm participants who provided genetic consent, for whom follow-up data were available in years 1–4, and whose genotype data passed quality-control procedures. The sample used for the weight-regain analyses consisted of 834 participants who lost at least 3% of their baseline weight at 1 year and attended at least one follow-up assessment in years 2–4, consistent with prior research in the DPP (8) and Look AHEAD trials (9,10).
Look AHEAD
Look AHEAD is a 16-site multicenter parallel arm RCT designed to determine whether weight loss achieved through lifestyle change of diet and physical activity reduces cardiovascular disease morbidity and mortality among 5,145 ethnically diverse overweight or obese adults with type 2 diabetes. The full details of the study are published elsewhere (11–14). Briefly, at baseline participants were randomized to either an intensive lifestyle intervention or diabetes support and education (DSE) arm. Both the lifestyle and DSE groups were provided one session of education on diabetes and cardiovascular risk. In addition, lifestyle participants received an intensive lifestyle program (focused on achieving ∼7% weight loss through calorie, fat gram, and physical activity goals) adapted from the DPP intervention. The lifestyle intervention included one individual and three group meetings per month for 6 months, followed by one individual and two group meetings per month through year 1. In years 2–4, lifestyle participants were seen individually at least monthly, contacted another time each month by telephone or e-mail, and offered a variety of group classes and campaigns, as in the DPP. The DSE group received the option of attending three sessions per year on nutrition, physical activity, and social support with no explicit weight-loss goals.
Look AHEAD Participants
Prior to initiating the study protocol, each participant provided written informed consent (84% consented to genetic analyses), and each study center obtained approval from its respective institutional review board. The analytic sample for the weight-change analyses consists of 3,906 DSE and lifestyle arm participants who provided genetic consent, for whom follow-up data were available in years 1–4, and whose genotype data passed quality-control procedures. The sample used for the weight-regain analyses consisted of 2,116 participants who lost at least 3% of their baseline weight at 1 year and attended at least one follow-up assessment in years 2–4, consistent with prior research in the DPP and Look AHEAD (8–10). Analyses for a small subset of SNPs examined here have been published previously in Look AHEAD (9,10). Both the DPP and Look AHEAD trials were conducted in accordance with the Declaration of Helsinki.
Neither the active intervention nor comparison arms of the DPP and Look AHEAD are identical. However, the active intervention in both studies represents considerably higher-intensity lifestyle interventions than were administered in the comparison arms. Therefore, for the sake of simplicity, we refer to the nonactive intervention arms of the DPP (placebo control) and Look AHEAD (DSE) as comparison and the active intervention arms as lifestyle from here on.
Genotyping
Ninety-one independent loci, characterized by 93 SNPs, identified or confirmed recently by the GIANT Consortium (5) were genotyped using the MetaboChip genotyping array (Illumina, San Diego, CA) in both studies. To ensure quality control, study participants with failed genotyping, sex inconsistency, or cryptic familial relatedness were excluded. SNPs with within-study genotyping call rates <95% or marked deviation from Hardy-Weinberg equilibrium (P < 1.2 × 10−4) in any ethnic group were also excluded. After quality control, the residual genotyping success rate for the 93 SNPs was >99.2% in the DPP and >99.7% in Look AHEAD (Supplementary Table 1).
Statistical Analysis
After excluding SNPs in linkage disequilibrium (r2 >0.30), EIGENSTRAT was used to compute principal components from all SNPs on the MetaboChip to control for population stratification in regression analyses. Four primary racial/ethnic groups were distinguished: non-Hispanic white, African American, Hispanic, and Asian.
Longitudinal trajectories of 1) weight change (baseline to 1-, 2-, 3-, and 4-year postrandomization) and 2) weight regain (year 1 to 2-, 3-, and 4-year postrandomization) among participants experiencing ≥3% weight loss from baseline to year 1 were first analyzed separately by study. Three-way interaction models of individual SNP markers (0, 1, and 2 copies of the minor allele; additive model) with study arm (lifestyle vs. comparison) and measurement time were estimated using the generalized least squares capabilities of S-Plus 8.2 (15). Three distinct types of SNP effects are presented, which can be interpreted as the effect per copy of the corresponding minor allele on 1) time-specific weight change within lifestyle, 2) time-specific weight change within the comparison arm, and 3) lifestyle versus comparison arm differences in relation to time-specific weight change. Longitudinal regression models for weight-change and -regain outcomes also included adjustment for age, sex, genetic ancestry (top three principal components), clinic site, and baseline weight (for weight-loss analyses) or year 1 weight (for regain analyses). With the exception of clinic site, all the aforementioned covariates were fully interacted with time, treatment, and time by treatment, so as to allow their effects to vary across study arm and/or time point. Correlation among repeated measures on study participants and variance heteroscedasticity across time points were accommodated using an unstructured covariance matrix whose parameters were estimated via restricted maximum likelihood.
Meta-analysis
Cross-Study Differences in SNP Effects
To determine if study-specific results could be pooled, we tested for DPP versus Look AHEAD (LA) differences in SNP effects using 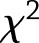 tests that combined information across all years of follow-up. Let
tests that combined information across all years of follow-up. Let 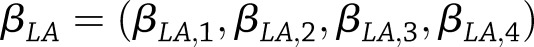 and
and 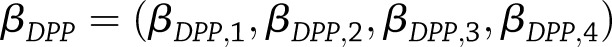 be the study-specific parameter vectors for a particular type of SNP effect from the weight-change analysis, with elements corresponding to year-specific parameters (years 2–4 only for weight-regain analysis). We tested
be the study-specific parameter vectors for a particular type of SNP effect from the weight-change analysis, with elements corresponding to year-specific parameters (years 2–4 only for weight-regain analysis). We tested 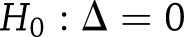 vs.
vs. 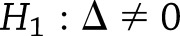 , where
, where 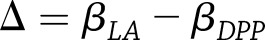 . If
. If 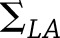 and
and 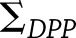 represent the covariance matrices of their estimates,
represent the covariance matrices of their estimates, 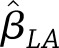 and
and 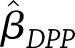 , respectively, then the estimated between-study difference is
, respectively, then the estimated between-study difference is 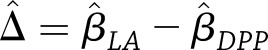 and the estimated variance of this difference is
and the estimated variance of this difference is 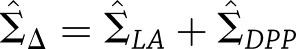 , as the estimates from the two studies are independent. Using these estimates, we used a
, as the estimates from the two studies are independent. Using these estimates, we used a 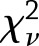 approximation of the distribution of the heterogeneity test statistic,
approximation of the distribution of the heterogeneity test statistic, 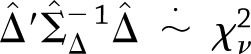 , where
, where 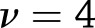 for weight loss and
for weight loss and 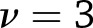 for weight regain.
for weight regain.
