Abstract
The RLK/Pelle class of proteins kinases is composed of over 600 members in Arabidopsis. Many of the proteins in this family are receptor-like kinases (RLK), while others have lost their extracellular domains and are found as cytoplasmic kinases. Proteins in this family that are RLKs have a variety of extracellular domains that drive function in a large number of processes, from cell wall interactions to disease resistance to developmental control. This review will briefly cover the major subclasses of RLK/Pelle proteins and their roles. In addition, two specific groups on RLKs will be discussed in detail, relating recent findings in Arabidopsis and how well these conclusions have been able to be translated to agronomically important species. Finally, some details on kinase activity and signal transduction will be addressed, along with the mystery of RLK/Pelle members lacking kinase enzymatic activity.
Overview
Protein kinases act to transfer phosphates to other proteins, often acting as switches to activate or inactivate proteins. Protein kinases are one of the largest protein families in plants, with over 1000 members in Arabidopsis and over 1400 members in rice (Dardick et al., 2007; Shiu and Bleecker, 2001a; Shiu and Bleecker, 2001b; Shiu and Bleecker, 2003). There are a large number of different groups of protein kinases, each with its own unique sequence and functional characteristics. These include MAPKs, MAPKKs, Rafs, CDPKs/CaMK, GSK3s, CDKs, NPH1s (Shiu and Bleecker, 2003) (Figure 1). These play many critical roles in plant development and physiology, including developmental patterning, hormone signaling, stress responses and disease resistance.
Figure 1.
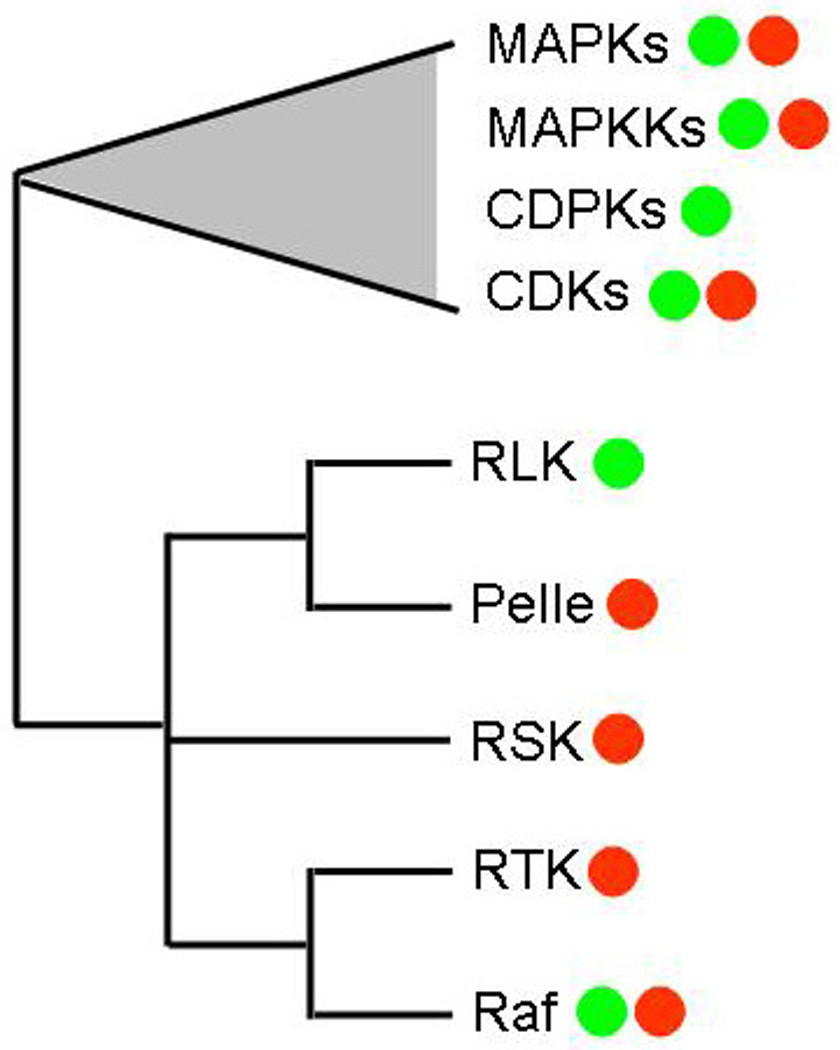
Phylogenetic relationships between major kinase families in plants and animals. Green dots indicate the clade contains representative genes in plant species, while a red dot indicate the clade contains representative genes in animal species. Adapted from (Shiu and Bleecker, 2001b).
This review will focus on proteins of the largest class of protein kinases, the RLK/Pelle family. This group is named for the receptor-like kinases (RLKs) structure commonly found within the family and the closest animals homologs which are the Pelle family of kinases (Shiu and Bleecker, 2001b) (Figure 1). For RLK/Pelle proteins kinases, over 600 are found in Arabidopsis and over 1000 rice, making up over 2% of each genome (Dardick et al., 2007; Shiu and Bleecker, 2003). Detailed analysis indicates that the RLK/Pelle family are a monophyletic group of genes – meaning that they all evolved from a common ancestral gene (see below).
Evolutionarily, the genes encoding the plant RLK/Pelle proteins are more closely related to many kinases-encoding genes in animals than to the plant proteins kinase in other classes such MAPKs, CDPKs, GSK3s and others (with the possible exception of genes coding for Rafs) (Figure 1). Interestingly the plant and animals genes forming a monophyletic group including the RLK/Pelle proteins are largely receptor-kinases, thus forming a receptor-kinase family that predates the split of plants and animals (Shiu and Bleecker, 2001b). While the ancestral kinase for this group was a serine/threonine protein kinase, tyrosine kinase activity also evolved within the animal receptor-tyrosine kinases (RTKs) within this group. Indeed, the plant RLK/Pelle protein kinase domains show similarity to motifs found in serine/threonine kinases as well as those found in tyrosine kinases. This may explain some recent findings on catalytic activity for these proteins (see below).
Because of the immense size of the RLK/Pelle kinase family in plants, this review will describe the different subclasses in only in broad terms. We will then focus on a two specific subclasses of kinase proteins to discuss functional relationships, evolutionary relationships, and how well findings in the model plant Arabidopsis have translated into understanding paralogous proteins in more agronomically important plants. Finally, we will address kinase catalytic activity, regulation, and kinases that are not in fact kinase.
RLK/Pelle subclasses
This very large family of over 600 proteins in Arabidopsis and 1000 in rice contains many transmembrane receptor-like kinases with various extracellular domains. For many of these subclasses, only one or a small number of proteins have been analyzed in significant detail. Thus, assuming that all members of a subclass have a similar developmental, physiological or cell biological role is premature. Nonetheless, knowing how proteins with similar domain structures function is a good starting point for considering the roles of previously uncharacterized proteins.
Several of the classes of RLK/Pelle proteins have been implicated in cell wall biogenesis and attachments, including proteins with extensin-like, lectin-like and epidermal-growth-factor-like extracellular domains (Ringli, 2010b; Seifert and Blaukopf, 2010). Extensins are hydroxyproline-rich proteins commonly found in plant cell walls where they play roles in cell wall structure (Cassab, 1998). The hydroxyprolines of extensins are often glycosylated and bound to wall components (Showalter, 1993). One version of the extensin-like RLKs is the family of proline-rich extensin-like receptor-kinases (PERKs), which appear to interact with pectin and effect cell wall deposition and cell growth (Bai et al., 2009; Nakhamchik et al., 2004). A second, smaller group of RLKs with extensin-like domains are found among the RLK/Pelle family and presumably also play a role in cell wall biogenesis, perhaps similar to the LRX1 extensin-like extracellular protein in Arabidopsis (Ringli, 2010a).
Another subclass of RLK/Pelle proteins with cell wall-associated function, the WAKs (wall-associated kinases), have extracellular domains that are cysteine-rich and share similarity to the epidermal growth factor receptor family in animals (He et al., 1999). WAKs are linked to cell growth, based on mutant and co-suppression phenotypes which exhibit defects in cell elongation and expansion (Kohorn et al., 2006; Wagner and Kohorn, 2001). WAKs can bind directly to pectin in the cell wall suggesting a role in linking cell expansion to the cell wall (Kohorn et al., 2009; Kohorn et al., 2006). WAKs have also been linked to metal tolerance and pathogen resistance (Hou et al., 2005; Li et al., 2009; Sivaguru et al., 2003).
Yet another RLK family associated with cell wall maintenance is the Catharanthus roseus-like RLKs (CrRLK1L). Evidence from mythically-inspired members such as FERONIA (FER), THESEUS1, HERKULES1 and HERKULES2 suggest a role in monitoring the integrity of the cell wall and providing response to challenges to integrity (Hematy et al., 2007). A separate studies suggests a role for these same genes in cell elongation as part of BRI1 steroid signaling (see below) (Guo et al., 2009). Interestingly, FER is also critical for the synergid interaction with the pollen during fertilization (Escobar-Restrepo et al., 2007). Two FER homologs, the genes encoding ANXUR1 and ANXUR2, are important for integrity of the pollen tube, and may control the timing of pollen rupture during fertilization (Boisson-Dernier et al., 2009; Miyazaki et al., 2009). More recently, FER was also shown to be important for the function of the small GTPase Rho during polarized growth of root hairs (Duan et al., 2010).
