Abstract
Lymph node (LN) metastasis is often an early event in the progression of malignant tumors and it contributes to the majority of cancer mortalities. MiRNAs play key roles in tumor metastasis. This study aimed to investigate the specific miRNAs as putative indicators of metastasis early diagnosis for non-small cell lung cancer (NSCLC). In this study, five NSCLC cases with LN metastasis and four cases without metastasis (NLN) were enrolled for Agilent Human miRNA array. The interested differentially expressed miRNA was validated by quantitative reverse transcription polymerase chain reaction (qRT-PCR) in the LN metastasis (n = 46) and NLN (n = 39) groups. The microarray results revealed that three miRNAs (miR-1260b, miR-423-3p, miR-23a-5p) were differentially expressed in LN metastasis group compared with NLN group. The expression of miR-1260b was tested by qRT-PCR and the mean relative expression fold change (2-ΔΔCt) in LN metastasis was significantly higher than that in the NLN group (3.942, 1.743 respectively, P = 1.179E-04). The patients with tumor-node-metastasis (TNM) stage III were identified more frequently in LN metastasis group (P = 1.772E-11) and with a higher expression level of miR-1260b (5.126, P = 1.147E-06). In addition, the LN metastasis cases were associated with a poorly differentiated degree (P = 0.007). The overexpression of miR-1260b in NSCLC with LN metastasis can be regarded as a specific signature for early progression and prognosis of NSCLC.
Keywords: non-small cell lung cancer, lymph node metastasis, miR-1260b
Lung cancer is currently the leading cause of death worldwide and 80% of patients are diagnosed as non-small cell lung cancer (NSCLC) [1,2]. The poor prognosis with a 5-year survival rate of 16% is due not only to the late stage diagnosis but also to the lack of effective treatments [3,4]. Metastasis and invasion are the main characteristics during the progression of malignant tumors, which contributes to the majority of cancer mortalities [5]. Lymph node (LN) metastasis is often an early event in epithelial tumor progression and mainly responsible for tumor recurrence [5].
MicroRNAs (miRNAs) are a large family of post-transcriptional regulators of gene expression with a length of 19-22 nucleotides [6,7]. Numerous reports cited that miRNAs play crucial functions in multiple cellular processes, such as cellular proliferation, apoptosis, differentiation and metabolism [7]. Human microRNA genes are frequently located at cancer-associated genomic regions or in fragile sites [8]. In recent studies, several miRNAs such as miR-145, miR-186 and the let-7 family were considered suppressors for the metastasis and epithelial mesenchymal transition (EMT) in lung cancer [2, 9, 10] while miR-10b and miR-155 were positively correlated with the NSCLC tumorous process [11, 12]. Therefore, there is a need to investigate the specific miRNAs as putative indicators for NSCLC metastasis more comprehensively and provide a sensitive method for the early diagnosis of NSCLC. The focus of this study is to identify differentially expressed miRNAs in NSCLC with LN metastasis through miRNA array and quantitative reverse transcription polymerase chain reaction (qRT-PCR).
METHODS AND MATERIALS
Patients and Tissue Specimens
Fresh frozen tissues obtained from five NSCLC cases with LN metastasis, four cases without LN (NLN) metastasis, and their paired para-carcinoma tissues were used for miRNA array analysis. For further validation, an additional 85 cases of NSCLC patients were enrolled (46 LN metastasis patients and 39 NLN paired with their para-carcinoma tissues). All patients were enrolled from the time period of Sep 2010 to Dec 2014 at Zhejiang University, Huzhou Centre Hospital (Huzhou, China) and Zhejiang Cancer Hospital (Hangzhou, China). The tissues were graded using the 7th International Union Against Cancer tumor, lymph node, and metastasis staging system [13]. All tissue specimens were surgically obtained from the diagnosed lung cancer patients, instantly frozen in liquid nitrogen, and stored at -80 °C until RNA extraction. None of these patients had received radiotherapy or chemotherapy before the operation. This study was approved by the Ethics Committee of Huzhou Centre Hospital and Zhejiang Cancer Hospital, and all participants provided informed consent.
