Abstract
Background
Conflicting data have been reported on the association between Toll-like receptor 4 (TLR4) +896A/G and +1196C/T polymorphisms and the risk of asthma. Therefore, we conducted this meta-analysis to clarify the effect of TLR4 +896A/G and +1196C/T polymorphisms on the risk of asthma.
Material/Methods
An electronic literature search was performed using PubMed, Embase, Web of Science, Chinese National Knowledge Infrastructure (CNKI), and Wanfang Data to find relevant studies. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to assess the strength of the associations. All statistical analyses were conducted using STATA software version 12.0.
Results
A total of 14 studies with 2873 asthma cases and 3110 controls were included. The pooled results indicated a significant association between TLR4 +1196C/T polymorphism and the risk of asthma (T vs. C: OR=0.79, 95%CI=0.63–0.99, P=0.04; TT+CT vs. CC: OR=0.76, 95%CI=0.59–0.96, P=0.03; CT vs. CC: OR=0.74, 95%CI=0.58–0.95, P=0.02). In subgroup analysis by ethnicity, TLR4 +1196C/T polymorphism was significantly associated with asthma risk in Asians (T vs. C: OR=0.73, 95%CI=0.54–0.98, P=0.04; TT+CT vs. CC: OR=0.70, 95%CI=0.51–0.96, P=0.03; CT vs. CC: OR=0.69, 95%CI=0.50–0.96, P=0.03), but not in whites. For TLR4 +896A/G polymorphism, no significant association was found between TLR4 +896A/G polymorphism and asthma risk under any genetic models.
Conclusions
The results of this meta-analysis suggest that T allele of the TLR4 +1196C/T might act as a protective factor against the development of asthma.
MeSH Keywords: Asthma; Meta-Analysis; Polymorphism, Genetic; Toll-Like Receptor 4
Background
Asthma is a chronic inflammatory airway disorder characterized by airway hyper-responsiveness, reversible airway obstruction, and airway remodeling [1,2]. It is commonly thought that asthma is a multi-factorial disease resulting from complex interactions between genetic predisposition and environmental factors [3]. Studies have shown that genetic influences on asthma are substantial, with heritability estimates ranging between 35% and 95% [4]. Notably, a large number of polymorphisms in genes positioned throughout the genome have been implicated in asthma causation, including the gene encoding Toll-like receptor 4 (TLR4) [5].
Toll-like receptors (TLRs) are a class of evolutionarily conserved membrane-bound pattern recognition receptors (PRRs) presented on the cell surface of innate immune cells. TLRs recognize pathogen-associated molecular patterns (PAMPs) exclusively expressed by microbial pathogens, and activate cellular signaling pathways to induce immune-response genes, including inflammatory cytokines [6]. Ten different human TLRs have been identified. TLR4, the best-studied TLR, is expressed on macrophages, dendritic cells, and other cell types, and mainly recognizes lipopolysaccharide (LPS) of gram-negative bacteria [7,8]. Interaction of LPS with TLR4 can activate the nuclear factor kappa B (NF-κB) signaling pathway, increase expression of inflammatory cytokines and disturb the Th1/Th2 balance in asthma [9]. TLR4 also regulates innate immune responses to respiratory syncytial virus infection, which is a risk factor for the development of asthma [10].
Human TLR4 gene is located on chromosome 9q32-q33 [11]. Two single-nucleotide polymorphisms (SNPs), TLR4 +896A/G (rs4986790, also known as Asp299Gly) and TLR4 +1196C/T (rs4986791, also known as Thr399Ile), have been demonstrated to modify the receptor’s response to endotoxin, which is an important trigger of asthma [12]. This genetically determined alteration in endotoxin responsiveness may be involved in the development of asthma. Several studies have evaluated the association of TLR4 +896A/G and +1196C/T polymorphisms with asthma [13–31]. However, due to the limitation of subjects, the results were inconsistent and controversial. A previous meta-analysis has shown a marginal association of TLR4 +896A/G with asthma, and no association between TLR4 +1196C/T polymorphism and asthma [32], but it did not cover all eligible studies. The exact correlation between TLR4 +896A/G and +1196C/T polymorphisms and asthma has not been entirely established. Therefore, we performed a meta-analysis including all eligible case-control studies to clarify and quantify the authentic effect of TLR4 +896A/G and +1196C/T polymorphisms on the risk of asthma.
Material and Methods
This study was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.
Literature search strategy
A literature search was performed using PubMed, Embase, Web of Science, Chinese National Knowledge Infrastructure (CNKI), and Wanfang Data from inception to July 2015. The search strings was: (“asthma” or “asthmatic”) and (“toll-like receptor 4” or “TLR4”) and (“mutation” or “polymorphism” or “variant”). The reference lists of the identified articles were also examined. No restrictions were applied for language, population, sample size, publication date, or type of report.
Inclusion and exclusion criteria
Studies were included in the meta-analysis that met the following criteria: 1) case-control study design, 2) evaluation of the association between the TLR4 +896A/G (Asp299Gly) and +1196C/T (Thr399Ile) polymorphisms and risk of asthma, 3) genotype frequency was available or sufficient data could be extracted to calculate odds ratios (ORs) and 95% confidence intervals (CIs), and 4) not animal studies. For overlapping studies, the most recent article or the one with the largest sample size was selected. Studies were excluded if they did not meet these inclusion criteria. Unpublished data were not considered.