Cross-Study Averages of SNP Effects
When no significant between-study heterogeneity was evident, pooled estimates of SNP effects across the DPP and Look AHEAD were obtained using matrix-weighted averages, which weigh study-specific estimates by their relative precision. Individual precision matrices are simply the inverses of the covariance matrices given above. On the basis of matrix-weighted averaging, the pooled estimate of each type of SNP effect was calculated as 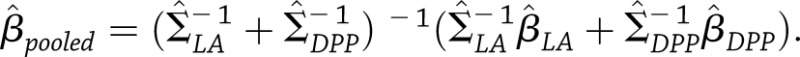 Alternatively, letting
Alternatively, letting 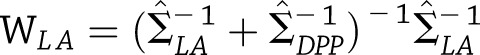 and
and 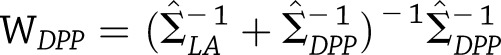 be the corresponding weight matrices, we see that
be the corresponding weight matrices, we see that 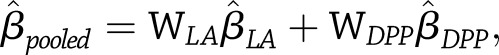 with estimated variance
with estimated variance 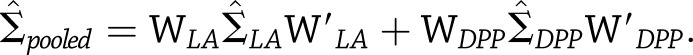
Univariate Tests of Pooled SNP Effects
To test whether the pooled estimate of a particular type of SNP effect was significantly different from zero at a specific time, 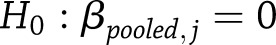 vs.
vs.
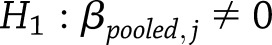 for
for 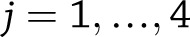 for weight change (or
for weight change (or 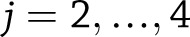 for weight regain), we used a normal approximation of the distribution of the pooled test statistic,
for weight regain), we used a normal approximation of the distribution of the pooled test statistic, 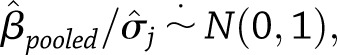 where
where 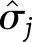 is the square root of the
is the square root of the 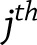 diagonal element of
diagonal element of  . Identical methodology was used to test the significance of within-study effects.
. Identical methodology was used to test the significance of within-study effects.
Multivariate Tests of Pooled SNP Effects
To test whether the pooled estimates of a particular SNP effect were significantly different from zero across all years of follow-up, 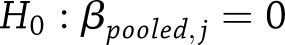 vs.
vs. 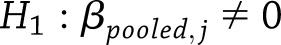 for
for 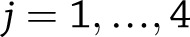 for weight change (or
for weight change (or 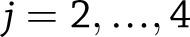 for weight regain), we used a
for weight regain), we used a 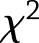 approximation of the distribution of the test statistic,
approximation of the distribution of the test statistic, 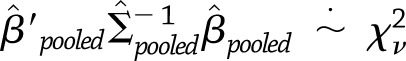 , where
, where 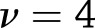 for weight change and
for weight change and 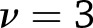 for weight regain. Identical methodology was used to test the significance of within-study effects.
for weight regain. Identical methodology was used to test the significance of within-study effects.
Study-wide Significance Threshold
In addition to the nominal P < 0.05 significance level, a multiplicity-adjusted significance threshold of P < 5.8 × 10−4 was derived via the Li and Ji method (16), taking into account the effective number of 88 uncorrelated markers under consideration (compared with 93 correlated ones). We note that this significance threshold corrects for the number of independent loci considered but assumes no prior probability of an effect. Hence, this may be a conservative estimate, given that these loci have known effects on BMI.
Results
Participant Characteristics
Tables 1 and 2 report the demographic characteristics for both the DPP and Look AHEAD participants. Table 1 describes the sample used in the weight-change analyses (years 1–4), while Table 2 reports characteristics for the subsample used in the weight-regain analyses (years 2–4). As previously reported in the full DPP (8) and Look AHEAD (11,12,17) cohorts, both short- and long-term weight loss was greater within the lifestyle arm, as was weight regain. Supplementary Table 2 reports the associations of each SNP with baseline BMI; 17 of these SNPs were nominally associated with BMI (P < 0.05). Two and seven SNPs showed evidence of multiyear heterogeneity for weight loss and weight regain, respectively, across the DPP and Look AHEAD (P < 0.05; Supplementary Tables 3 and 4).
Table 1.