The LysM domain is conserved across prokaryotes and eukaryotes. The various LysM domains recognize peptidoglycans and other oligosaccharides. A group of RLK/Pelle family proteins have LysM domains that fall into different categories (Zhang et al., 2009; Zhang et al., 2007). The diversity of different LysM domains suggest multiple recognition targets. Legume isoforms of RLKs with LysM-containing extracellular domains recognize symbiotic bacterial signals that trigger plant responses to facilitate the formation of nodules for nitrogen fixation (Arrighi et al., 2006; Limpens et al., 2003; Madsen et al., 2003; Mulder et al., 2006; Radutoiu et al., 2003). The LysM domains in these receptors recognize lipochitin-oligosaccharide signals to mediate plant-bacterial interactions (Radutoiu et al., 2007).
Another unusual domain found among RLK/Pelle proteins is similar to tumor necrosis factor receptor domain. The original protein identified with this motif was the Crinkly4 (CR4) RLK in maize (Becraft et al., 1996), which also contains an RCC1-like domain, similar to a large group of plant and animal proteins (Hadjebi et al., 2008). CR4 is important for epidermal patterning in the maize epidermis and aleurone. ACR4, encoded by the Arabidopsis ortholog of CR4, is an epidermal-specific proteins that mediates several aspects of epidermal patterning, in addition to integument development in ovules (Gifford et al., 2003; Gifford et al., 2005).
The S-domain is present in the extracellular domains of approximately 40 different Arabidopsis RLKs. The function of this domain is best characterized in the Brassica S-receptor kinase (SRK), which it critical for pollen recognition during self-incompatibility (Goring and Rothstein, 1992; Stein et al., 1991; Takasaki et al., 2000). The S-domain is a cysteine-rich domain comprised of a number of distinct motifs, including an EGF repeat, an agglutinin motif, and a PAN motif (Haffani et al., 2004; Tordai et al., 1999). The similarity between the agglutinin motif and lectins raises once again the possibility of carbohydrate and/or glycoprotein interactions seen or suspected in many other RLK/Pelle family members. SRKs are essential to recognize self pollen in self-incompatible Brassica species, and presumably do so through the S-domain. The pollen factor recognized by SRK is SCR (for S-locus cysteine rich protein) (Schopfer et al., 1999) which binds to SRK in an allele-specific manner (Kachroo et al., 2001). Critically, SRK and SCR are linked together in a single genetic locus so that the SRK remains specific for self pollen (Casselman et al., 2000). Given the large number of S-domain RLKs in Arabidopsis and the lack of self-incompatibility, these receptors presumably have multiple functions outside of self recognition. The broad expression of many S-domain RLKs in many tissues and their induction linked to pathogenesis suggest possible roles in both developmental control and disease responses (Dwyer et al., 1994) (Pastuglia et al., 1997; Pastuglia et al., 2002). The S-domain shares its cysteine-rich characteristics with the DUF26 domain found in the extracellular region of a similarly large class of RLKs that are also termed cysteine-rich RLKs (CRKs) (Haffani et al., 2004; Shiu and Bleecker, 2003). The presence of cysteines has been hypothesized to facilitate a role for these receptors in oxidative response. Indeed, several members are induced by oxidative stress and pathogen infection (Wrzaczek et al., 2010). Several analyses have identified roles in pathogen defense and programmed cell death for these receptors (Chen et al., 2003; Chen et al., 2004; Chen, 2001).
Another relatively smaller class of RLK/Pelle members contain thaumatin-like domains. A representative member that has been investigated is PR5K, which, intriguingly, resembles the pathogenesis-related protein 5 (PR5), suggesting a role in pathogenesis (Wang et al., 1996).
By far the largest class, and likely the ancestral version of the plant RLK/Pelle family, is the leucine-rich repeat (LRR)-RLK, with over 200 members in Arabidopsis. Their large numbers have lead to extensive investigations of the LRR-RLK proteins across many different biological contexts. Leucine-rich repeats are 24 residue motifs that have a high portion of leucine and other hydrophobic residues. Each repeat forms a “loop”, with a beta-sheet face and an alpha-helical face. The hydrophobic residues form an internal core for each loop. The number of LRRs in each LRR-RLK varies from just one or two to more than twenty-five. Flanking the sequence of LRRs are often pairs of cysteine residues. Disulfide-linkage between the cysteines forms a “cap” to prevent exposure of the LRR hydrophobic core (Kolade et al., 2006). The beta-sheet faces of each repeat line up in LRR proteins to form a pocket, often capable of protein interactions. While one of the original LRR receptors in animals, Toll, binds to a protein ligand, other animal LRR receptors binds to a variety of lipid, carbohydrate and RNA ligands (Barton and Medzhitov, 2002; Weber et al., 2003).
LRR-RLKs, as might be expected from their large numbers, have been linked to a myriad of biological functions, from pathogen recognition and disease resistance (Gomez-Gomez and Boller, 2000; Song et al., 1995) to steroid signaling (Li and Chory, 1997; Li et al., 2002; Nam and Li, 2002) to stem cell control (Clark et al., 1997) to embryo patterning (Nodine et al., 2007) to epidermal patterning (Kwak et al., 2005; Shpak et al., 2004; Torii et al., 1996). Ligands for several of these receptors have been identified, and they include endogenous proteins, sulfonated peptides, steroids and pathogen-derived peptide elicitors (Gomez-Gomez and Boller, 2002; Matsubayashi et al., 2002). Because of the size and diversity in function of the LRR-RLK family, this review will focus in detail on two subgroups to describe what is known about these factors in Arabidopsis and how this knowledge has translated to agronomic species.
Many of the RLK/Pelle subgroups classified by their extracellular domain each form a monophyletic group (Shiu and Bleecker, 2003), suggesting they evolved from a single event that replaced an LRR extracellular domain with the motif that now characterizes the subclade. These include the PERK, WAK and Lectin-like RLKs. Other subgroups, such as the S-domain and CrRLKs have multiple origins.
The RLK/Pelle family also contains a large number of cytoplasmic kinases lacking both a transmembrane and an extracellular domain. The cytoplasmic kinases have evolved repeatedly from various transmembrane versions of the protein (Shiu and Bleecker, 2003). There are large subclades that are comprised only of cytoplasmic kinases.
Finally, for many of the RLK/Pelle subgroups, there are receptor-like proteins (RLPs) lacking an intracellular kinase domain that share the same extracellular domain. In addition, some motifs are also found as secreted proteins lacking both a transmembrane domain and a cytoplasmic kinase domain. Some of these RLPs appear to have functions independent of any RLK, such as the Cf family of proteins from tomato that provide fungal resistance (Dixon et al., 1996; Jones et al., 1994). Others appear to function alongside their RLK counterpart. CLAVATA2 (CLV2), an LRR-RLP, acts together with the LRR-RLK CLV1 to control meristem size (Jeong et al., 1999; Kayes and Clark, 1998). The secreted S-domain protein SLG works in concert with the S-domain SRK to provide self-incompatibility (Giranton et al., 1995; Takasaki et al., 2000). Thus, while the RLPs lack the kinase domain that provides functional and evolutionary relationships between the RLK/Pelle family of proteins, understanding RLP function may provide a window into their cognate RLKs.
The BRI1 clade
Studies in Arabidopsis
The BRI1 (BRASSINOSTEROID INSENSTITIVE1) locus was originally identified in an EMS mutagenesis screen selecting for insensitivity to brassinosteroid signaling normally inhibiting root growth in wild-type Arabidopsis. (Clouse et al., 1996). Brassinosteroids are hormones important for plant growth and development and plants that cannot perceive, process or synthesize BRs exhibit strong dwarf phenotypes. Brassinosteroids play broad roles in many aspects of plant physiology including pathogenesis, stress responses, and cell elongation (Kim et al., 2010; Noguchi et al., 1999). bri1 mutant plant growth is stunted and, unlike brassinosteroid biosynthesis mutants, is not rescued by exogenous brassinolide. Additional loss-of-functions screens have led to the identification of many alleles of BRI1. The severe phenotypes of bri1 null alleles imply that BRI1 itself provides the bulk of brassinosteroid perception in Arabidopsis (Li and Chory, 1997).
BRI1 is a member of the LRR-RLK class of RLK/Pelle proteins. The BRI1 extracellular domain contains 25 LRRs (Li and Chory, 1997). This domain of tandem LRRs is broken up by a 70 amino acid “island” of non-LRR sequence. Like many LRR-RLKs, the island of non-LRR sequence is located between the fourth and fifth LRR counting from the transmembrane domain. Presumably, this conserved organization of LRR domains is important for how the extracellular domains function to relay signaling. Critically, the island sequence plus an adjacent LRR are necessary and sufficient for direct binding to brassinolide (Kinoshita et al., 2005). Whether other “islands” in various LRR-RLKs act as ligand binding domains remains to be determined.
Given the severe phenotypes of bri1 mutants and their strong insensitivity to brassinosteroids, one might assume that BRI1 is the only brassinosteroid receptor. BRI1 forms a monophyletic group with three other Arabidopsis genes BRL1 (BRI1-like), BRL2 (also called VASCULAR HIGHWAY1), BRL3. All four genes share the same basic structure including 25 LRR repeats and 70 amino acid “island” responsible for binding BRs in BRI1. BRL1 and BRL3 share 47% amino acid identity with BRI1 while BRL2 shares 45% amino acid identity with BRI1. BRL1, BRL2 and BRL3 are all expressed in the vasculature and the corresponding mutants have vascular related defects (Cano-Delgado et al., 2004; Clay and Nelson, 2002). Interestingly, BRL1 and BRL3 are also brassinosteroid receptors, while BRL2 is not (Cano-Delgado et al., 2004; Zhou et al., 2004). This conclusion is based on direct brassinosteroid binding, cross-complementation, and enhancement of the bri1 mutant phenotype. Thus, establishing the identity of bona fide brassinosteroid receptors has to be determined experimentally and not simply based on sequence similarity or phylogeny.