Data extraction
The following data were extracted: year of publication, first author, country, ethnicity, age, atopic status, detection methods, sample size, and genotype frequencies in cases and controls. Two investigators independently extracted data. Disagreements were resolved by discussion and consensus.
Statistical analysis
All the statistical analyses were conducted using STATA software version 12.0 (STATA Corporation, College Station, TX, USA). We first evaluated Hardy-Weinberg equilibrium (HWE) in the control group for each study using the chi-square test, and it was considered statistically significant when P<0.05. A statistical test was performed based on the Q statistic to assess heterogeneity. The P>0.10 of the Q test indicated a lack of heterogeneity among studies. If heterogeneity was observed among studies, the random-effects model was used. Otherwise, the fixed-effects model was adopted. The strength of association between TLR4 +896A/G and +1196C/T polymorphisms and the risk of asthma was assessed by calculating ORs with 95% CIs. Stratified analysis was performed by ethnicity, if possible. Potential publication bias was assessed by Begg’s rank correlation test and Egger’s linear regression test, and P<0.05 was considered significant publication bias.
Results
Study characteristics
The study selection process was shown in Figure 1. In total, 72 English articles and 119 Chinese articles met search criteria after we initially searched PubMed, Embase, Web of Science, CNKI, and Wanfang Data. After reading titles and abstracts, 131 articles were excluded because they did not refer to TLR4 gene polymorphisms and asthma risk. The remaining 60 articles were identified for full-text view; 22 were excluded because they investigated other gene polymorphisms, 16 were excluded because they were reviews, 2 were not case-control studies, and 1 was repeated. A total of 19 relevant studies investigating the TLR4 +896A/G and +1196C/T polymorphisms and asthma for meta-analysis were identified. However, 5 studies were monomorphic at site and were excluded from this meta-analysis [15,21,22,29,30]. Finally, 14 case-control studies were included in the present meta-analysis. The characteristics of the selected studies are listed in Table 1, with a total of 13 studies of 2649 asthma cases and 2542 controls for investigating +896A/G polymorphism and 7 studies of 1443 asthma cases and 1693 controls for +1196C/T polymorphism. As for ethnicity, 11 studies investigated white populations [13,17,19,20,23–28,31], 2 studies investigated Asian populations [16,18], and 1 study investigated the U.S. population [14]. Deviation from HWE was detected in the controls of 2 eligible studies [13,16]. The genotype frequencies of TLR4 +896A/G and +1196C/T polymorphisms in each study are presented in Table 2.
Figure 1.
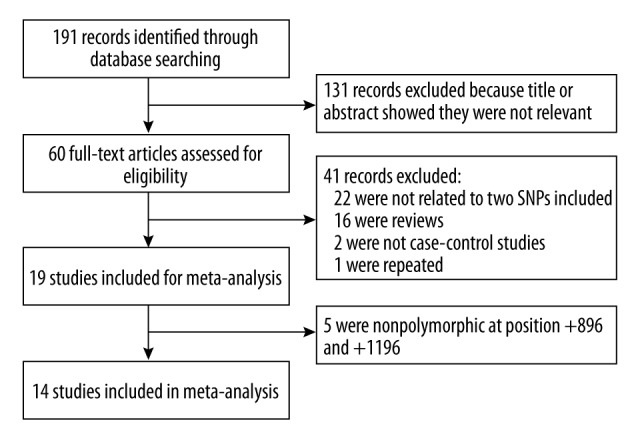
The process of study selection.
Table 1.
Characteristics of the studies included in the meta-analysis.
| Study | Year | Country | Ethnicity | Age | Atopic status | Cases | Controls | Genotyping method |
|---|---|---|---|---|---|---|---|---|
| +896A/G | ||||||||
| Yang | 2004 | UK | Caucasian | Adults | Mixed | 185 | 179 | ARMS-PCR |
| Adjers | 2005 | Finland | Caucasian | Adults | Mixed | 243 | 401 | TaqMan |
| Liu | 2005 | China | Asian | Mixed | Atopic | 197 | 156 | PCR-RFLP |
| Larocca | 2006 | Venezuela | American | Mixed | Mixed | 100 | 100 | PCR-RFLP |
| Smit | 2007 | Denmark | Caucasian | Adults | Mixed | 100 | 87 | TaqMan |
| Carvalho | 2008 | UK | Caucasian | Adults | Atopic | 14 | 80 | Bi-PASA |
| Lachheb | 2008 | Tunisia | Caucasian | Children | Mixed | 210 | 224 | PCR-RFLP |
| Voronko | 2011 | Russia | Caucasian | Mixed | Atopic | 283 | 227 | MALDI-TOF-MS |
| Zaborowski | 2011 | Poland | Caucasian | Adults | Mixed | 106 | 159 | PCR-RFLP |
| Hussein | 2012 | Egypt | Caucasian | Children | Mixed | 500 | 251 | ARMS-PCR |
| Sahin | 2014 | Turkey | Caucasian | Adults | Mixed | 131 | 75 | RT-PCR |
| Sinha | 2014 | India | Asian | Mixed | Mixed | 481 | 483 | PCR-RFLP |
| Bahrami | 2015 | Iran | Caucasian | Adults | Mixed | 99 | 120 | PCR-RFLP |
| +1196C/T | ||||||||
| Liu | 2005 | China | Asian | Mixed | Atopic | 197 | 156 | PCR-RFLP |
| Larocca | 2006 | Venezuela | American | Mixed | Mixed | 100 | 100 | PCR-RFLP |
| Smit | 2007 | Denmark | Caucasian | Adults | Mixed | 100 | 87 | PCR-SSP |
| Lachheb | 2008 | Tunisia | Caucasian | Children | Mixed | 210 | 224 | PCR-RFLP |
| Smit | 2009 | France | Caucasian | Adults | Mixed | 224 | 568 | Taqman and Illumina Golden Gate assays |
| Sahin | 2014 | Turkey | Caucasian | Adults | Mixed | 131 | 75 | RT-PCR |
| Sinha | 2014 | India | Asian | Mixed | Mixed | 481 | 483 | PCR-RFLP |
ARMS-PCR – amplification refractory mutation system-polymerase chain reaction; PCR-RFLP – polymerase chain reaction-restriction fragment length polymorphism; Bi-PASA – bidirectional polymerase chain reaction amplification of specific alleles; MALDI-TOF-MS – matrix-assisted laser desorption/ionization time of flight mass spectrometry; RT-PCR – real time polymerase chain reaction; PCR-SSP – polymerase chain reaction with sequence-specific primers.