Look AHEAD and DPP (weight-loss sample) participant characteristics
| Look AHEAD |
DPP |
|||||
|---|---|---|---|---|---|---|
| Characteristics | Total (N = 3,906) | Comparison (N = 1,892) | Lifestyle (N = 2,014) | Total (N = 1,824) | Comparison (N = 907) | Lifestyle (N = 917) |
| Women (%) |
2,251 (57.6) |
1,081 (57.1) |
1,170 (58.1) |
1,241 (68.0) |
623 (68.7) |
618 (67.4) |
| Successful weight losers (%)† |
2,116 (54.2) |
475 (25.1) |
1,641 (81.5) |
834 (45.7) |
201 (22.2) |
633 (69.0) |
| Age (years) |
59 (6.8) |
59.1 (6.8) |
58.9 (6.8) |
50.7 (10.9) |
50.7 (10.4) |
50.7 (11.4) |
| Ethnicity (%) |
||||||
| African American |
586 (15.0) |
280 (14.8) |
306 (15.2) |
371 (20.3) |
190 (21.0) |
181 (19.7) |
| American Indian/Alaskan Nativea |
81 (2.1) |
41 (2.2) |
40 (2.0) |
56 (3.1) |
28 (3.1) |
28 (3.1) |
| Asian/Pacific Islander |
26 (0.7) |
10 (0.5) |
16 (0.8) |
89 (4.9) |
37 (4.1) |
52 (5.7) |
| Non-Hispanic white |
2,747 (70.3) |
1,350 (71.4) |
1,397 (69.4) |
991 (54.3) |
499 (55.0) |
492 (53.7) |
| Hispanic/Latino |
392 (10.0) |
173 (9.1) |
219 (10.9) |
317 (17.4) |
153 (16.9) |
164 (17.9) |
| Other (multiple) |
74 (1.9) |
38 (2.0) |
36 (1.8) |
— |
— |
— |
| BMI (kg/m2) |
36.1 (5.9) |
36.2 (5.8) |
36 (6.1) |
34.1 (6.7) |
34.3 (6.7) |
34.0 (6.7) |
| Weight (kg) |
||||||
| Overall Sample |
||||||
| Baseline |
101.8 (19.2) |
102.2 (18.7) |
101.4 (19.6) |
94.6 (20.2) |
94.8 (19.9) |
94.4 (20.5) |
| Year 1 |
96.7 (19.4) |
101.4 (18.9) |
92.4 (18.9) |
90.8 (20.5) |
94.3 (20.4) |
87.3 (19.9) |
| Year 2 |
97.6 (19.4) |
100.9 (18.8) |
94.6 (19.4) |
91.6 (20.6) |
94.7 (20.3) |
88.5 (20.5) |
| Year 3 |
98.3 (19.5) |
100.8 (19.1) |
96.0 (19.6) |
92.8 (21.2) |
95.1 (20.4) |
90.5 (21.8) |
| Year 4 |
98.4 (19.6) |
100.6 (19.2) |
96.3 (19.7) |
94.1 (21.3) |
95.6 (18.4) |
92.7 (23.6) |
| Women |
||||||
| Baseline |
96.0 (17.5) |
96.4 (17.4) |
95.6 (17.7) |
92.9 (20.4) |
93.1 (20.1) |
92.7 (20.7) |
| Year 1 |
91.4 (17.9) |
95.6 (17.6) |
87.5 (17.3) |
89.2 (20.7) |
92.5 (20.9) |
86.0 (20.1) |
| Year 2 |
92.3 (18.0) |
95.1 (17.6) |
89.7 (18.0) |
90.1 (20.8) |
93.0 (20.6) |
87.0 (20.5) |
| Year 3 |
92.8 (17.9) |
94.7 (17.4) |
91.1 (18.1) |
91.7 (21.5) |
93.4 (20.5) |
90.0 (22.2) |
| Year 4 |
92.7 (17.9) |
94.5 (17.7) |
91.1 (18.0) |
94.7 (22.5) |
95.9 (18.8) |
93.5 (25.5) |
| Men |
||||||
| Baseline |
109.7 (18.6) |
109.8 (17.7) |
109.6 (19.3) |
98.3 (19.3) |
98.5 (18.9) |
98.1 (19.8) |
| Year 1 |
104.0 (19.0) |
109.0 (17.8) |
99.2 (18.9) |
94.1 (19.4) |
98.2 (18.7) |
90.2 (19.3) |
| Year 2 |
104.9 (18.8) |
108.4 (17.6) |
101.4 (19.4) |
94.9 (19.8) |
98.4 (19.0) |
91.5 (20.1) |
| Year 3 |
105.8 (19.2) |
108.9 (18.2) |
102.8 (19.6) |
95.0 (20.6) |
98.5 (19.7) |
91.7 (20.9) |
| Year 4 | 106.1 (19.1) | 108.8 (18.1) | 103.6 (19.7) | 93.0 (18.8) | 94.9 (17.9) | 91.2 (19.5) |
All variables summarized in mean (SD) format, unless indicated otherwise.
aThe number of American Indian participants included in this study is lower than in the parent Look AHEAD trial due to lack of institutional review board approval.
†This is the weight-regain analysis subsample that achieved at least 3% weight loss at year 1 (see the separate demographics table for this subsample in Table 2).
Table 2.
Look AHEAD and DPP (weight-regain sample) participant characteristics
| Look AHEAD |
DPP |
|||||
|---|---|---|---|---|---|---|
| Characteristics | Total (N = 2,116) | Comparison (N = 475) | Lifestyle (N = 1,641) | Total (N = 834) | Comparison (N = 201) | Lifestyle (N = 633) |
| Women (%) |
1,224 (58.0) |
291 (61.3) |
933 (56.9) |
544 (65.2) |
139 (69.2) |
405 (64.0) |