The functional relationships between the genes do match evolutionary relationships, as BRL1 and BRL3 are the most closely related genes to BRI1 in Arabidopsis (Figure 2). Interestingly, most of the gene duplications within this clade are ancient events. This is based on the observations that there are monocot versions of BRI1, BRL1/3 and BRL2. Thus all of the duplications except the BLR1/BRL3 duplication predate the split of monocots and dicots. The relatively ancient organization of this gene family increases the chances of finding functional orthologs in other plant species. It also points out how rarely gene duplication events are maintained within this clade.
Figure 2.
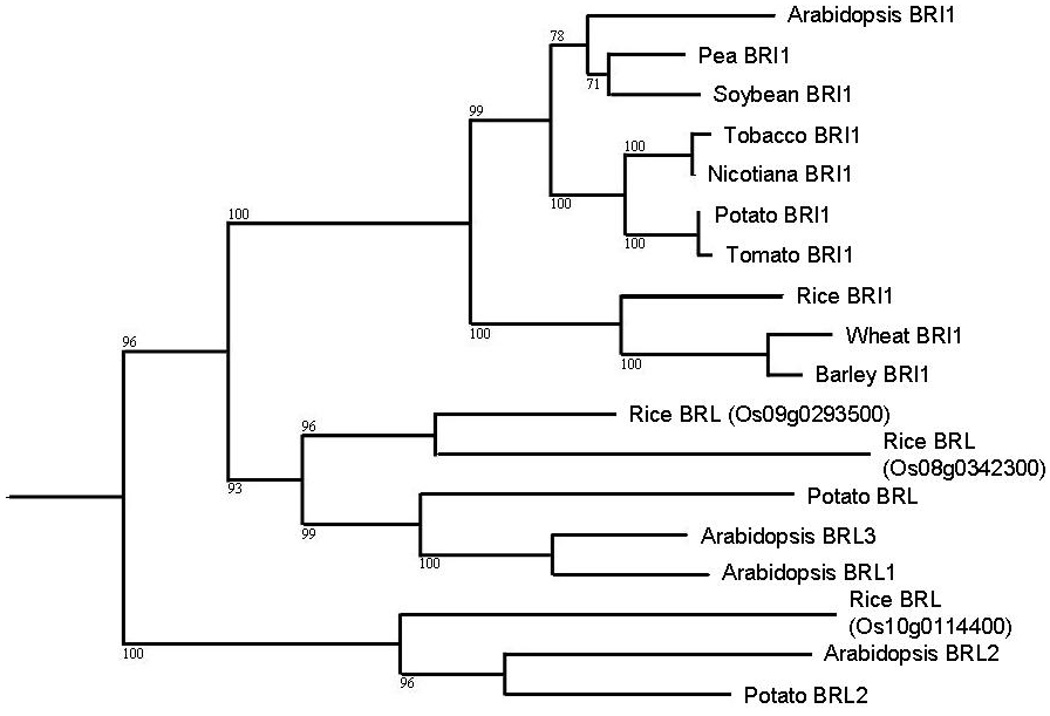
Phylogenetic relationships between BRI1 and related genes from selected angiosperms. The tree was generated by ClustalX using the BLAST results from residues 857–1157 of the Arabidopsis BRI1 kinase domain and viewed with NJplot. Genbank accession numbers for protein sequences are as follows: Arabidopsis BRI1 NP_195650, Pea BRI1 BAC99050, Soybean BRI1 ACJ37420, Tobacco BRI1 ABR18799, Nicotiana BRI1 ABO27628, Potato BRI1 ABO27627, Tomato BRI1 AAN85409, Rice BRI1 Os08g0342300, Wheat BRI1 ABG43096, Barley BRI1 BAD01654, Rice BRL Os09g0293500, Rice BRL Os08g0342300, Potato BRLunannotated, Arabidopsis BRL3 NP_187946, Arabidopsis BRL1 NP_175957, Rice BRL2 Os10g0114400, Arabidopsis BRL2 NP_178304, Potato BRL2unannotated.
Extending results to agronomic species
BRI1 orthologs have been identified and characterized in a number of agronomic species. bri1 mutants have properties that may be useful in crop sciences such as increased grain production in rice and constitutive activation of cold tolerance genes or the larger leaf and plant size from constitutive brassinosteroid signaling (Kim et al., 2010; Zhao et al., 2002). Furthermore, the dwarf stature of plants with attenuated BRI1 signaling could provide protection against lodging. This is the case in the rice ortholog, OsBRI1. The dwarf and brassinosteroid-insensitive phenotypes of the rice d61 mutants were shown to be the result of lesions in the rice OsBRI1 (Yamamuro et al., 2000). Mutations in this gene cause erect leaves and this allows for more effective light capture for photosynthesis and denser planting (Morinaka et al., 2006). Pilot studies show increased biomass from these lines with attenuated brassinosteroid signaling. Similar results are seen for mutants with attenuated brassinosteroid biosynthesis, suggesting a real mechanism for increasing yields (Sakamoto et al., 2006).
In barley, the BRI1 ortholog, HvBRI1, is the cause of a well-known dwarf mutation. The Japanese dwarf barley carrying what was known as the “uzu” gene were shown to contain a missense substitution in a conserved residue in the kinase domain encoded by HvBRI1 (Chono et al., 2003). These plants have reduced sensitivity to brassinosteroids and provide a mechanism to introduce agronomically beneficial dwarfism in various barley cultivars (Honda et al., 2003).
The pea BRI1 ortholog, LKA, retains a key role in brassinosteroid signaling (Nomura et al., 2003), providing mutant phenotypes consistent with reduction in brassinosteroid signaling.
An interesting and controversial aspect of BRI1 function in agronomic plants comes from analysis of BRI1 paralogs in solanaceae, including potato, and tomato. Potato and tomato use the peptide hormone systemin in response to wounding while the sister subtribe nicotianoideae, which includes tobacco, does not (McGurl et al., 1992; Pearce et al., 1991; Ryan, 2000). SR160 was originally identified as the systemin receptor with a radiolabeled systemin and was later identified as a tomato BRI1 paralog (Scheer and Ryan, 1999; Scheer and Ryan, 2002). Like rice, barley and pea, this tomato BRI1 ortholog corresponded to an existing dwarf mutants that was the result of lesions within the coding sequence (Montoya et al., 2002). These studies suggested that the solanaceae BRI1s were dual brassinosteroid/systemin receptors. Further evidence came from experiments indicating that systemin signaling could be engineered in tobacco by expression of the tomato BRI1 (Scheer et al., 2003). However, later experiments indicated that the BRI1 orthlogs were not required for solanaceae defense-related systemin signaling in vivo, nor was the BRI1 ortholog mutant altered in systemin responses (Holton et al., 2007; Malinowski et al., 2009). The data are conflicting, but suggest that while solanaceae BRI1 receptors may be able to bind systemin, they may not be physiologically relevant to defense responses.
The CLV1 clade
Studies in Arabidopsis
CLAVATA1 (CLV1) plays a central role in meristem regulation and was the first component of the pathway identified (Clark et al., 1997). Subsequent research from many labs has found both upstream ligands, receptor complexes, downstream signaling mediators, and the ultimate target of CLV1 function in the Arabidopsis shoot meristem (see below). Loss of CLV1 signaling leads to an enlargement of the shoot and flower meristems, resulting in more rapid production of leaves and flowers, thicker stems, and larger fruits (Clark et al., 1993).
The minimal CLV1 clade in Arabidopsis is composed of four genes, - CLV1, BAM1, BAM2 and BAM3 (DeYoung et al., 2006) (Figure 3). Critically, the gene duplication event that gave rise to CLV1 and BAM genes predates the monocot/dicot split, as there are genes in rice that pair with CLV1 and genes in rice that pair with BAM. This suggests that many of the extant LRR-RLKs were present in early angiosperms, which, if true, would make establishing orthologs between model species and agronomic species easier. It also echoes the BRI1 gene family, in that most of the duplications are ancient and pointing out the scarcity of maintained gene duplications over 200 million years of evolution.
Figure 3.