Table 2.
Distribution of TLR4 +896A/G and +1196C/T polymorphisms among patients and controls.
| Study | Cases | Controls | HWE(P) | ||||
|---|---|---|---|---|---|---|---|
| +896A/G | AA | AG | GG | AA | AG | GG | |
| Yang | 155 | 30 | 0 | 159 | 19 | 1 | 0.603 |
| Adjers | 202 | 39 | 2 | 334 | 64 | 3 | 0.973 |
| Liu | 161 | 29 | 7 | 128 | 23 | 5 | 0.006 |
| Larocca | 91 | 9 | 0 | 92 | 8 | 0 | 0.677 |
| Smit | 91 | 8 | 1 | 78 | 9 | 0 | 0.611 |
| Carvalho | 12 | 2 | 0 | 70 | 10 | 0 | 0.551 |
| Lachheb | 209 | 1 | 0 | 223 | 0 | 1 | < 0.01 |
| Voronko | 245 | 31 | 7 | 200 | 27 | 0 | 0.341 |
| Zaborowski | 94 | 12 | 0 | 142 | 17 | 0 | 0.476 |
| Hussein | 434 | 64 | 2 | 223 | 27 | 1 | 0.851 |
| Sahin | 122 | 9 | 0 | 71 | 4 | 0 | 0.812 |
| Sinha | 390 | 87 | 4 | 381 | 95 | 7 | 0.699 |
| Bahrami | 85 | 14 | 0 | 104 | 16 | 0 | 0.434 |
| +1196C/T | CC | CT | TT | CC | CT | TT | |
| Liu | 191 | 4 | 2 | 150 | 5 | 1 | < 0.01 |
| Larocca | 95 | 4 | 1 | 94 | 6 | 0 | 0.757 |
| Smit | 93 | 6 | 1 | 77 | 10 | 0 | 0.570 |
| Lachheb | 209 | 0 | 1 | 221 | 2 | 1 | < 0.01 |
| Smit | 198 | 26 | 0 | 492 | 76 | 0 | 0.087 |
| Sahin | 120 | 11 | 0 | 71 | 4 | 0 | 0.812 |
| Sinha | 408 | 68 | 5 | 384 | 92 | 7 | 0.581 |
HWE – Hardy-Weinberg equilibrium.
Main results of the meta-analysis
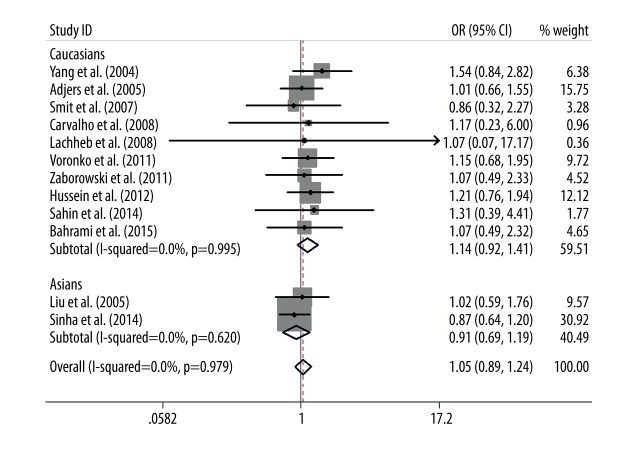
The main results of the relationship between TLR4 +896A/G and +1196C/T polymorphisms and asthma risk are listed in Table 3. Overall, no significant association between TLR4 +896A/G polymorphism and asthma risk was found in the allele model (G vs. A: OR=1.05, 95% CI=0.90–1.23, P=0.51), the dominant model (GG + AG vs. AA: OR=1.05, 95% CI=0.89–1.24, P=0.56), or the codominant models (AG vs. AA: OR=1.04, 95% CI=0.88–1.23, P=0.64). After categorizing subjects into different subgroups on the basis of ethnicity, the results remained non-significant (Figure 2). The recessive model (GG vs. AG+AA) and codominant model (GG vs. AA) were not performed due to the low frequency of the GG genotype in cases and controls.