| Age (years) |
59.2 (6.8) |
59.3 (6.7) |
59.2 (6.9) |
51.7 (11.0) |
50.5 (9.6) |
52.1 (11.4) |
| Ethnicity (%) |
||||||
| African American |
291 (13.8) |
61 (12.8) |
230 (14.0) |
136 (16.3) |
33 (16.4) |
103 (16.3) |
| American Indian/Alaskan Nativea |
40 (1.9) |
11 (2.3) |
29 (1.8) |
17 (2.0) |
2 (1.0) |
15 (2.4) |
| Asian/Pacific Islander |
14 (0.7) |
1 (0.2) |
13 (0.8) |
45 (5.4) |
5 (2.5) |
40 (6.3) |
| Non-Hispanic white |
1,518 (72.0) |
350 (74.0) |
1,168 (71.0) |
486 (58.3) |
129 (64.2) |
357 (56.4) |
| Hispanic/Latino |
220 (10.4) |
43 (9.1) |
177 (10.8) |
150 (18.0) |
32 (15.9) |
118 (18.6) |
| Other (multiple) |
33 (1.6) |
9 (1.9) |
24 (1.5) |
— |
— |
— |
| BMI (kg/m2) |
36.1 (6.1) |
36.9 (6.0) |
35.9 (6.1) |
33.7 (6.4) |
34.5 (6.7) |
33.4 (6.3) |
| Weight (kg) |
||||||
| Overall |
||||||
| Baseline |
102.0 (19.8) |
103.8 (20.1) |
101.4 (20.0) |
93.7 (19.9) |
95.5 (20.2) |
93.1 (19.8) |
| Year 1 |
91.8 (18.5) |
96.7 (18.8) |
90.4 (18.2) |
84.9 (18.5) |
88.8 (19.4) |
83.6 (18.0) |
| Women |
||||||
| Baseline |
95.9 (17.8) |
97.6 (18.4) |
95.3 (17.6) |
91.4 (20.0) |
93.2 (19.4) |
90.8 (20.2) |
| Year 1 |
86.6 (16.8) |
91.0 (17.2) |
85.2 (16.4) |
82.8 (18.6) |
86.5 (19.1) |
81.5 (18.3) |
| Men |
||||||
| Baseline |
110.3 (19.0) |
113.4 (18.9) |
109.5 (20.0) |
97.9 (19.0) |
100.7 (21.0) |
97.2 (18.4) |
| Year 1 |
98.9 (18.4) |
105.8 (17.6) |
97.1 (18.2) |
88.9 (17.5) |
94.0 (19.4) |
87.5 (16.7) |
| Weight regain (kg)† |
||||||
| Overall |
||||||
| Year 2 |
2.4 (5.2) |
0.6 (6.4) |
2.9 (4.6) |
2.0 (4.2) |
2.7 (4.9) |
1.7 (4.0) |
| Year 3 |
3.7 (7.2) |
1 (8.1) |
4.5 (6.7) |
3.3 (5.8) |
4.2 (6.4) |
3.0 (5.6) |
| Year 4 |
4.2 (8.0) |
1.4 (8.5) |
5.1 (7.7) |
4.3 (7.7) |
4.6 (9.2) |
4.2 (7.2) |
| Women |
||||||
| Year 2 |
2.1 (5.3) |
0.2 (6.7) |
2.7 (4.6) |
2.1 (4.4) |
2.9 (5.0) |
1.8 (4.2) |
| Year 3 |
3.4 (7.1) |
0.4 (8.9) |
4.4 (6.1) |
3.5 (6.2) |
4.2 (6.9) |
3.3 (6.0) |
| Year 4 |
3.8 (8.1) |
0.7 (9.2) |
4.8 (7.5) |
4.9 (8.4) |
4.4 (10.0) |
5.1 (7.7) |
| Men |
||||||
| Year 2 |
2.7 (5.0) |
1.3 (6.0) |
3.0 (4.7) |
1.7 (3.8) |
2.2 (4.7) |
1.6 (3.6) |
| Year 3 |
4.1 (7.4) |
2.0 (6.7) |
4.7 (7.4) |
2.8 (4.9) |
4.2 (5.3) |
2.4 (4.8) |
| Year 4 | 4.9 (7.8) | 2.4 (7.2) | 5.5 (7.8) | 3.3 (6.1) | 5.2 (5.9) | 2.9 (6.2) |
All variables summarized in mean (SD) format, unless indicated otherwise.
aThe number of American Indian participants included in this study is lower than the parent Look AHEAD trial due to lack of institutional review board approval.
†Weight regain calculated as Yj weight – Y1 weight (kg) for j = 2, 3, 4.
Weight Loss in Combined Analysis of the DPP and Look AHEAD
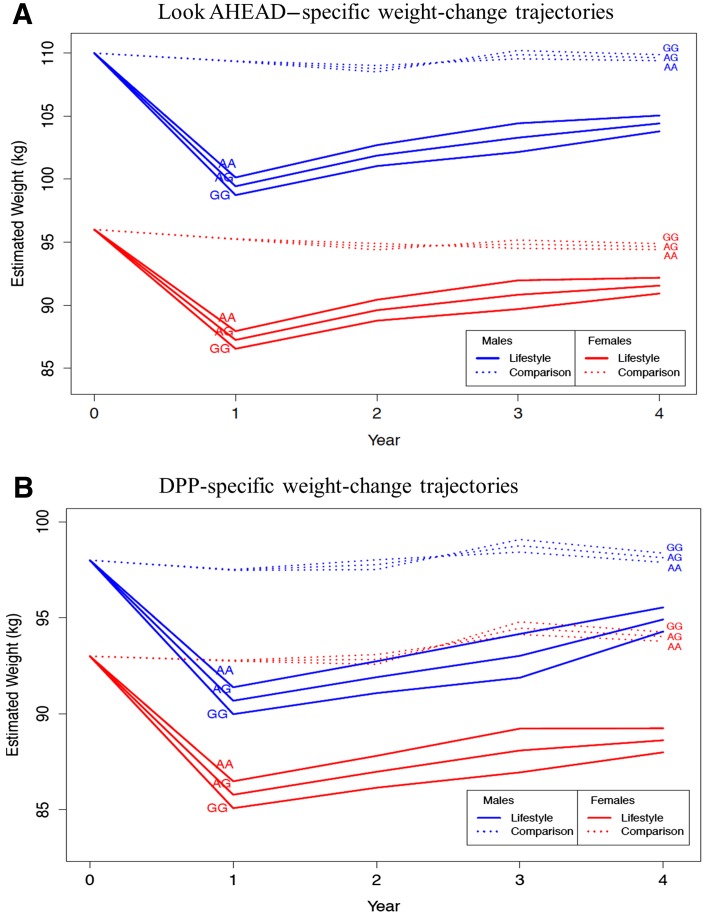
The intronic rs1885988 variant (at MTIF3 and a close proxy for GIANT marker rs12016871) modified weight-loss response to lifestyle intervention (Table 3). The effect within the combined interventions reached statistical significance in year 3 (P = 2 × 10−4 for year 3 pooled effect within lifestyle arms), after multiple-test correction. Each copy of the minor G allele was associated with a mean −1.14 kg (95% CI −1.75, −0.53) lower weight in the lifestyle arm versus a mean 0.33 kg (−0.30, 0.95) higher weight in the comparison arm (P = 0.30), resulting in a nominally significant interaction (P = 9 × 10−4 for SNP×treatment interaction at year 3). Hence, the mean differences in year-3 weight change between the lifestyle intervention and comparison arms were estimated at −1.48 kg (−2.35, −0.61) between AA homozygotes and AG heterozygotes, and −2.96 kg (−4.71, −1.22) between AA and GG homozygotes.
Table 3.