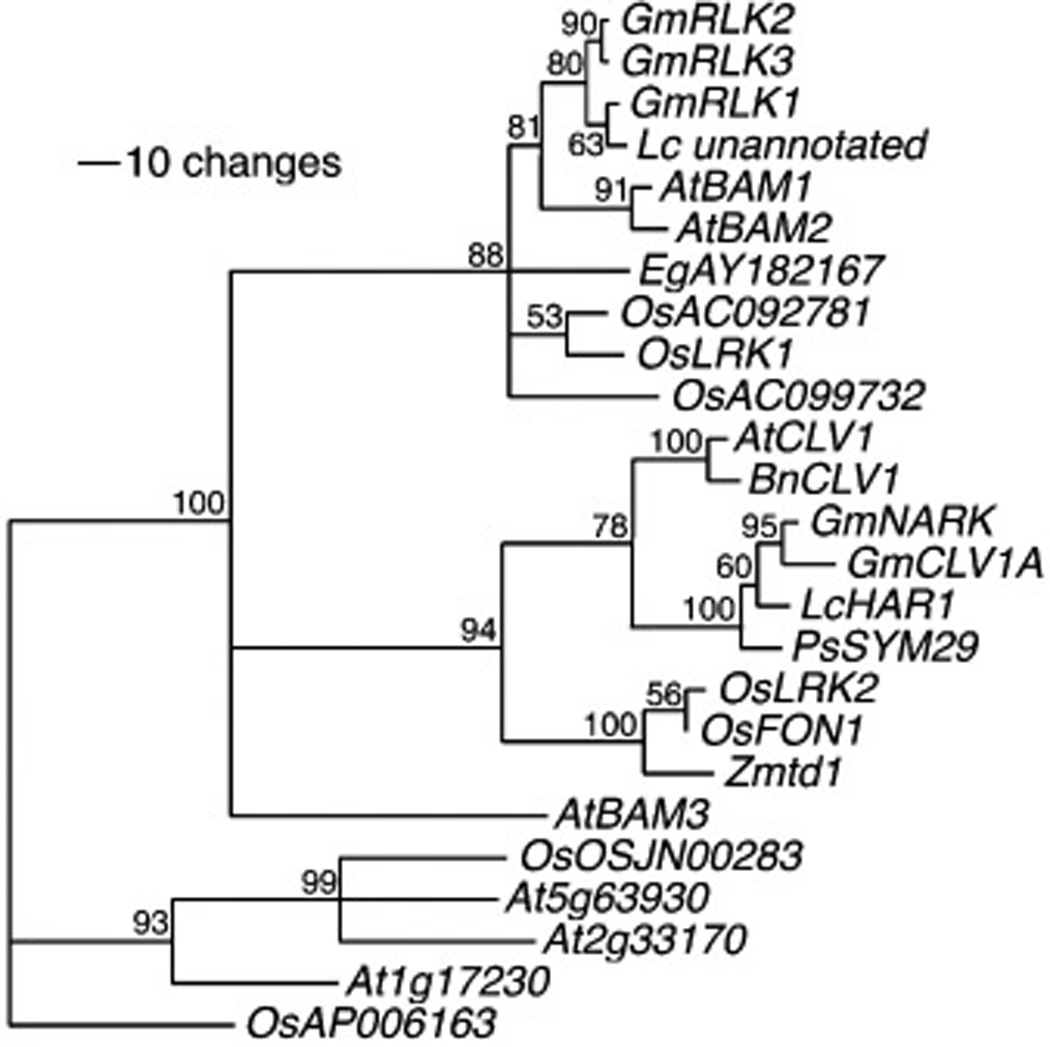
Phylogenetic relationships between CLV1 and related genes from selected angiosperms. From (DeYoung et al., 2006).
While CLV1 and BAM receptors have nearly equivalent function at the biochemical level (see below), they have very different roles developmentally driven primarily by where they are expressed (DeYoung et al., 2006; DeYoung and Clark, 2008). CLV1 is expressed in the meristem center where it acts to repress transcription of the meristem-promoting WUS transcription factor (Schoof et al., 2000). As a result, clv1 mutants develop larger meristems, with many accompanying defects as a result (Clark et al., 1993). In the center of the meristem, CLV1 function is weakly redundant with BAM receptors. The very low levels of BAM expression in the meristem center explain why clv1 null alleles have visible, albeit weak, phenotypes.
Because of redundancy, most of the clv1 mutants that have been identified to date are dominant-negative alleles containing missense mutations within the kinase domain (Diévart et al., 2003). The mechanism of dominant-negative function evolves from the nature of the CLV1 receptor complex, which is composed of CLV1/CLV1 and CLV1/BAM multimers (see below).
Outside of the meristem, BAM receptors regulate a large number of developmental events, linked to broad expression of the BAM receptors. These include vascular patterning, leaf morphology, and the development of anthers and other floral organs (DeYoung et al., 2006). Male sterility in bam1 bam2 double mutants can be traced back to early defects in asymmetric cell fate specification during early anther development (Hord et al., 2006). Critically, expressing CLV1 within these developing organs can complement the bam1 bam2 mutant defects, indicating that the different developmental pathways under BAM and CLV1 control use the same signaling pathway and kinase activity.
Extending results to agronomic species
Many CLV1-paralogous genes have been studied in a large number of species. These have revealed instances of strong functional conservation as well as the evolution of novel, agronomically-critical functions.
In both maize and rice, the genes most closely related to CLV1 have retained orthologous function. The maize gene thick tassel dwarf1 (td1) is the homolog most closely related to CLV1. td1 mutants have phenotypes similar to clv1 mutants – namely enlargement of the shoot and flower meristems (Bommert et al., 2005). Similarly in rice, the closest CLV1 homolog FLORAL ORGAN NUMBER1 (FON1) limits meristem size as well (Suzaki et al., 2004). fon1 mutations only affect flower meristem size – presumably the other related LRR-RLKs in rice may mask the function of FON1 in shoot meristem size.
Additional evidence suggests that the entire CLV1 signaling pathway is functionally conserved between monocots and Arabidopsis. In Arabidopsis, CLV2 acts in a parallel receptor complex that appears to work in concert with CLV1 (see below) and, as such, clv2 mutants developed enlarged shoot and flower meristems (Kayes and Clark, 1998). Mutations in the maize CLV2 homolog, fasciated ear2, lead to enlarged ear inflorescence meristems, indicating a conserved function (Taguchi-Shiobara et al., 2001). In Arabidopsis, CLV3 encodes the ligand for CLV1 and CLV2 (Fletcher et al., 1999). As a result, clv3 mutants also developed enlarged meristems. Critically the CLV3 homologs in rice, FON4 and FON2, also act to limit meristem size (Chu et al., 2006; Suzaki et al., 2006). A third CLV3-like protein, FOS1, controls meristem size redundantly with FON2 and FON4, but may do so in a separate FON1-independent pathway (Suzaki et al., 2009). In total, these findings suggest a relatively ancient role for the CLV signaling pathway in shoot meristem function that is conserved across angiosperms. Given that enlarged meristems have accompanied domestication of multiple plant species, CLV1 orthologs are logical targets for controlling stem and fruit size. Mutations in single CLV pathway genes may have relatively weak effects presumably related to genetic redundancy (e.g., clv1 null alleles) or may only effect a subset of tissues (e.g., FON2 mutations only alter rice flower meristem development). Thus, a challenge in translating model system results to agronomic species is sorting through potentially functional homologs to identify the genes most important for control of that particular tissue or cell type. Analysis of gene expression patterns, to identify the isoforms most likely to play major roles in specific tissues, will likely be essential in many cases.
A critical divergence in the function of CLV1 homologs has occurred in legumes. The CLV1 homologs GmNARK in soybeans, PsSYM29 in pea, MtSUNN in Medicago truncatula and LcHAR1 in lotus have all evolved a new function (Krusell et al., 2002; Nishimura et al., 2002; Schnabel et al., 2005; Searle et al., 2003). The expression of these genes is broader than CLV1 and they have a novel physiological role specific to legumes. Mutations in these genes result in defects in nodulations in the roots, which are specialized symbiotic bacterial interactions that serve to fix nitrogen. In the RLK mutants, the nodulation lacks proper control, forming a much higher than normal number of nodules referred to as hypernodulation. Interestingly, the receptors appear to function in the leaves to produce signals to the root to control the nodulation process (Lin et al., 2010). This RLK-dependent signal to the root is a small organic molecule (Lin et al., 2010).
RLK/Pelle protein kinase activity and signal transduction pathways
To date most of the characterized RLK/Pelle class kinase domain exhibit serine/threonine protein phosphorylation. This activity is normally tested in vitro with E. coli-expressed kinase domain, often measured by autophosphorylation and/or artificial substrate phosphorylation.
The most detailed analysis of kinase domain phosphorylation has been conducted on the BRI1 brassinosteroid receptor. Detailed analysis of autophosphorylation sites in vitro were first performed for the BRI1 kinase domain alone expressed in E. coli (Oh et al., 2000). Even though the assay was entirely in vitro with just the BRI1 kinase domain, the sites identified were largely later confirmed by analysis of BRI1 in vivo (Wang et al., 2005a). These include multiple sites in the juxtamembrane domain (between the transmembrane and kinase catalytic domain), the kinase domain, primarily the activation loop (see above) and the C-terminal tail. Substitutions at the activation loop sites were shown the be essential for both catalytic activity and in vivo function (Wang et al., 2005a). Several of these sites effect BRI1 function in vivo when mutated. The biggest surprise came from recent characterization of BRI1 tyrosine phosphorylation both in vivo and in vitro (Oh et al., 2009). This indicates that BRI1 is a dual-specificity kinase, with both serine/threonine and tyrosine kinase activity. Only two tyrosines were detected phosphorylated on BRI1, one is essential for catalytic activity, while the other effects BRI1 function in regulating development (Oh et al., 2009; Wang et al., 2005a; Wang et al., 2008).
BRI1 kinase activity is regulated by several factors (Figure 4). First, BRI1 contains a C-terminal auto-inhibitory domain (Wang et al., 2005b). Removal of this domain leads to a hyperactive BRI1. The function of this auto-inhibitory domain is controlled by its phosphorylation status. Another BRI1 control is exerted by a regulatory protein, BKI1, which acts to prevent BRI1 activation (Wang and Chory, 2006). Upon BRI1 ligand binding, BKI1 is phosphorylated by BRI1 and dissociates from the BRI1 homodimers, leaving BRI1 free to interact with its signaling partner BAK1 (Li et al., 2002; Nam and Li, 2002). BAK1, also known as SERK3 (Russinova et al., 2004), is a LRR-RLK as well, with five LRRs positioned between leucine zipper and proline-motifs. Detailed phosphorylation analysis has revealed sequential trans-phosphorylation events between BRI1 and BAK1 that activate signaling (Li et al., 2002; Nam and Li, 2002; Wang et al., 2005a; Wang et al., 2008) (see (Kim and Wang, 2010) for detailed review of phosphorylation sites).