Table 3.
Summary ORs and 95%CI of TLR4 +896A/G and +1196C/T polymorphisms and asthma risk.
| Subgroup | Genetic model | Genotype/Allele | Type of model | Heterogeneity | Test of association | |||
|---|---|---|---|---|---|---|---|---|
| I2 | P | OR | 95%CI | P | ||||
| +896A/G | ||||||||
| Overall | Dominant model | GG+AG vs. AA | Fixed | 0.0% | 0.99 | 1.05 | 0.89–1.24 | 0.56 |
| Codominant model | AG vs. AA | Fixed | 0.0% | 0.97 | 1.04 | 0.88–1.23 | 0.64 | |
| Allele model | G vs. A | Fixed | 0.0% | 0.97 | 1.05 | 0.90–1.23 | 0.51 | |
| Caucasians | Dominant model | GG+AG vs. AA | Fixed | 0.0% | 1.00 | 1.14 | 0.92–1.41 | 0.22 |
| Codominant model | AG vs. AA | Fixed | 0.0% | 0.96 | 1.12 | 0.90–1.39 | 0.32 | |
| Allele model | G vs. A | Fixed | 0.0% | 0.99 | 1.16 | 0.95–1.42 | 0.15 | |
| Asians | Dominant model | GG+AG vs. AA | Fixed | 0.0% | 0.62 | 0.91 | 0.69–1.19 | 0.48 |
| Codominant model | AG vs. AA | Fixed | 0.0% | 0.74 | 0.92 | 0.69–1.22 | 0.56 | |
| Allele model | G vs. A | Fixed | 0.0% | 0.52 | 0.91 | 0.71–1.16 | 0.43 | |
| +1196C/T | ||||||||
| Overall | Dominant model | TT+CT vs. CC | Fixed | 0.0% | 0.83 | 0.76 | 0.59–0.96 | 0.03 |
| Codominant model | CT vs. CC | Fixed | 0.0% | 0.75 | 0.74 | 0.58–0.95 | 0.02 | |
| Allele model | T vs. C | Fixed | 0.0% | 0.87 | 0.79 | 0.63–0.99 | 0.04 | |
| Caucasians | Dominant model | TT+CT vs. CC | Fixed | 0.0% | 0.52 | 0.84 | 0.57–1.24 | 0.39 |
| Codominant model | CT vs. CC | Fixed | 0.0% | 0.40 | 0.82 | 0.55–1.22 | 0.33 | |
| Allele model | T vs. C | Fixed | 0.0% | 0.65 | 0.87 | 0.60–1.26 | 0.46 | |
| Asians | Dominant model | TT+CT vs. CC | Fixed | 0.0% | 0.84 | 0.70 | 0.51–0.96 | 0.03 |
| Codominant model | CT vs. CC | Fixed | 0.0% | 0.89 | 0.69 | 0.50–0.96 | 0.03 | |
| Allele model | T vs. C | Fixed | 0.0% | 0.67 | 0.73 | 0.54–0.98 | 0.04 | |
Figure 2.
Forest plot of the association between TLR4 +896A/G polymorphism and asthma risk by ethnicity stratification under the dominant model (GG+AG vs. AA).
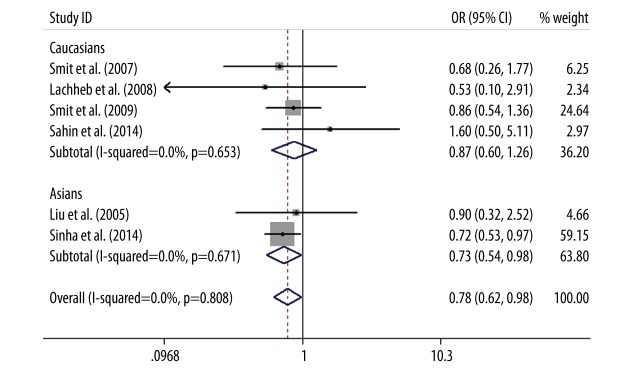
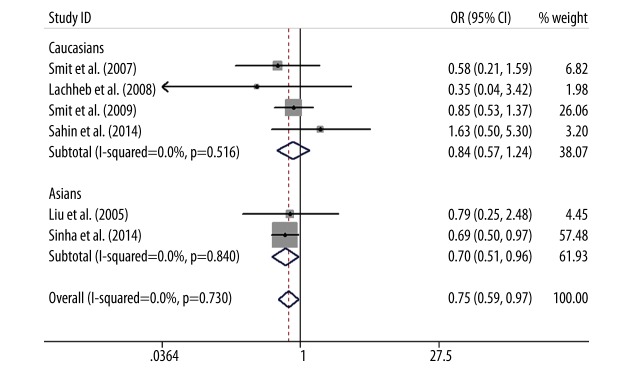
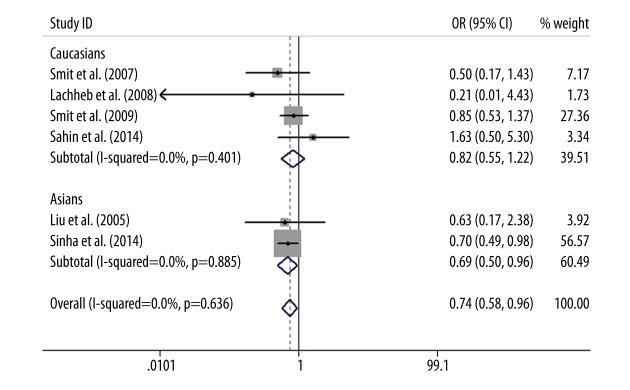
As for TLR4 +1196C/T polymorphism, a protective association was found between TLR4 +1196C/T polymorphism and asthma in the allele model (T vs. C: OR=0.79, 95%CI=0.63–0.99, P=0.04), the dominant model (TT+CT vs. CC: OR=0.76, 95%CI=0.59–0.96, P=0.03), and the codominant model (CT vs. CC: OR=0.74, 95%CI=0.58–0.95, P=0.02). This association was not examined via the recessive model (TT vs. CT+CC) or codominant model (TT vs. CC) due to the low frequency of the TT genotype in cases and controls. The subgroup analysis by ethnicity showed that the T allele of TLR4 +1196C/T polymorphism was a significant protective gene for the development of asthma in Asians (T vs. C: OR=0.73, 95%CI=0.54–0.98, P=0.04; TT+CT vs. CC: OR=0.70, 95%CI=0.51–0.96, P=0.03; CT vs. CC: OR=0.69, 95%CI=0.50–0.96, P=0.03), but there was no statistically significant difference in whites (T vs. C: OR=0.87, 95%CI=0.60–1.26, P=0.46; TT+CT vs. CC: OR=0.84, 95%CI=0.57–1.24, P=0.39; CT vs. CC: OR=0.82, 95%CI=0.55–1.22, P=0.33) (Figures 3–5).
Figure 3.
Forest plot of the TLR4 +1196C/T polymorphism associated with asthma risk by ethnicity stratification under the allele model (T vs. C).
Figure 4.
Forest plot of the TLR4 +1196C/T polymorphism associated with asthma risk by ethnicity stratification under the dominant model (TT+CT vs. CC).
Figure 5.
Forest plot of the TLR4 +1196C/T polymorphism associated with asthma risk by ethnicity stratification under the codominant model (CT vs. CC).
Test of heterogeneity, sensitivity analysis, and publication bias