Highlighted loci results for MTIF3 (rs1885988) and weight loss over 4 years in the full sample and FUBP1 (rs12401738) and weight regain across years 2–4 among those who lost 3% or more of their baseline weight at year 1
| Variant | Effect | Year | Individual study results |
Meta-analysis |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Look AHEAD |
DPP |
Differences in estimates (LA − DPP) |
Matrix-weighted averages (LA + DPP) |
|||||||
| PE (95% CI) | P | PE (95% CI) | P | PE (95% CI) | P | PE (95% CI) | P | |||
|
MTIF3 (rs1885988) |
Lifestyle |
1 |
−0.73 (−1.30, −0.15) |
0.009 |
−0.66 (−1.45, 0.13) |
0.22 |
−0.07 (−1.04, 0.91) |
0.62 |
−0.70 (−1.16, −0.24) |
0.002 |
| 2 |
−0.95 (−1.62, −0.27) |
−0.65 (−1.55, 0.25) |
−0.30 (−1.42, 0.83) |
−0.83 (−1.37, −0.29) |
||||||
| 3 |
−1.18 (−1.93, −0.42) |
−1.10 (−2.14, −0.06) |
−0.08 (−1.36, 1.21) |
−1.14 (−1.75, −0.53) |
||||||
| 4 |
−0.54 (−1.33, 0.25) |
−1.48 (−3.03, 0.08) |
0.94 (−0.80, 2.68) |
−0.63 (−1.30, 0.05) |
||||||
| Comparison |
1 |
−0.36 (−0.94, 0.22) |
0.09 |
0.68 (−0.15, 1.51) |
0.34 |
−1.04 (−2.05, −0.03) |
0.26 |
−0.02 (−0.50, 0.45) |
0.11 |
|
| 2 |
−0.69 (−1.36, −0.01) |
0.60 (−0.34, 1.54) |
−1.29 (−2.44, −0.13) |
−0.25 (−0.80, 0.29) |
||||||
| 3 |
−0.06 (−0.82, 0.70) |
1.10 (−0.02, 2.21) |
−1.15 (−2.50, 0.20) |
0.33 (−0.30, 0.95) |
||||||
| 4 |
−0.07 (−0.87, 0.72) |
0.73 (−1.13, 2.60) |
−0.81 (−2.83, 1.22) |
0.24 (−0.46, 0.94) |
||||||
| Lifestyle - Comparison |
1 |
−0.37 (−1.18, 0.44) |
0.08 |
−1.34 (−2.48, −0.20) |
0.048 |
0.97 (−0.43, 2.38) |
0.62 |
−0.69 (−1.36, −0.03) |
0.004 |
|
| 2 |
−0.26 (−1.21, 0.69) |
−1.25 (−2.55, 0.05) |
0.99 (−0.62, 2.60) |
−0.59 (−1.36, 0.17) |
||||||
| 3 |
−1.12 (−2.19, −0.05) |
−2.20 (−3.72, −0.67) |
1.08 (−0.78, 2.94) |
−1.48 (−2.35, −0.61) |
||||||
| 4 |
−0.46 (−1.58, 0.66) |
−2.21 (−4.63, 0.21) |
1.75 (−0.92, 4.42) |
−0.85 (−1.82, 0.12) |
||||||
| FUBP1 (rs12401738) | Lifestyle |
2 |
−0.12 (−0.51, 0.27) |
0.24 |
0.51 (−0.04, 1.06) |
0.27 |
−0.63 (−1.30, 0.05) |
0.33 |
0.09 (−0.23, 0.41) |
0.2 |
| 3 |
0.06 (−0.48, 0.59) |
0.70 (−0.16, 1.57) |
−0.65 (−1.66, 0.37) |
0.26 (−0.19, 0.72) |
||||||
| 4 |
−0.36 (−0.96, 0.25) |
0.41 (−1.04, 1.86) |
−0.76 (−2.33, 0.80) |
−0.16 (−0.70, 0.38) |
||||||
| Comparison |
2 |
−0.81 (−1.55, −0.07) |
0.13 |
−0.91 (−1.92, 0.10) |
0.32 |
0.10 (−1.15, 1.35) |
0.99 |
−0.84 (−1.44, −0.25) |
0.03 |
|
| 3 |
−1.12 (−2.13, −0.10) |
−1.22 (−2.77, 0.32) |
0.11 (−1.74, 1.96) |
−1.15 (−2.00, −0.30) |
||||||
| 4 |
−0.93 (−2.07, 0.21) |
−0.98 (−3.84, 1.89) |
0.05 (−3.04, 3.13) |
−0.96 (−1.98, 0.06) |
||||||
| Lifestyle - Comparison | 2 |
0.69 (−0.14, 1.52) |
0.18 | 1.42 (0.28, 2.56) |
0.08 | −0.73 (−2.14, 0.69) |
0.79 | 0.94 (0.27, 1.61) |
0.01 | |
| 3 |
1.17 (0.02, 2.32) |
1.93 (0.16, 3.69) |
−0.76 (−2.87, 1.35) |
1.42 (0.46, 2.38) |
||||||
| 4 | 0.58 (−0.71, 1.86) | 1.39 (−1.82, 4.6) | −0.81 (−4.27, 2.65) | 0.80 (−0.35, 1.96) | ||||||
Point estimates (PE) and 95% CI represent weight change (kg) per minor allele (MTIF3-rs1885988 G allele; FUBP1-rs12401738 A allele, respectively). Longitudinal regression models adjusted for baseline weight, age, sex, ancestry (principal components 1–3), and study site. P values were obtained using multiyear χ2 tests for the corresponding effect estimate. LA, Look AHEAD.
Longitudinal analyses (Fig. 1) showed that rs1885988 was consistently associated with weight loss within the lifestyle but not the comparison arms (P = 2.4 × 10−3 and P = 0.11 for multiyear tests of pooled SNP effects, respectively), leading to a consistent pattern of between-arm differences in weight change in favor of the lifestyle intervention (P = 4.3 × 10−3 for multiyear test of pooled SNP×treatment interaction). No other SNP×lifestyle interaction effects on weight loss showed similarly consistent patterns across all 4 years of follow-up, although several were nominally significant (Supplementary Table 3).
Figure 1.
Model-based estimates of MTIF3 genotype effects on weight change among 60-year-old participants following lifestyle and control intervention in the Look AHEAD (A) and the DPP (B) trials. Baseline weight chosen to be representative of males and females in each study (see Table 1). Ancestry-informative principal components set at study-specific means.
Weight Regain in Combined Analysis of the DPP and Look AHEAD
No SNP×treatment arm interactions reached statistical significance in longitudinal analysis after correction for multiple testing. The strongest SNP×treatment interaction was for the FUBP1-DNAJB4 rs12401738 (P = 0.014 for multiyear test of pooled SNP×treatment interaction) (Table 3). Treatment arm–specific analyses showed that the minor allele at FUBP1-DNAJB4 rs12401738 was consistently associated with weight regain within the comparison but not the lifestyle arms (P = 0.03 and P = 0.20 for multiyear test of pooled SNP effects, respectively) (Table 3). Specifically, each copy of the minor A allele was associated with lesser weight regain mean −0.84 kg (95% CI −1.44, −0.25) in year 2 (P = 5.3 × 10−3), −1.15 kg (−2.00, −0.30) in year 3 (P = 7.7 × 10−3), and −0.96 kg (−1.98, −0.06) in year 4 (P = 0.065) among participants within the comparison arm. Many other pooled SNP×treatment interaction effects on weight regain were nominally significant within either the DPP or the Look AHEAD (Supplementary Table 4).