Figure 4.
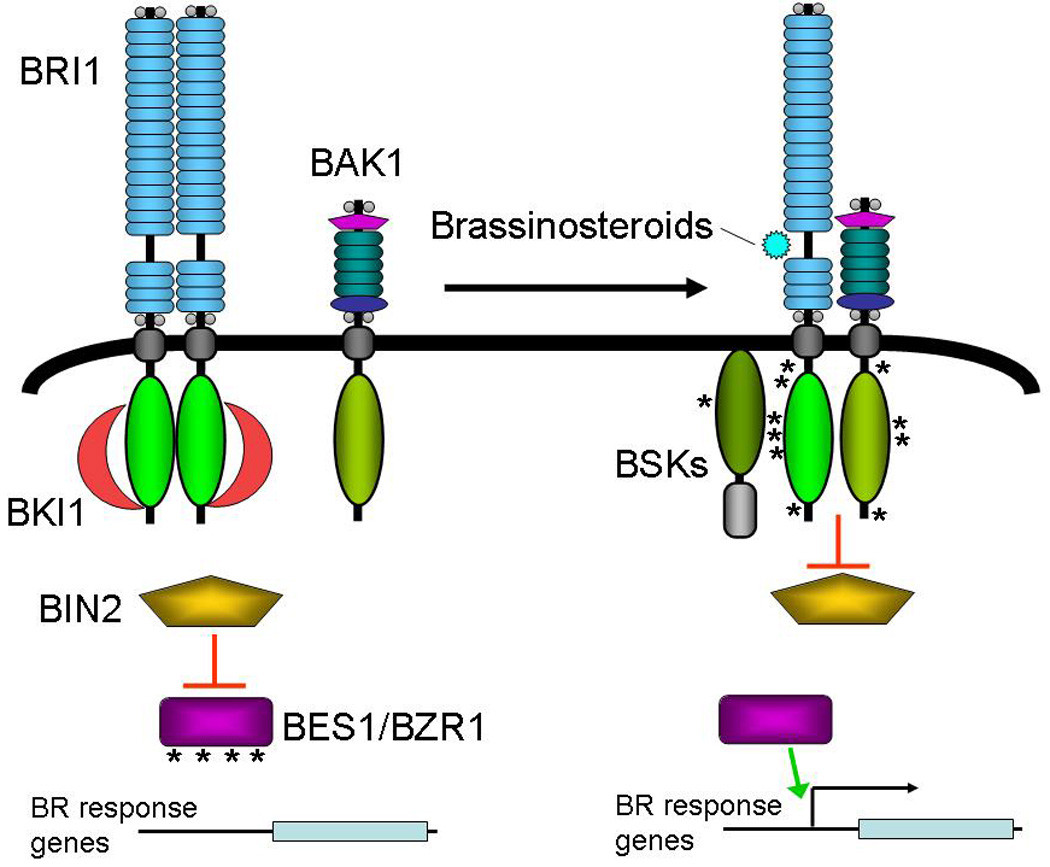
Model for brassinosteroid signaling. In the absence of signaling, BRI1 forms constitutive homodimers held inactive by BKI1 and the BRI1 autoinhibitory domain.BIN2 actively represses BES1 and BZR1 through direct phosphorylation. Binding of brassinosteroids to the BRI1 extracellular domains relieves BKI1 inhibition and allows BRI1-BAK1 interaction and sequential trans-phosphorylation. BSKs facilitate BRI1 repression of BIN2, derepressing BES1 and BZR1, allow for brassinosteroid-responsive transcriptional control. Asterisks indicate protein phosphorylation. See text for explanation and citations.
Downstream of these paired receptor-kinases are a series of other kinases. A trio of cytoplasmic kinases that are also members of the RLK/Pelle family mediate signaling from the BRI1/BAK1 complex. These BR-signaling kinases (BSK1, BSK2 and BSK3) are membrane-associated despite the lack of transmembrane domain (Tang et al., 2008). BRI1 phosphorylates BSKs, presumably regulating their function.
The key cytoplasmic switch controlled by brassinosteroid responses is the GSK3-class kinase BIN2 (Li and Nam, 2002; Li et al., 2001). BIN2 constitutively represses brassinosteroid responses by phosphorylating the transcription factors BES1 and BZR1 (He et al., 2002; Yin et al., 2002; Zhao et al., 2002). When BIN2 is inactivated by BRI1 signaling, BES1 and BZR1 accumulate in the nucleus and regulate the transcription of response elements (see (Li, 2010) for a detailed review of transcriptional brassinosteroid responses).
CLV1 signaling has also been extensively characterized. Like BRI1, CLV1 forms constitutive homodimers (Bleckmann et al., 2010; Guo et al., 2010; Zhu et al., 2010) (Figure 5). In addition to homodimers, CLV1 forms heteromultimers with the redundant BAM receptors, which explains the dominant-negative nature of the majority of clv1 alleles by poisoning complexes with BAM (Guo et al., 2010). These dominant-negative lesions cluster around the putative activation segment – a region in many protein kinases that lies across the active site and controls kinase activity. Phosphorylation within the activation loop of the activation segment drives a catalytically active confirmation (Lochhead, 2009; Nolen et al., 2004). Consistent with the nature of these missense clv1 alleles, over-expression of kinase-dead CLV1 has a similar dominant-negative effect (Williams et al., 1997). Another observation that points to the high level of functional specificity even within the core kinase catalytic domain came from characterization of CLV1/BRI1 chimeric receptors. Here, swapping just the core kinase catalytic domains lead to inactive proteins with dominant-negative characteristics (Diévart et al., 2003; Diévart et al., 2006).
Figure 5.
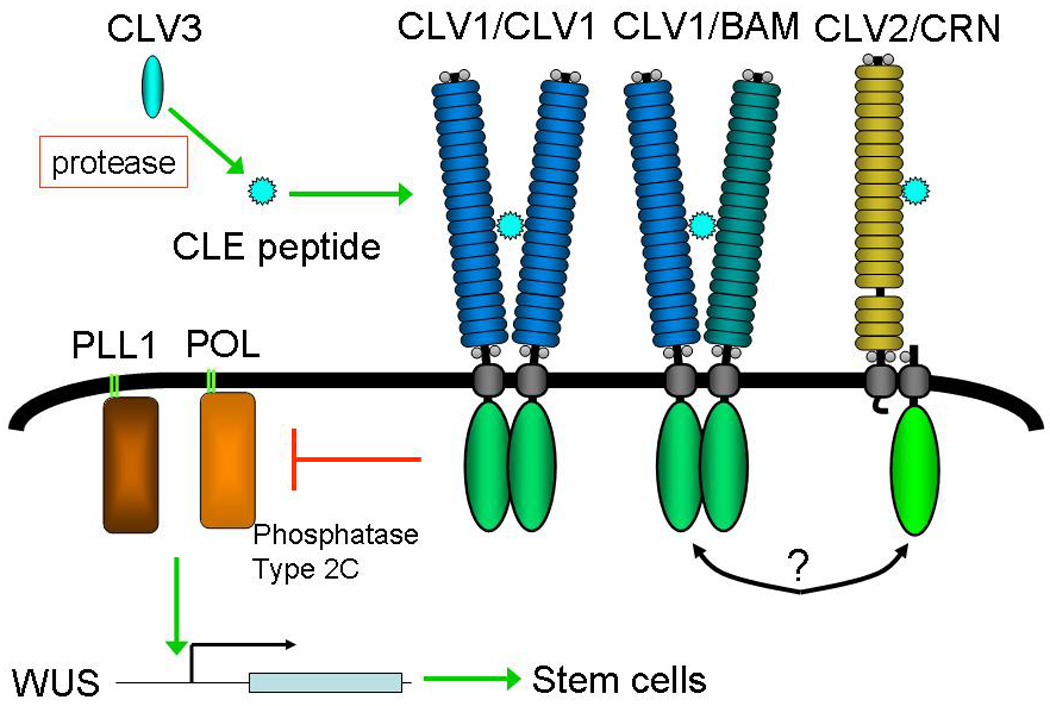
Model for CLV1 signaling. CLV3 is processed by a serine protease to create the active ligand (Ito et al., 2006; Ni et al., 2010). Mature CLV3 binds to CLV1, BAM and CLV2 receptors. The CLV1 and CLV2 complexes may interact with each other. Receptor activation leads to repression of POL/PLL1, which are positive regulators of WUS transcription.
The CLV1 and BAM receptors directly bind to the mature form of the CLV3 polypeptide ligand (Guo et al., 2010; Kondo et al., 2008; Ogawa et al., 2008). However, the most unique aspect of CLV1 signaling is the presence of a separate receptor complex composed of the CLV2 LRR-RLP and the transmembrane putative kinase CORYNE (CRN) (Bleckmann et al., 2010; Guo et al., 2010; Zhu et al., 2010). While CLV2 lacks a cytoplasmic domain, and CRN lacks an extracellular domain, together these proteins form a potential signaling receptor (Kayes and Clark, 1998; Muller et al., 2008). In this case, signaling is engendered by the ability of CLV2 to also bind directly to mature CLV3 with an affinity similar to that of CLV1 and BAM (Guo et al., 2010). In otherwise wild-type plants, both the CLV1/CLV1 and CLV2/CRN complexes are required for signaling, based on the strong phenotypic effects of both clv1 and clv2 mutations. Thus CLV1 signaling requires parallel activation of two complexes by the same polypeptide ligand.