There was no significant heterogeneity between any studies when analyzing the association of TLR4 +896A/G and +1196C/T polymorphisms and asthma risk in all genetic models, so we used fixed-effects models. Sensitivity analyses were conducted by altering the statistical models. No material alterations were detected, indicating that our results were statistically robust. Exclusion of the HWE-deviated studies did not meaningfully change the pooled estimates (data not shown). The funnel plot was used to evaluate publication bias, and there was no obvious asymmetry. Furthermore, no significant publication bias was detected by Begg’s test and Egger’s test (all P>0.05).
Discussion
Asthma is a chronic inflammatory airway disorder with complex etiologies involving both genetic and environmental contributions. Several candidate genes, such as TLR4, CD14, STAT6, ADAM33, and IL-13, have been reported to be associated with asthma susceptibility [18,33–36]. TLR4 is a principal receptor for LPS. Recognition of LPS by TLR4 plays a crucial role in the activation of subsequent immune and inflammatory responses against invaders. Numerous studies have indicated the role of TLR4 in the pathogenesis of asthma [37–40]. TLR4 is up-regulated in patients with asthma/allergic rhinitis [37]. Bortolatto et al. demonstrated that LPS impair the development of Th2 immunity, signaling via TLR4 and MyD88 molecules and via the IL-12/IFN-γ axis, and the synthetic TLR4 agonists protect against allergic asthma development [41]. Genetic polymorphisms of TLR4 gene have been demonstrated to be associated with diminished airway responsiveness to inhaled LPS [42], and to be closely involved in the susceptibility to many diseases, including asthma, juvenile spondyloarthritis, inflammatory bowel disease, and systemic lupus erythematosus [18,43–45].
Two co-segregating single-nucleotide polymorphisms, +896 A/G and +1196C/T, in human TLR4 gene that result in amino acid changes in the extracellular domain of the TLR4 protein have been widely studied. Environmental endotoxins are important triggers of asthma. These 2 variants have been reported to be associated with a blunted response to inhaled endotoxin on bronchial challenge testing and a reduced systemic inflammatory response to low-dose inhaled endotoxin [42,46]. Non-carriers of these polymorphisms have been found to be more frequently affected by asthma [12]. Several case-control studies have evaluated the association of TLR4 +896A/G and +1196C/T polymorphisms with asthma susceptibility. However, the results remain controversial. A previous meta-analysis has shown a marginal association of TLR4 +896A/G with asthma, and no association between TLR4 +1196C/T polymorphism and asthma [32]. However, the previous meta-analysis did not cover all eligible studies related to asthma. Therefore, to obtain a more precise conclusion we conducted this meta-analysis including all eligible case-control studies.
On the basis of 7 case-control studies including 1443 asthma cases and 1693 controls, the present meta-analysis found that TLR4 +1196C/T polymorphism might be a protective factor against the development of asthma. This result is different from the previous meta-analysis. The discrepancy stemmed from the fact that Tizaoui and coworkers only included 4 studies with 541 asthma cases and 486 controls. Next, we conducted the stratified analysis by ethnicity. A significant protective association between TLR4 +1196C/T polymorphism and asthma was detected in Asians. However, we found no significant relationship between TLR4 +1196C/T polymorphism and asthma in whites. These results suggest that the effect of TLR4 +1196C/T polymorphism on asthma risk might be influenced by ethnicity. More studies should be performed based on different ethnic groups.