Discussion
Many loci have been robustly associated with anthropometric indices of obesity through genome-wide association studies (GWAS) meta-analyses, revealing multiple biologic pathways (18,19). However, as these studies focused exclusively on cross-sectional epidemiological data, little is known of whether these variants influence weight change or modify the response to weight-loss interventions. Here, we sought to address these two clinically relevant questions by examining the effects of these variants on weight loss and weight regain in two large RCTs of lifestyle modification, one in people with prediabetes (DPP) and the other in patients diagnosed with type 2 diabetes prior to randomization (Look AHEAD). We found that of nearly 100 loci examined, variant rs1885988 at MTIF3, a close proxy for GIANT variant rs12016871, appeared to modify the effects of the lifestyle interventions on weight loss, reaching study-wide statistical significance within the lifestyle intervention arms in year 3 (P = 2 × 10−4) and demonstrating consistently beneficial effects across all 4 years of follow-up (P = 2.4 × 10−3). As no similar benefit was observed within the comparison arms, this led to a nominally significant pooled SNP×treatment interaction across all 4 years of follow-up (P = 4.3 × 10−3). For weight regain, no SNPs modified treatment responses at a level reaching study-wide statistical significance. The strongest effect was observed for an SNP near FUBP1-DNAJB4 (rs12401738), which appeared to modify the effects of the lifestyle interventions in both studies (Pinteraction = 0.014) by slowing weight regain within the comparison but not the lifestyle interventions.
MTIF3 encodes a 29-kDa nuclear-encoded protein that promotes the formation of the initiation complex on the mitochondrial 55S ribosome (20,21). The mitochondrial ribosome is responsible for the synthesis of 13 of the inner mitochondrial membrane proteins and its regulation is essential for ATP synthesis, energy balance, and modulation of reactive oxygen species production in the mitochondria by the electron transport chain (21). Using the HaploReg interface (http://www.broadinstitute.org/mammals/haploreg/haploreg.php) to access the ENCODE database, we looked up the functional properties of our lead SNP (rs1885988). Although this is an intronic variant, it is 411 bp from a triallelic missense SNP with a deoxyribonuclease (DNAse) peak, with which it is in perfect linkage disequilibrium (r2 = 1; D′ = 1) in the 1000 Genomes database. Thus, the rs1885988 variant tags a chromatin site that is sensitive to transcription factor binding and hence likely regulates gene expression. We further queried the transcriptional properties of this variant in RegulomeDB (http://regulome.stanford.edu/index), which characterized the variant as a DNAse peak site in immune-regulating T cells (score 5).
A recent publication in Look AHEAD (22) examined the relationships between obesity gene variants, including MTIF3-rs12016871 and diet preference, but no other published studies have examined variation at this locus and lifestyle. Here, the minor G allele was associated with lifestyle-elicited weight loss in both trials and with weight regain in the DPP but not Look AHEAD. The minor G allele has previously been associated with higher BMI (18,19). Thus, carriers of the MTIF3 obesity-inducing allele appear to benefit more from intensive lifestyle interventions than noncarriers whether they have prediabetes or overt type 2 diabetes. In a recently published, cross-sectional meta-analysis of 32 gene×dietary pattern interactions for BMI (N >65,000) (23), the locus with the strongest signal of an interaction with a healthy diet was a close proxy of our MTIF3 variant. The obvious differences in study design make extrapolation of those cross-sectional data to the clinical trial context difficult. However, the fact that the MTIF3 locus was top ranked in both studies strengthens the credibility of our findings and reinforces MTIF3 as a plausible candidate locus for gene×lifestyle interactions in obesity.
FUBP1 encodes a single-stranded DNA and RNA binding protein that regulates gene transcription, stability, and splicing (24,25), particularly for the C-MYC oncogene (26). Mutations at FUPB1 are especially common in oligodendroglioma (27), a rare form of brain cancer. DNAJB4 is preferentially expressed in heart, skeletal muscle, and pancreas and encodes a 337 amino acid heat-inducible protein (28). Expression of DNAJB4 is associated with non–small-cell lung cancer survival (29). No publication to our knowledge has reported on the variation at FUBP1 or DNAJB4 and lifestyle factors in obesity or metabolic disease. In the current analyses, the minor A allele at the FUBP1-DNAJB4 rs12401738 variant appeared to slow weight regain within the comparison arms in both trials, but not within the lifestyle arms. In the GIANT meta-analysis, the minor A allele was associated with higher BMI.
The FTO locus (proximal to rs9960939), which has been shown previously to interact with lifestyle factors, was not associated with weight loss directly or via treatment interactions in either the DPP (30) or Look AHEAD (10). Previously in the DPP, the FTO rs9939609 variant predicted a greater increase in subcutaneous adipose tissue in the placebo group compared with lifestyle intervention at year 1, but no significant genotype×treatment interaction was observed for weight loss. In Look AHEAD, several SNPs in high linkage disequilibrium showed a nominal association with weight regain at year 4 among those who had lost 3% or more of their baseline weight at year 1, but there was no effect on weight loss. The previously reported additive effect on weight change of FTO rs1421085 was weakened to nonsignificance in this report (from year 1–4 in the comparison arm, P = 0.10; treatment arm interaction, P = 0.10), likely due to a different subsample of Look AHEAD, including a greater representation of Native and Hispanic Americans with available MetaboChip genotyping (vs. IBC chip genotyping presented in the prior article), and to differences in analytic methods, predominantly the derivation and adjustment for new principal components from MetaboChip data to statistically adjust for ancestry. We note, however, that the effect of the lead SNP on weight regain from the prior article, FTO rs3751812, was maintained in this more diverse group (from year 1–4 in the comparison arm, P = 0.02; treatment arm interaction, P = 0.03). Thus, it appears that any impact of the obesity-associated region of FTO on weight loss following clinical intervention is weak and that larger studies designed to examine effects within racial/ethnic groups or with more detailed measurements of body composition will be needed for the effects reported here to be confirmed.