CLV1 signaling acts by repressing the activity of a pair of downstream protein phosphatases, POLTERGEIST (POL) and PLL1 (Song and Clark, 2005; Song et al., 2006; Yu et al., 2003). While there is no evidence of direct interaction between the receptor complexes and POL/PLL1, the phosphatases are plasma membrane localized by the attachments of lipids to at the N-terminus of the proteins. In addition, the catalytic activity of POL/PLL1 may be regulated by the membrane lipid phosphatidylinositol 4-phosphate (Gagne and Clark, 2010). POL/PLL1 act to promote expression of the transcription factor WUS which promotes stem cell specification (Mayer et al., 1998; Schoof et al., 2000; Song et al., 2006). Further complicating the CLV signaling pathway is the recent identification of a CLV3-responsive, but CLV1-independent parallel pathway represented by the RPK2/TOAD2 RLKs (Kinoshita et al., 2010). Whether this pathway acts through POL/PLL1 is unknown.
RLK/Pelle proteins that are not functional kinases
Perhaps the most intriguing members of the RLK/Pelle family are those that appear to have retain high sequence identity with other family members, yet have lost kinase catalytic activity. The first member characterized in this fashion was the STRUBBELIG (SUB) RLK, also identified as the SCRAMBLED (SCM) RLK (Chevalier et al., 2005; Kwak et al., 2005). SUB was characterized by the effects of mutants on the development of ovules and other organs, while SCM was identified by the effect of mutations on the patterning of hair and non-hair cells in the Arabidopsis root.
Critically, Schneitz and co-workers noted that the catalytic loop of SUB (subdomain VIa) lack two key residues conserved among functional kinases (Chevalier et al., 2005). When tested in vitro, the SUB kinase domain lacked phosphotransfer activity. This is not a definitive test given that enzymatic activity in vivo might differ from that in vitro. However, a final test in which SUB with additional, kinase-dead mutations, was used to rescue the sub mutant phenotype demonstrated that kinase activity is unnecessary for SUB function. Taken together, these experiments indicate that SUB contains a catalytically inactive kinase domain. This is not to say that the kinase domain is dispensable, as SUB isoforms lacking the kinase domain do not function in vivo (Chevalier et al., 2005). In a further twist, even cataltically functional kinase domains have been found to be dispensable for RLK function (Xu et al., 2008).
Further bioinformatic analysis has revealed that upwards of 20% of the Arabidopsis RLKs contain non-conserved substitutions in key catalytic residues and may be dead kinases (Castells and Casacuberta, 2007). The remarkable aspect of these known and putative dead kinases is that their kinase domains retain such a high degree of sequence conservation. While the extracellular domains have evolved dramatic divergences in sequences and even domains, even the dead kinase have been under select constraints. This suggests that the kinase dead RLK/Pelle members still require much of the kinase domain fold and structure to carry out signaling.
Acknowledgements
Research in the authors’ lab on receptor-kinase signaling is supported by a National Institutes of Health grant (1R01GM62962-01A1) to SEC.
References
- Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Gherardi M, Huguet T, et al. The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Zhang G, Zhou Y, Zhang Z, Wang W, Du Y, Wu Z, Song CP. Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca signalling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J. 2009;60:314–327. doi: 10.1111/j.1365-313X.2009.03956.x. [DOI] [PubMed] [Google Scholar]
- Barton GM, Medzhitov R. Toll-like receptors and their ligands. Curr Top Microbiol Immunol. 2002;270:81–92. doi: 10.1007/978-3-642-59430-4_5. [DOI] [PubMed] [Google Scholar]
- Becraft PW, Stinard PS, McCarty DR. CRINKLY4 - a TNFR-like receptor kinase involved in maize epidermal differentiation. Science. 1996;273:1406–1409. doi: 10.1126/science.273.5280.1406. [DOI] [PubMed] [Google Scholar]
- Bleckmann A, Weidtkamp-Peters S, Seidel CA, Simon R. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 2010;152:166–176. doi: 10.1104/pp.109.149930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Roy S, Kritsas K, Grobei MA, Jaciubek M, Schroeder JI, Grossniklaus U. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development. 2009;136:3279–3288. doi: 10.1242/dev.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P, Lunde C, Nardmann J, Vollbrecht E, Running M, Jackson D, Hake S, Werr W. thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development. 2005;132:1235–1245. doi: 10.1242/dev.01671. [DOI] [PubMed] [Google Scholar]
- Cano-Delgado A, Yin Y, Yu C, Vafeados D, Mora-Garcia S, Cheng JC, Nam KH, Li J, Chory J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development. 2004;131:5341–5351. doi: 10.1242/dev.01403. [DOI] [PubMed] [Google Scholar]
- Cassab GI. Plant Cell Wall Proteins. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:281–309. doi: 10.1146/annurev.arplant.49.1.281. [DOI] [PubMed] [Google Scholar]
- Casselman AL, Vrebalov J, Conner JA, Singhal A, Giovannoni J, Nasrallah ME, Nasrallah JB. Determining the physical limits of the Brassica S locus by recombinational analysis. Plant Cell. 2000;12:23–33. doi: 10.1105/tpc.12.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells E, Casacuberta JM. Signalling through kinase-defective domains: the prevalence of atypical receptor-like kinases in plants. J Exp Bot. 2007;58:3503–3511. doi: 10.1093/jxb/erm226. [DOI] [PubMed] [Google Scholar]
- Chen K, Du L, Chen Z. Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol Biol. 2003;53:61–74. doi: 10.1023/B:PLAN.0000009265.72567.58. [DOI] [PubMed] [Google Scholar]
- Chen K, Fan B, Du L, Chen Z. Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol Biol. 2004;56:271–283. doi: 10.1007/s11103-004-3381-2. [DOI] [PubMed] [Google Scholar]
- Chen Z. A superfamily of proteins with novel cysteine-rich repeats. Plant Physiol. 2001;126:473–476. doi: 10.1104/pp.126.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier D, Batoux M, Fulton L, Pfister K, Yadav RK, Schellenberg M, Schneitz K. STRUBBELIG defines a receptor kinase-mediated signaling pathway regulating organ development in Arabidopsis. Proc Natl Acad Sci U S A. 2005;102:9074–9079. doi: 10.1073/pnas.0503526102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chono M, Honda I, Zeniya H, Yoneyama K, Saisho D, Takeda K, Takatsuto S, Hoshino T, Watanabe Y. A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant Physiol. 2003;133:1209–1219. doi: 10.1104/pp.103.026195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Qian Q, Liang W, Yin C, Tan H, Yao X, Yuan Z, Yang J, Huang H, Luo D, et al. The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 2006;142:1039–1052. doi: 10.1104/pp.106.086736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Clay NK, Nelson T. VH1, a provascular cell-specific receptor kinase that influences leaf cell patterns in Arabidopsis. Plant Cell. 2002;14:2707–2722. doi: 10.1105/tpc.005884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardick C, Chen J, Richter T, Ouyang S, Ronald P. The rice kinase database. A phylogenomic database for the rice kinome. Plant Physiol. 2007;143:579–586. doi: 10.1104/pp.106.087270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, Clark SE. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 2006;45:1–16. doi: 10.1111/j.1365-313X.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, Clark SE. BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics. 2008;180:895–904. doi: 10.1534/genetics.108.091108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diévart A, Dalal M, Tax FE, Lacey AD, Huttly A, Li J, Clark SE. CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell. 2003;15:1198–1211. doi: 10.1105/tpc.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diévart A, Hymes MJ, Li J, Clark SE. Brassinosteroid-independent function of BRI/CLV1 chimeric receptors. Functional Plant Biology. 2006;33:1–8. doi: 10.1071/FP06080. [DOI] [PubMed] [Google Scholar]
- Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JDG. The tomato Cf-2 disease resistance locus comprises 2 functional genes encoding leucine-rich repeat proteins. Cell. 1996;84:451–459. doi: 10.1016/s0092-8674(00)81290-8. [DOI] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci U S A. 2010;107:17821–17826. doi: 10.1073/pnas.1005366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer KG, Kandasamy MK, Mahosky DI, Acciai J, Kudish BI, Miller JE, Nasrallah ME, Nasrallah JB. A superfamily of S locus-related sequences in Arabidopsis: diverse structures and expression patterns. Plant Cell. 1994;6:1829–1843. doi: 10.1105/tpc.6.12.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science. 2007;317:656–660. doi: 10.1126/science.1143562. [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Gagne JM, Clark SE. The Arabidopsis stem sell factor POLTERGEIST is membrane localized and phospholipid stimulated. Plant Cell. 2010;22:729–743. doi: 10.1105/tpc.109.068734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean S, Ingram GC. The Arabidopsis ACR4 gene plays a role in cell layer organisation during ovule integument and sepal margin development. Development. 2003;130:4249–4258. doi: 10.1242/dev.00634. [DOI] [PubMed] [Google Scholar]
- Gifford ML, Robertson FC, Soares DC, Ingram GC. ARABIDOPSIS CRINKLY4 function, internalization, and turnover are dependent on the extracellular crinkly repeat domain. Plant Cell. 2005;17:1154–1166. doi: 10.1105/tpc.104.029975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giranton JL, Ariza MJ, Dumas C, Cock JM, Gaude T. The S locus receptor kinase gene encodes a soluble glycoprotein corresponding to the SKR extracellular domain in Brassica oleracea. Plant J. 1995;8:827–834. doi: 10.1046/j.1365-313x.1995.8060827.x. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T. Flagellin perception: a paradigm for innate immunity. Trends Plant Sci. 2002;7:251–256. doi: 10.1016/s1360-1385(02)02261-6. [DOI] [PubMed] [Google Scholar]