For TLR4 +896A/G polymorphism, no significant correlation was observed between TLR4 +896A/G polymorphism and asthma risk. The results remained non-significant after subgroup analysis by ethnicity. Our results are consistent with previous meta-analyses [32,47,48]. Therefore, TLR4 +896A/G polymorphism seemed not to be associated with the risk of asthma development. However, a study conducted in Turkish children with asthma observed that both TLR4 +896A/G and +1196C/T polymorphisms were statistically more frequent in the mild asthma group [49]. A strong association between TLR4 +896A/G and asthma course has been found in a Russian study, which reported that the minor G allele was associated with moderate/severe asthma [26]. In addition, the G allele has been suggested to be significantly associated with moderate-severe asthma compared to mild asthma in an Egyptian population study [27]. These findings indicate that TLR4 +896A/G polymorphism might be associated with the severity of asthma, but not susceptibility to asthma.
Several limitations in this study should be addressed. Firstly, the number of studies and subjects included in the present meta-analysis were relatively small. Secondly, only published studies with sufficient data were included, so the possibility of publication bias cannot be completely ruled out, even though funnel plot and Egger’s test did not detect publication bias. Thirdly, the frequencies of GG genotype and TT genotype were low, which may undermine the findings. Moreover, the potential interactions between gene-gene and gene-environment during development of asthma were not conducted due to a lack of original data. Considering these limitations, the results of the meta-analysis should be interpreted with caution. Well-designed case-control studies with larger sample sizes and different population characteristics are needed to confirm these results.
Conclusions
In summary, this meta-analysis suggests that the T allele of the TLR4 +1196C/T might act as a protective factor against the development of asthma, but there was no significant association between TLR4 +896A/G polymorphism and risk of asthma. Larger well-designed studies based on different ethnic groups should be performed to confirm our findings.
Footnotes
Source of support: National Natural Science Foundation of China (Grant No. 81070045) and the Key Clinical Project for Affiliated Hospital of Ministry of Public Health of China (Grant No.111)
Conflicts of interest
None of the authors have any conflicts of interest to declare.
References
- 1.Nakagome K, Nagata M. Pathogenesis of airway inflammation in bronchial asthma. Auris Nasus Larynx. 2011;38:555–63. doi: 10.1016/j.anl.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Keglowich LF, Borger P. The three a’s in asthma – airway smooth muscle, airway remodeling & angiogenesis. Open Respir Med J. 2015;9:70–80. doi: 10.2174/1874306401509010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harb H, Renz H. Update on epigenetics in allergic disease. J Allergy Clin Immunol. 2015;135:15–24. doi: 10.1016/j.jaci.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshikawa T, Kanazawa H, Fujimoto S, Hirata K. Epistatic effects of multiple receptor genes on pathophysiology of asthma – its limits and potential for clinical application. Med Sci Monit. 2014;20:64–71. doi: 10.12659/MSM.889754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu MH, Zhang P, Huang X. Toll-like receptors in innate immunity and infectious diseases. Front Med China. 2010;4:385–93. doi: 10.1007/s11684-010-0600-x. [DOI] [PubMed] [Google Scholar]
- 7.Hemmi H, Akira S. TLR signalling and the function of dendritic cells. Chem Immunol Allergy. 2005;86:120–35. doi: 10.1159/000086657. [DOI] [PubMed] [Google Scholar]
- 8.Ferwerda B, McCall MB, Verheijen K, et al. Functional consequences of toll-like receptor 4 polymorphisms. Mol Med. 2008;14:346–52. doi: 10.2119/2007-00135.Ferwerda. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams LK, Ownby DR, Maliarik MJ, Johnson CC. The role of endotoxin and its receptors in allergic disease. Ann Allergy Asthma Immunol. 2005;94:323–32. doi: 10.1016/S1081-1206(10)60983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diez-Domingo J, Perez-Yarza EG, Melero JA, et al. Social, economic, and health impact of the respiratory syncytial virus: a systematic search. BMC Infect Dis. 2014;14:544. doi: 10.1186/s12879-014-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rock FL, Hardiman G, Timans JC, et al. A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci USA. 1998;95:588–93. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Werner M, Topp R, Wimmer K, et al. TLR4 gene variants modify endotoxin effects on asthma. J Allergy Clin Immunol. 2003;112:323–30. doi: 10.1067/mai.2003.1648. [DOI] [PubMed] [Google Scholar]