Although a handful of potential gene×treatment interaction effects are evident in these two large RCTs, one of the key findings is that the vast majority of GWAS-derived obesity-associated loci do not appear to convey clinically meaningful effects on weight loss or weight regain. This is key because many anticipate that modern population genetics research will help prevent or treat diabetes and obesity (31). Although discoveries made through GWAS meta-analyses help elucidate biological pathways, the use of individual obesity-associated SNPs derived through GWAS is unlikely to help clinicians optimize the delivery of weight-loss interventions through targeted intervention. One possible reason that GWAS-derived loci do not serve this role well is because GWAS meta-analyses are conventionally performed without accounting for interactions with lifestyle factors and by ranking loci based solely on marginal effect P values, which may bias against the discovery of loci that modify the effects of lifestyle interventions (32).
The DPP and Look AHEAD are the largest existing RCTs of lifestyle intervention in people who are either at high-risk of developing or who have already developed type 2 diabetes, respectively; nevertheless, even with ∼6,000 participants this analysis is likely underpowered to detect effects as small as those observed in large observational meta-analyses. However, it is unlikely these effects are clinically relevant and failing to detect them here is thus of little consequence in the context of our clinically oriented objectives. It is also important to bear in mind some key differences between the DPP and Look AHEAD trials, not least that the former focuses on people with prediabetes and the latter on people who have already developed the disease, some who are on pharmacotherapy known to influence weight. Differences in intervention protocols also exist. These factors may inhibit the detection of gene×treatment interactions. Moreover, it is likely that gene×diet interactions are nutrient specific; as such, it may be that dietary regimes that focus on different elements of the diet from the DPP and Look AHEAD interventions, such as carbohydrate or salt intake, might yield interaction effects with the variants studied here. Last, although the test of heterogeneity between the DPP and Look AHEAD suggests that there were no statistically significant differences in interaction effects between studies, it is likely that with only two studies included in this analysis the heterogeneity test was underpowered, and, as such, the absence of a significant heterogeneity test statistic should be interpreted with caution.
In conclusion, we assessed the effects on weight change of 91 established BMI-associated loci in two large RCTs of intensive lifestyle modification. The strongest association with weight loss across studies was MTIF3-rs1885988. Although studying BMI-associated variants derived from cross-sectional observational studies has, in this instance, provided few new insights into the genetics of behavioral weight loss and weight regain, future studies focused on genome-wide hypothesis-free discovery efforts may yield more promising results.
Supplementary Material
Article Information
Acknowledgments. The investigators gratefully acknowledge the commitment and dedication of all participants in the DPP and Look AHEAD, without whom this work would not have been possible. The investigators also thank Tibor V. Varga (Lund University, Malmö, Sweden) for advice on functional analyses.
Funding. The DPP and Look AHEAD analyses were funded by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grants R01 DK072041-02 (to K.A.J. and J.C.F. [W.C.K. and P.W.F. are co-investigators]) and DK056992-14S1, respectively. S.E.K. is supported in part by the Department of Veterans Affairs. J.C.F. is supported by a Doris Duke Charitable Foundation Clinical Scientist Development Award. P.W.F. was supported by grants from the Swedish Heart-Lung Foundation, the Novo Nordisk Foundation, the Swedish Research Council, EXODIAB, and the Swedish Diabetes Association.
DPP: The NIDDK of the National Institutes of Health (NIH) provided funding to the clinical centers and the coordinating center for the design and conduct of the study and collection, management, analysis, and interpretation of the data. The Southwestern American Indian Centers were supported directly by the NIDDK and the Indian Health Service. The General Clinical Research Center Program of the National Center for Research Resources supported data collection at many of the clinical centers. Funding for data collection and participant support was also provided by the NIH Office of Research on Minority Health, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the National Institute on Aging, the Centers for Disease Control and Prevention, the NIH Office of Research on Women's Health, the Department of Veterans Affairs, and the American Diabetes Association. Bristol-Myers Squibb and Parke-Davis provided medication. This research was also supported, in part, by the Intramural Research Program of the NIDDK. LifeScan, Health O Meter, Hoechst Marion Roussel Inc., Merck-Medco Managed Care, LLC, Merck & Co., Inc., Nike Sports Marketing, SlimFast Foods Co., and Quaker Oats Co. donated materials, equipment, or medicines for concomitant conditions. McKesson BioServices, Matthews Media Group, Inc., and the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. provided support services under subcontract with the coordinating center.
The opinions expressed are those of the investigators and do not necessarily reflect the views of the Indian Health Service or other funding agencies. A complete list of centers, investigators, and staff can be found in the Supplementary Appendix online.
Duality of Interest. L.M.D. has a financial interest in Omada Health, a company that develops online behavior-change programs, with a focus on diabetes. The interests of L.M.D. were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict-of-interest policies. P.W.F. was supported by grants from Novo Nordisk and is a member of scientific advisory boards for Eli Lilly & Co. and Sanofi Aventis. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. G.D.P., J.C.F., J.M.M., and P.W.F. designed the analyses and wrote the manuscript. G.D.P., Q.P., B.E., and K.A.J. undertook statistical analyses. L.M.D., L.E.W., S.E.K., R.R.W., and W.C.K. coordinated data collection. S.A., M.H., L.C., and P.F. undertook coordination tasks. All authors read and critically appraised the manuscript. G.D.P. and P.W.F. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 74th Scientific Sessions of the American Diabetes Association, San Francisco, CA, 13–17 June 2014.
Footnotes
Clinical trial reg. nos. NCT00004992 (Diabetes Prevention Program), and NCT00017953 (Look AHEAD), clinicaltrials.gov.
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db15-0441/-/DC1.
A list of the Diabetes Prevention Program Research Group and Look AHEAD Research Group investigators is provided in the Supplementary Data online.