- Goring DR, Rothstein SJ. The S-locus receptor kinase gene in self-incompatible Barssica napus line encodes a functional serine/threonine kinase. Plant Cell. 1992;4:1273–1281. doi: 10.1105/tpc.4.10.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Li L, Ye H, Yu X, Algreen A, Yin Y. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2009;106:7648–7653. doi: 10.1073/pnas.0812346106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Han L, Hymes M, Denver R, Clark SE. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 2010;63:899–900. doi: 10.1111/j.1365-313X.2010.04295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjebi O, Casas-Terradellas E, Garcia-Gonzalo FR, Rosa JL. The RCC1 superfamily: from genes, to function, to disease. Biochim Biophys Acta. 2008;1783:1467–1479. doi: 10.1016/j.bbamcr.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Haffani YZ, Silva NF, Goring DR. Receptor kinase signalling in plants. Can. J. Bot. 2004;82:1–15. [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He ZH, Cheeseman I, He D, Kohorn BD. A cluster of five cell wall-associated receptor kinase genes, Wak1-5, are expressed in specific organs of Arabidopsis. Plant Mol Biol. 1999;39:1189–1196. doi: 10.1023/a:1006197318246. [DOI] [PubMed] [Google Scholar]
- Hematy K, Sado PE, Van Tuinen A, Rochange S, Desnos T, Balzergue S, Pelletier S, Renou JP, Hofte H. A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr Biol. 2007;17:922–931. doi: 10.1016/j.cub.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Holton N, Cano-Delgado A, Harrison K, Montoya T, Chory J, Bishop GJ. Tomato BRASSINOSTEROID INSENSITIVE1 is required for systemin-induced root elongation in Solanum pimpinellifolium but is not essential for wound signaling. Plant Cell. 2007;19:1709–1717. doi: 10.1105/tpc.106.047795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda I, Zeniya H, Yoneyama K, Chono M, Kaneko S, Watanabe Y. Uzu mutation in barley (Hordeum vulgare L.) reduces the leaf unrolling response to brassinolide. Biosci Biotechnol Biochem. 2003;67:1194–1197. doi: 10.1271/bbb.67.1194. [DOI] [PubMed] [Google Scholar]
- Hord CL, Chen C, Deyoung BJ, Clark SE, Ma H. The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell. 2006;18:1667–1680. doi: 10.1105/tpc.105.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Tong H, Selby J, Dewitt J, Peng X, He ZH. Involvement of a cell wall-associated kinase, WAKL4, in Arabidopsis mineral responses. Plant Physiol. 2005;139:1704–1716. doi: 10.1104/pp.105.066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell. 1999;11:1925–1934. doi: 10.1105/tpc.11.10.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JD. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science. 1994;266:789–793. doi: 10.1126/science.7973631. [DOI] [PubMed] [Google Scholar]
- Kachroo A, Schopfer CR, Nasrallah ME, Nasrallah JB. Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science. 2001;293:1824–1826. doi: 10.1126/science.1062509. [DOI] [PubMed] [Google Scholar]
- Kayes JM, Clark SE. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development. 1998;125:3843–3851. doi: 10.1242/dev.125.19.3843. [DOI] [PubMed] [Google Scholar]
- Kim SY, Kim BH, Lim CJ, Lim CO, Nam KH. Constitutive activation of stress-inducible genes in a brassinosteroid-insensitive 1 (bri1) mutant results in higher tolerance to cold. Physiol Plant. 2010;138:191–204. doi: 10.1111/j.1399-3054.2009.01304.x. [DOI] [PubMed] [Google Scholar]
- Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, Simon R, Yamaguchi-Shinozaki K, Fukuda H, Sawa S. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development. 2010;137:3911–3920. doi: 10.1242/dev.048199. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Cano-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- Kohorn BD, Johansen S, Shishido A, Todorova T, Martinez R, Defeo E, Obregon P. Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J. 2009;60:974–982. doi: 10.1111/j.1365-313X.2009.04016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD, Kobayashi M, Johansen S, Friedman HP, Fischer A, Byers N. Wall-associated kinase 1 (WAK1) is crosslinked in endomembranes, and transport to the cell surface requires correct cell-wall synthesis. J Cell Sci. 2006;119:2282–2290. doi: 10.1242/jcs.02968. [DOI] [PubMed] [Google Scholar]
- Kolade OO, Bamford VA, Ancillo Anton G, Jones JD, Vera P, Hemmings AM. In vitro characterization of the cysteine-rich capping domains in a plant leucine rich repeat protein. Biochim Biophys Acta. 2006;1764:1043–1053. doi: 10.1016/j.bbapap.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Kondo T, Nakamura T, Yokomine K, Sakagami Y. Dual assay for MCLV3 activity reveals structure-activity relationship of CLE peptides. Biochem Biophys Res Commun. 2008;377:312–316. doi: 10.1016/j.bbrc.2008.09.139. [DOI] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, Duc G, Kaneko T, Tabata S, de Bruijn F, et al. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature. 2002;420:422–426. doi: 10.1038/nature01207. [DOI] [PubMed] [Google Scholar]
- Kwak SH, Shen R, Schiefelbein J. Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science. 2005;307:1111–1113. doi: 10.1126/science.1105373. [DOI] [PubMed] [Google Scholar]
- Li H, Zhou SY, Zhao WS, Su SC, Peng YL. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol Biol. 2009;69:337–346. doi: 10.1007/s11103-008-9430-5. [DOI] [PubMed] [Google Scholar]
- Li J. Regulation of the nuclear activities of brassinosteroid signaling. Curr Opin Plant Biol. 2010;13:540–547. doi: 10.1016/j.pbi.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- Lin YH, Ferguson BJ, Kereszt A, Gresshoff PM. Suppression of hypernodulation in soybean by a leaf-extracted, NARK- and Nod factor-dependent, low molecular mass fraction. New Phytol. 2010;185:1074–1086. doi: 10.1111/j.1469-8137.2009.03163.x. [DOI] [PubMed] [Google Scholar]
- Lochhead PA. Protein kinase activation loop autophosphorylation in cis: overcoming a Catch-22 situation. Sci Signal. 2009;2:4. doi: 10.1126/scisignal.254pe4. [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- Malinowski R, Higgins R, Luo Y, Piper L, Nazir A, Bajwa VS, Clouse SD, Thompson PR, Stratmann JW. The tomato brassinosteroid receptor BRI1 increases binding of systemin to tobacco plasma membranes, but is not involved in systemin signaling. Plant Mol Biol. 2009;70:603–616. doi: 10.1007/s11103-009-9494-x. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Ogawa M, Morita A, Sakagami Y. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science. 2002;296:1470–1472. doi: 10.1126/science.1069607. [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- McGurl B, Pearce G, Orozco-Cardenas M, Ryan CA. Structure, expression, and antisense inhibition of the systemin precursor gene. Science. 1992;255:1570–1573. doi: 10.1126/science.1549783. [DOI] [PubMed] [Google Scholar]
- Miyazaki S, Murata T, Sakurai-Ozato N, Kubo M, Demura T, Fukuda H, Hasebe M. ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr Biol. 2009;19:1327–1331. doi: 10.1016/j.cub.2009.06.064. [DOI] [PubMed] [Google Scholar]
- Montoya T, Nomura T, Farrar K, Kaneta T, Yokota T, Bishop GJ. Cloning the tomato curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signaling. Plant Cell. 2002;14:3163–3176. doi: 10.1105/tpc.006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaka Y, Sakamoto T, Inukai Y, Agetsuma M, Kitano H, Ashikari M, Matsuoka M. Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 2006;141:924–931. doi: 10.1104/pp.106.077081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder L, Lefebvre B, Cullimore J, Imberty A. LysM domains of Medicago truncatula NFP protein involved in Nod factor perception. Glycosylation state, molecular modeling and docking of chitooligosaccharides and Nod factors. Glycobiology. 2006;16:801–809. doi: 10.1093/glycob/cwl006. [DOI] [PubMed] [Google Scholar]
- Muller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 2008;20:934–946. doi: 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhamchik A, Zhao Z, Provart NJ, Shiu SH, Keatley SK, Cameron RK, Goring DR. A comprehensive expression analysis of the Arabidopsis proline-rich extensin-like receptor kinase gene family using bioinformatic and experimental approaches. Plant Cell Physiol. 2004;45:1875–1881. doi: 10.1093/pcp/pch206. [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J. BRI1/BAK1, a Receptor Kinase Pair Mediating Brassinosteroid Signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- Ni J, Guo Y, Hartsell J, Clark SE. Characterization of a CLE processing activity. Plant Mol. Bio. 2010 doi: 10.1007/s11103-010-9708-2. in press. [DOI] [PubMed] [Google Scholar]
- Nishimura R, Hayashi M, Wu GJ, Kouchi H, Imaizumi-Anraku H, Murakami Y, Kawasaki S, Akao S, Ohmori M, Nagasawa M, et al. HAR1 mediates systemic regulation of symbiotic organ development. Nature. 2002;420:426–429. doi: 10.1038/nature01231. [DOI] [PubMed] [Google Scholar]
- Nodine MD, Yadegari R, Tax FE. RPK1 and TOAD2 are two receptor-like kinases redundantly required for arabidopsis embryonic pattern formation. Dev Cell. 2007;12:943–956. doi: 10.1016/j.devcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Nomura T, Bishop GJ, Kaneta T, Reid JB, Chory J, Yokota T. The LKA gene is a BRASSINOSTEROID INSENSITIVE 1 homolog of pea. Plant J. 2003;36:291–300. doi: 10.1046/j.1365-313x.2003.01863.x. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science. 2008;319:294. doi: 10.1126/science.1150083. [DOI] [PubMed] [Google Scholar]