- 13.Lachheb J, Dhifallah IB, Chelbi H, et al. Toll-like receptors and CD14 genes polymorphisms and susceptibility to asthma in Tunisian children. Tissue Antigens. 2008;71:417–25. doi: 10.1111/j.1399-0039.2008.01011.x. [DOI] [PubMed] [Google Scholar]
- 14.Larocca N, DeSanctis J, Toro F, Moreno D. F.23. Polymorphisms of Toll-Like Receptor 2 and 4 Genes in Asthma and COPD. Clini Immunol. 2006;119(Suppl):S58–59. [Google Scholar]
- 15.Liang XH, Cheung W, Heng CK, Wang DY. Absence of the toll-like receptor 4 gene polymorphisms Asp299Gly and Thr399Ile in Singaporean Chinese. Ther Clin Risk Manag. 2005;1:243–46. [PMC free article] [PubMed] [Google Scholar]
- 16.Liu RM, Wu JM, Cui TP, Li YR. Asp299Gly and Thr399Ile polymorphisms in the toll-like receptor 4 and the susceptivity of allergic asthma and IgE level. Chin J Microbiol Immunol. 2005;25:94–97. [in Chinese] [Google Scholar]
- 17.Sahin F, Yildiz P, Kuskucu A, et al. The effect of CD14 and TLR4 gene polymorphisms on asthma phenotypes in adult Turkish asthma patients: a genetic study. BMC Pulm Med. 2014;14:20. doi: 10.1186/1471-2466-14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinha S, Singh J, Jindal SK, et al. Role of TLR4 C>1196T (Thr399Ile) and TLR4 A>896G (Asp299Gly) polymorphisms in a North Indian population with asthma: a case-control study. Int J Immunogenet. 2014;41:463–71. doi: 10.1111/iji.12115. [DOI] [PubMed] [Google Scholar]
- 19.Smit LA, Bongers SI, Ruven HJ, et al. Atopy and new-onset asthma in young Danish farmers and CD14, TLR2, and TLR4 genetic polymorphisms: a nested case-control study. Clin Exp Allergy. 2007;37:1602–8. doi: 10.1111/j.1365-2222.2007.02831.x. [DOI] [PubMed] [Google Scholar]
- 20.Smit LA, Siroux V, Bouzigon E, et al. CD14 and toll-like receptor gene polymorphisms, country living, and asthma in adults. Am J Respir Crit Care Med. 2009;179:363–68. doi: 10.1164/rccm.200810-1533OC. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Simayi M, Wushouer Q, et al. Association between polymorphisms in ADAM33, CD14, and TLR4 with asthma in the Uygur population in China. Genet Mol Res. 2014;13:4680–90. doi: 10.4238/2014.June.18.11. [DOI] [PubMed] [Google Scholar]
- 22.Zhang HL, Chen ZM, Ni LY, et al. Analysis of the single nucleotide polymorphisms (SNPs) of the human Toll-like receptor 4 (TLR4) gene and CD14 gene in Chinese Han children in Wenzhou. Chin J Pract Pediatr. 2007;22:591–94. [in Chinese] [Google Scholar]
- 23.Yang IA, Barton SJ, Rorke S, et al. Toll-like receptor 4 polymorphism and severity of atopy in asthmatics. Genes Immun. 2004;5:41–45. doi: 10.1038/sj.gene.6364037. [DOI] [PubMed] [Google Scholar]
- 24.Adjers K, Karjalainen J, Pessi T, et al. Epistatic effect of TLR4 and IL4 genes on the risk of asthma in females. Int Arch Allergy Immunol. 2005;138:251–56. doi: 10.1159/000088726. [DOI] [PubMed] [Google Scholar]
- 25.Carvalho A, Pasqualotto AC, Pitzurra L, et al. Polymorphisms in toll-like receptor genes and susceptibility to pulmonary aspergillosis. J Infect Dis. 2008;197:618–21. doi: 10.1086/526500. [DOI] [PubMed] [Google Scholar]
- 26.Voron’ko OE, Dmitrieva-Zdorova EV, Latysheva EA, et al. CARD15 and TLR4 genes polymorphisms in atopic bronchial asthma. Mol Biol (Mosk) 2011;45:831–39. [in Russian] [PubMed] [Google Scholar]
- 27.Hussein YM, Awad HA, Shalaby SM, et al. Toll-like receptor 2 and Toll-like receptor 4 polymorphisms and susceptibility to asthma and allergic rhinitis: a case-control analysis. Cell Immunol. 2012;274:34–38. doi: 10.1016/j.cellimm.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Bahrami H, Daneshmandi S, Heidarnazhad H, Pourfathollah AA. Lack of Association between Toll Like Receptor-2 and Toll Like Receptor-4 Gene Polymorphisms and Other Feature in Iranian Asthmatics Patients. Iran J Allergy Asthma Immunol. 2015;14:48–54. [PubMed] [Google Scholar]
- 29.Noguchi E, Nishimura F, Fukai H, et al. An association study of asthma and total serum immunoglobin E levels for Toll-like receptor polymorphisms in a Japanese population. Clin Exp Allergy. 2004;34:177–83. doi: 10.1111/j.1365-2222.2004.01839.x. [DOI] [PubMed] [Google Scholar]
- 30.Sun YF, Chang XY, Sun WJ. Correlation study of gene polymorphism of TNF-α and TLR4 to bronchial asthma of Han nationality in Neimenggu area. J Clini Pulm Med. 2013;18:662–25. [in Chinese[ [Google Scholar]
- 31.Zaborowski T, Wojas-Krawczyk K, Krawczyk P, et al. The effect of CD14 and TLR4 gene polymorphisms on the occurrence of atopic and non-atopic asthma. Adv Clin Exp Med. 2011;20:413–21. [Google Scholar]