References
- 1.Campos DB, Palin MF, Bordignon V, Murphy BD. The ‘beneficial’ adipokines in reproduction and fertility. Int J Obes 2008;32:223–231 [DOI] [PubMed] [Google Scholar]
- 2.Kuzawa CW. Adipose tissue in human infancy and childhood: an evolutionary perspective. Am J Phys Anthropol 1998;107(Suppl. 27):177–209 [DOI] [PubMed] [Google Scholar]
- 3.Min J, Chiu DT, Wang Y. Variation in the heritability of body mass index based on diverse twin studies: a systematic review. Obes Rev 2013;14:871–882 [DOI] [PMC free article] [PubMed]
- 4.Speliotes EK, Willer CJ, Berndt SI, et al.; MAGIC; Procardis Consortium . Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010;42:937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Locke AE, Kahali B, Berndt SI, et al.; LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium . Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knowler WC, Barrett-Connor E, Fowler SE, et al.; Diabetes Prevention Program Research Group . Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 2002;346:393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubin RR, Fujimoto WY, Marrero DG, et al.; DPP Research Group . The Diabetes Prevention Program: recruitment methods and results. Control Clin Trials 2002;23:157–171 [DOI] [PubMed] [Google Scholar]
- 8.Delahanty LM, Pan Q, Jablonski KA, et al.; Diabetes Prevention Program Research Group . Genetic predictors of weight loss and weight regain after intensive lifestyle modification, metformin treatment, or standard care in the Diabetes Prevention Program. Diabetes Care 2012;35:363–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCaffery JM, Papandonatos GD, Huggins GS, et al.; Genetic Subgroup of Look AHEAD; Look AHEAD Research Group . Human cardiovascular disease IBC chip-wide association with weight loss and weight regain in the Look AHEAD trial. Hum Hered 2013;75:160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCaffery JM, Papandonatos GD, Huggins GS, et al.; Genetic Subgroup of Look AHEAD, Look AHEAD Research Group . . FTO predicts weight regain in the Look AHEAD clinical trial. Int J Obes 2013;37:1545–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pi-Sunyer X, Blackburn G, Brancati FL, et al.; Look AHEAD Research Group . Reduction in weight and cardiovascular disease risk factors in individuals with type 2 diabetes: one-year results of the Look AHEAD trial. Diabetes Care 2007;30:1374–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wing RR; Look AHEAD Research Group . Long-term effects of a lifestyle intervention on weight and cardiovascular risk factors in individuals with type 2 diabetes mellitus: four-year results of the Look AHEAD trial. Arch Intern Med 2010;170:1566–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan DH, Espeland MA, Foster GD, et al.; Look AHEAD Research Group . Look AHEAD (Action for Health in Diabetes): design and methods for a clinical trial of weight loss for the prevention of cardiovascular disease in type 2 diabetes. Control Clin Trials 2003;24:610–628 [DOI] [PubMed] [Google Scholar]
- 14.Bray G, Gregg E, Haffner S, et al.; Look Ahead Research Group . Baseline characteristics of the randomised cohort from the Look AHEAD (Action for Health in Diabetes) study. Diab Vasc Dis Res 2006;3:202–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.TIBCO Software, Inc. TIBCO Spotfire SPLUS 8.2 for Solaris/Linux User’s Guide (Version 8.2). Seattle, WA, TIBCO Software, Inc., 2010 [Google Scholar]
- 16.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 2005;95:221–227 [DOI] [PubMed] [Google Scholar]
- 17.Wing RR, Bolin P, Brancati FL, et al.; Look AHEAD Research Group . Cardiovascular effects of intensive lifestyle intervention in type 2 diabetes. N Engl J Med 2013;369:145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Sayed Moustafa JS, Froguel P. From obesity genetics to the future of personalized obesity therapy. Nat Rev Endocrinol 2013;9:402–413 [DOI] [PubMed] [Google Scholar]
- 19.Herrera BM, Lindgren CM. The genetics of obesity. Curr Diab Rep 2010;10:498–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christian BE, Spremulli LL. Evidence for an active role of IF3mt in the initiation of translation in mammalian mitochondria. Biochemistry 2009;48:3269–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behrouz B, Vilariño-Güell C, Heckman MG, et al.. Mitochondrial translation initiation factor 3 polymorphism and Parkinson’s disease. Neurosci Lett 2010;486:228–230 [DOI] [PubMed] [Google Scholar]
- 22.McCaffery JM, Papandonatos GD, Peter I, et al.; Genetic Subgroup of Look AHEAD; Look AHEAD Research Group . Obesity susceptibility loci and dietary intake in the Look AHEAD Trial. Am J Clin Nutr 2012;95:1477–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nettleton JA, Follis JL, Ngwa JS, et al.. Gene × dietary pattern interactions in obesity: analysis of up to 68 317 adults of European ancestry. Hum Mol Genet 2015;24:4728–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Wang Z, Zhou X, et al.. Far upstream element-binding protein 1 and RNA secondary structure both mediate second-step splicing repression. Proc Natl Acad Sci U S A 2013;110:E2687–E2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Chen QM. Far upstream element binding protein 1: a commander of transcription, translation and beyond. Oncogene 2013;32:2907–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsiao HH, Nath A, Lin CY, Folta-Stogniew EJ, Rhoades E, Braddock DT. Quantitative characterization of the interactions among c-myc transcriptional regulators FUSE, FBP, and FIR. Biochemistry 2010;49:4620–4634 [DOI] [PubMed] [Google Scholar]
- 27.Sahm F, Koelsche C, Meyer J, et al.. CIC and FUBP1 mutations in oligodendrogliomas, oligoastrocytomas and astrocytomas. Acta Neuropathol 2012;123:853–860 [DOI] [PubMed] [Google Scholar]
- 28.Hoe KL, Won M, Chung KS, et al.. Isolation of a new member of DnaJ-like heat shock protein 40 (Hsp40) from human liver. Biochim Biophys Acta 1998;1383:4–8 [DOI] [PubMed] [Google Scholar]
- 29.Tsai MF, Wang CC, Chang GC, et al.. A new tumor suppressor DnaJ-like heat shock protein, HLJ1, and survival of patients with non-small-cell lung carcinoma. J Natl Cancer Inst 2006;98:825–838 [DOI] [PubMed] [Google Scholar]
- 30.Franks PW, Jablonski KA, Delahanty LM, et al.; Diabetes Prevention Program Research Group . Assessing gene-treatment interactions at the FTO and INSIG2 loci on obesity-related traits in the Diabetes Prevention Program. Diabetologia 2008;51:2214–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy MI. Genomics, type 2 diabetes, and obesity. N Engl J Med 2010;363:2339–2350 [DOI] [PubMed] [Google Scholar]
- 32.Franks PW. Gene × environment interactions in type 2 diabetes. Curr Diab Rep 2011;11:552–561 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.