- Oh MH, Ray WK, Huber SC, Asara JM, Gage DA, Clouse SD. Recombinant brassinosteroid insensitive 1 receptor-like kinase autophosphorylates on serine and threonine residues and phosphorylates a conserved peptide motif in vitro. Plant Physiol. 2000;124:751–766. doi: 10.1104/pp.124.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MH, Wang X, Kota U, Goshe MB, Clouse SD, Huber SC. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:658–663. doi: 10.1073/pnas.0810249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuglia M, Roby D, Dumas C, Cock JM. Rapid induction by wounding and bacterial infection of an S gene family receptor-like kinase gene in Brassica oleracea. Plant Cell. 1997;9:49–60. doi: 10.1105/tpc.9.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuglia M, Swarup R, Rocher A, Saindrenan P, Roby D, Dumas C, Cock JM. Comparison of the expression patterns of two small gene families of S gene family receptor kinase genes during the defence response in Brassica oleracea and Arabidopsis thaliana. Gene. 2002;282:215–225. doi: 10.1016/s0378-1119(01)00821-6. [DOI] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA. A polypeptide from tomato leaves induces would-inducible proteinase inhibitor proteins. Science. 1991;253:895–898. doi: 10.1126/science.253.5022.895. [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, Albrektsen AS, James EK, Thirup S, Stougaard J. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. Embo J. 2007;26:3923–3935. doi: 10.1038/sj.emboj.7601826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli C. The hydroxyproline-rich glycoprotein domain of the Arabidopsis LRX1 requires Tyr for function but not for insolubilization in the cell wall. Plant J. 2010a;63:662–669. doi: 10.1111/j.1365-313X.2010.04270.x. [DOI] [PubMed] [Google Scholar]
- Ringli C. Monitoring the outside: cell wall-sensing mechanisms. Plant Physiol. 2010b;153:1445–1452. doi: 10.1104/pp.110.154518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Cano-Delgado A, Yin Y, Chory J, de Vries SC. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1) Plant Cell. 2004;16:3216–3229. doi: 10.1105/tpc.104.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CA. The systemin signaling pathway: differential activation of plant defensive genes. Biochim Biophys Acta. 2000;1477:112–121. doi: 10.1016/s0167-4838(99)00269-1. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, et al. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol. 2006;24:105–109. doi: 10.1038/nbt1173. [DOI] [PubMed] [Google Scholar]
- Scheer JM, Pearce G, Ryan CA. Generation of systemin signaling in tobacco by transformation with the tomato systemin receptor kinase gene. Proc Natl Acad Sci U S A. 2003;100:10114–10117. doi: 10.1073/pnas.1432910100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer JM, Ryan CA. A 160-kD systemin receptor on the surface of lycopersicon peruvianum suspension-cultured cells. Plant Cell. 1999;11:1525–1536. doi: 10.1105/tpc.11.8.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer JM, Ryan CA., Jr The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc Natl Acad Sci U S A. 2002;99:9585–9590. doi: 10.1073/pnas.132266499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel E, Journet EP, de Carvalho-Niebel F, Duc G, Frugoli J. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol Biol. 2005;58:809–822. doi: 10.1007/s11103-005-8102-y. [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Schopfer CR, Nasrallah ME, Nasrallah JB. The male determinant of self-incompatibility in Brassica. Science. 1999;286:1697–1700. doi: 10.1126/science.286.5445.1697. [DOI] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science. 2003;299:109–112. doi: 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]
- Seifert GJ, Blaukopf C. Irritable walls: the plant extracellular matrix and signaling. Plant Physiol. 2010;153:467–478. doi: 10.1104/pp.110.153940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Plant receptor-like kinase gene family: diversity, function, and signaling. Sci STKE. 2001a;2001:RE22. doi: 10.1126/stke.2001.113.re22. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci U S A. 2001b;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132:530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM. Structure and function of plant cell wall proteins. Plant Cell. 1993;5:9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, Berthiaume CT, Hill EJ, Torii KU. Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development. 2004;131:1491–1501. doi: 10.1242/dev.01028. [DOI] [PubMed] [Google Scholar]
- Sivaguru M, Ezaki B, He ZH, Tong H, Osawa H, Baluska F, Volkmann D, Matsumoto H. Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol. 2003;132:2256–2266. doi: 10.1104/pp.103.022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SK, Clark SE. POL and related phosphatases are dosage-sensitive regulators of meristem and organ development in Arabidopsis. Dev Biol. 2005;285:272–284. doi: 10.1016/j.ydbio.2005.06.020. [DOI] [PubMed] [Google Scholar]
- Song SK, Lee MM, Clark SE. POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells. Development. 2006;133:4691–4698. doi: 10.1242/dev.02652. [DOI] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Stein JC, Howlett B, Boyes DC, Nasrallah ME, Nasrallah JB. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl. Acad. Sci. 1991;88:8816–8820. doi: 10.1073/pnas.88.19.8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T, Ohneda M, Toriba T, Yoshida A, Hirano HY. FON2 SPARE1 redundantly regulates floral meristem maintenance with FLORAL ORGAN NUMBER2 in rice. PLoS Genet. 2009;5:e1000693. doi: 10.1371/journal.pgen.1000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T, Sato M, Ashikari M, Miyoshi M, Nagato Y, Hirano HY. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development. 2004;131:5649–5657. doi: 10.1242/dev.01441. [DOI] [PubMed] [Google Scholar]
- Suzaki T, Toriba T, Fujimoto M, Tsutsumi N, Kitano H, Hirano HY. Conservation and diversification of meristem maintenance mechanism in Oryza sativa: Function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 2006;47:1591–1602. doi: 10.1093/pcp/pcl025. [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D. The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev. 2001;15:2755–2766. doi: 10.1101/gad.208501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki T, Hatakeyama K, Suzuki G, Watanabe M, Isogai A, Hinata K. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature. 2000;403:913–916. doi: 10.1038/35002628. [DOI] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordai H, Banyai L, Patthy L. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999;461:63–67. doi: 10.1016/s0014-5793(99)01416-7. [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. The Arabidopsis ERECTA Gene Encodes a Putative Receptor Protein Kinase with Extracellular Leucine-Rich Repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner TA, Kohorn BD. Wall-Associated Kinases Are Expressed throughout Plant Development and Are Required for Cell Expansion. Plant Cell. 2001;13:303–318. doi: 10.1105/tpc.13.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell. 2005a;17:1685–1703. doi: 10.1105/tpc.105.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Wang X, Li X, Meisenhelder J, Hunter T, Yoshida S, Asami T, Chory J. Autoregulation and Homodimerization Are Involved in the Activation of the Plant Steroid Receptor BRI1. Dev Cell. 2005b;8:855–865. doi: 10.1016/j.devcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Wang XQ, Zafian P, Choudhuary M, Lawton M. The PRK5 receptor protein kinase from Arabidopsis thaliana is structurally related to a family of plant defense proteins. Proc. Natl. Acad. Sci. USA. 1996;93:2598–2602. doi: 10.1073/pnas.93.6.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AN, Tauszig-Delamasure S, Hoffmann JA, Lelievre E, Gascan H, Ray KP, Morse MA, Imler JL, Gay NJ. Binding of the Drosophila cytokine Spatzle to Toll is direct and establishes signaling. Nat Immunol. 2003;4:794–800. doi: 10.1038/ni955. [DOI] [PubMed] [Google Scholar]
- Williams RW, Wilson JM, Meyerowitz EM. A possible role for kinase-associated protein phosphatase in the arabidopsis CLAVATA1 signaling pathway. Proc. Natl. Acad. Sci. USA. 1997;94:10467–10472. doi: 10.1073/pnas.94.19.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M, Brosche M, Salojarvi J, Kangasjarvi S, Idanheimo N, Mersmann S, Robatzek S, Karpinski S, Karpinska B, Kangasjarvi J. Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 2010;10:95. doi: 10.1186/1471-2229-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SL, Rahman A, Baskin TI, Kieber JJ. Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell. 2008;20:3065–3079. doi: 10.1105/tpc.108.063354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 2000;12:1591–1606. doi: 10.1105/tpc.12.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Yu LP, Miller AK, Clark SE. POLTERGEIST Encodes a Protein Phosphatase 2C that Regulates CLAVATA Pathways Controlling Stem Cell Identity at Arabidopsis Shoot and Flower Meristems. Curr Biol. 2003;13:179–188. doi: 10.1016/s0960-9822(03)00042-3. [DOI] [PubMed] [Google Scholar]
- Zhang XC, Cannon SB, Stacey G. Evolutionary genomics of LysM genes in land plants. BMC Evol Biol. 2009;9:183. doi: 10.1186/1471-2148-9-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XC, Wu X, Findley S, Wan J, Libault M, Nguyen HT, Cannon SB, Stacey G. Molecular evolution of lysin motif-type receptor-like kinases in plants. Plant Physiol. 2007;144:623–636. doi: 10.1104/pp.107.097097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Peng P, Schmitz RJ, Decker AD, Tax FE, Li J. Two Putative BIN2 Substrates Are Nuclear Components of Brassinosteroid Signaling. Plant Physiol. 2002;130:1221–1229. doi: 10.1104/pp.102.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Wang H, Walker JC, Li J. BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 2004;40:399–409. doi: 10.1111/j.1365-313X.2004.02214.x. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Wang Y, Li R, Song X, Wang Q, Huang S, Jin JB, Liu CM, Lin J. Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J. 2010;61:223–233. doi: 10.1111/j.1365-313X.2009.04049.x. [DOI] [PubMed] [Google Scholar]