- 32.Tizaoui K, Kaabachi W, Hamzaoui K, Hamzaoui A. Association of single nucleotide polymorphisms in toll-like receptor genes with asthma risk: a systematic review and meta-analysis. Allergy Asthma Immunol Res. 2015;7:130–40. doi: 10.4168/aair.2015.7.2.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang YN, Li YJ, Li H, et al. Association of CD14 C159T polymorphism with atopic asthma susceptibility in children from Southeastern China: a case-control study. Genet Mol Res. 2015;14:4311–17. doi: 10.4238/2015.April.30.3. [DOI] [PubMed] [Google Scholar]
- 34.Al-Muhsen S, Vazquez-Tello A, Jamhawi A, et al. Association of the STAT-6 rs324011 (C2892T) variant but not rs324015 (G2964A), with atopic asthma in a Saudi Arabian population. Hum immunol. 2014;75:791–95. doi: 10.1016/j.humimm.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 35.Zihlif M, Zihlif N, Obeidat NM, et al. Association between ADAM33 polymorphisms and susceptibility with adult and childhood asthma among Jordanians. Genet Test Mol Biomarkers. 2014;18:767–74. doi: 10.1089/gtmb.2014.0190. [DOI] [PubMed] [Google Scholar]
- 36.Shazia M, Kanza M, Mehwish I, et al. IL-13 gene polymorphisms and their association with atopic asthma and rhinitis in Pakistani patients. Iran J Allergy Asthma Immunol. 2013;12:391–96. [PubMed] [Google Scholar]
- 37.Aryan Z, Holgate ST, Radzioch D, Rezaei N. A new era of targeting the ancient gatekeepers of the immune system: toll-like agonists in the treatment of allergic rhinitis and asthma. Int Arch Allergy Immunol. 2014;164:46–63. doi: 10.1159/000362553. [DOI] [PubMed] [Google Scholar]
- 38.Liu CF, Drocourt D, Puzo G, et al. Innate immune response of alveolar macrophage to house dust mite allergen is mediated through TLR2/-4 co-activation. PLoS One. 2013;8:e75983. doi: 10.1371/journal.pone.0075983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vawda S, Mansour R, Takeda A, et al. Associations between inflammatory and immune response genes and adverse respiratory outcomes following exposure to outdoor air pollution: a HuGE systematic review. Am J Epidemiol. 2014;179:432–42. doi: 10.1093/aje/kwt269. [DOI] [PubMed] [Google Scholar]
- 40.Lee SH, Kang MJ, Yu HS, et al. Association between recent acetaminophen use and asthma: modification by polymorphism at TLR4. J Korean Med Sci. 2014;29:662–68. doi: 10.3346/jkms.2014.29.5.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bortolatto J, Borducchi E, Rodriguez D, et al. Toll-like receptor 4 agonists adsorbed to aluminium hydroxide adjuvant attenuate ovalbumin-specific allergic airway disease: role of MyD88 adaptor molecule and interleukin-12/interferon-gamma axis. Clin Exp Allergy. 2008;38:1668–79. doi: 10.1111/j.1365-2222.2008.03036.x. [DOI] [PubMed] [Google Scholar]
- 42.Arbour NC, Lorenz E, Schutte BC, et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25:187–91. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 43.Marija P, Mandica V, Lovro L, et al. Single nucleotide polymorphism of toll-like receptor 4 (TLR4) is associated with juvenile spondyloarthritis in Croatian population. Clin Rheumatol. 2015 doi: 10.1007/s10067-015-2952-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 44.Manolakis AC, Kapsoritakis AN, Kapsoritaki A, et al. Readressing the role of Toll-like receptor-4 alleles in inflammatory bowel disease: colitis, smoking, and seroreactivity. Dig Dis Sci. 2013;58:371–80. doi: 10.1007/s10620-012-2348-4. [DOI] [PubMed] [Google Scholar]
- 45.Rupasree Y, Naushad SM, Rajasekhar L, et al. Association of TLR4 (D299G, T399I), TLR9–1486T>C, TIRAP S180L and TNF-alpha promoter (–1031, –863, –857) polymorphisms with risk for systemic lupus erythematosus among South Indians. Lupus. 2015;24:50–57. doi: 10.1177/0961203314549792. [DOI] [PubMed] [Google Scholar]
- 46.Michel O, LeVan TD, Stern D, et al. Systemic responsiveness to lipopolysaccharide and polymorphisms in the toll-like receptor 4 gene in human beings. J Allergy Clin Immunol. 2003;112:923–29. doi: 10.1016/j.jaci.2003.05.001. [DOI] [PubMed] [Google Scholar]
- 47.Chen S. Association between the TLR4 +896A>G (Asp299Gly) polymorphism and asthma: a systematic review and meta-analysis. J Asthma. 2012;49:999–1003. doi: 10.3109/02770903.2012.738270. [DOI] [PubMed] [Google Scholar]
- 48.Yao Y, Ren X, He L, et al. TLR4 +896A>G (Asp299Gly) polymorphism is not associated with asthma: a update meta-analysis. Int J Clin Exp Med. 2014;7:5358–61. [PMC free article] [PubMed] [Google Scholar]
- 49.Sackesen C, Karaaslan C, Keskin O, et al. The effect of polymorphisms at the CD14 promoter and the TLR4 gene on asthma phenotypes in Turkish children with asthma. Allergy. 2005;60:1485–92. doi: 10.1111/j.1398-9995.2005.00874.x. [DOI] [PubMed] [Google Scholar]