miRNA array
The total RNA was extracted using mirVana™ miRNA Isolation Kit (Cat # AM1560, Ambion, Austin, TX, US) following the manufacturer’s instructions and the RNA quality and quantity were measured by Agilent Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA, US). The Agilent Human miRNA array (8*60K, V16.0, Agilent Technologies) [14,15] that contains more than 1205 probes was used to explore the aberrantly expressed mature miRNAs in NSCLC with LN metastasis. The miRNA in total RNA was labeled by Cy-3 using miRNA Complete Labeling and Hyb Kit (Cat # 5190-0456, Agilent technologies, Santa Clara, CA, US). The labeled Cy-3 RNA were then hybridized with a slide containing capture probes in the hybridization oven (Cat # G2545A, Agilent technologies, Santa Clara, CA, US) at 55°C, 20 rpm for 20 hr. After hybridization, the slides were washed with Gene Expression Wash Buffer Kit (Cat #5188-5327, Agilent technologies, Santa Clara, CA, US) in staining dishes (Cat # 121, Thermo Shandon, Waltham, MA, US). The sign collection and analysis were performed by Agilent Microarray Scanner (Cat # G2565CA, Agilent technologies, Santa Clara, CA, US) and Feature Extraction software 10.7 (Agilent technologies, Santa Clara, CA, US). The obtained raw data were normalized by Quantile algorithm with Gene Spring Software 12.6 (Agilent technologies, Santa Clara, CA, US).
Total RNA isolation
Total RNAs were extracted from NSCLC tissues and para-carcinoma tissues by Trizol (Invitrogen, USA) according to the manufacturer’s protocol. RNA quality and quantity were measured by Bio-Rad SmartSpec Plus Spectrophotometer (Bio-Rad, USA). The RNA integrity and purity were analyzed by gel electrophoresis and the ratio of the absorbances at 260 nm and 280 nm (A260/A280≥1.8).
Validation analysis by qRT-PCR
2 μg of total RNA was subjected to reverse transcription (RT) with All-in-One™ miRNA qRT-PCR Detection Kit (No. AOMD-Q050, GeneCopoeia, USA). The prepared RT mixture was incubated at 37 °C for 60 min then inactivated at 85 °C for 5 min. The synthesized cDNA was amplified using the All-in-One™ qPCR Mix (No. AOPR-0200, GeneCopoeia, USA) on the 7500 real-time PCR System (Applied Biosystems, USA). PCR reactions were prepared following the manufacturer’s instructions. The quantitative PCR cycling profile was as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 20 s, and primer extension at 72 °C for 30 s, then a melting curve (90 °C for 10 s, 60 °C for 30 s, and 90 °C 10 s). For normalization, U48 rather than U6 was used as an internal reference gene according to our preliminary experiments that benefited from its stability and sensitivity [16]. The primers used for amplification were supplied by All-in-One™ qPCR Primer (GeneCopoeia, USA). Water was used as a negative and quality control, and each sample was measured in duplicates.
Data analysis
The miRNA array data were analyzed in two steps. Firstly, differentially expressed miRNAs were obtained by comparing cancer and matched para-carcinoma tissues in LN metastasis and NLN groups respectively with paired-samples t test. The selected miRNAs were pooled together and analyzed with independent samples t test if they were differentially expressed between LN metastasis and NLN. The obtained differentially expressed miRNAs were validated by qPCR in additional NSCLC patients (46 LN metastasis and 39 NLN). The fold changes of expression between cancer and para-carcinoma tissues were determined by the comparative Ct method (2-ΔΔCt). Comparison of fold change values between the two groups was performed with the independent samples t test. Clinical characteristics were assessed by the chi-square test and Student’s t test with SPSS 16.0 (SPSS Inc., USA). A P value < 0.05 was considered to be statistically significant.
RESULTS
Differentially expressed miRNAs in NSCLC with LN metastasis
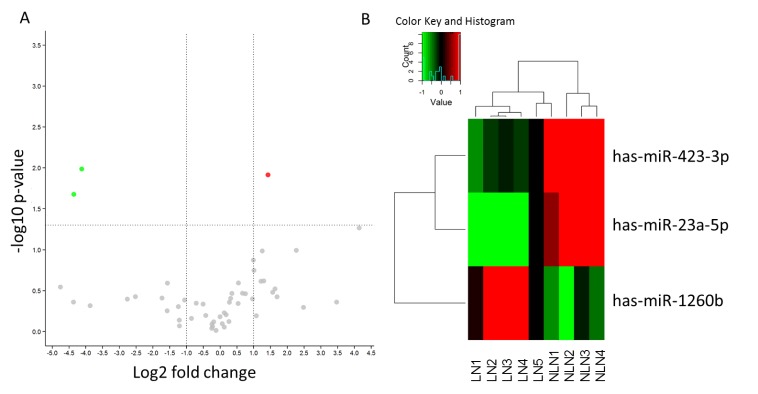
MiRNA microarray analysis was performed to evaluate the miRNA expression profiles between LN metastasis and NLN groups in NSCLC. We set the cut-off level as fold change > 2 and P value < 0.05. The original intensities data were normalized using the Quantile method. We found 39 and 18 differentially expressed miRNAs in the LN and NLN groups respectively by comparing NSCLC and their matched para-carcinoma tissues. Six miRNAs were found in both the two groups. After removing the reduplicative miRNAs, we analyzed the 51-pooled miRNAs for the comparison of the LN metastasis and NLN groups, and finally found 3 differentially expressed miRNAs (Fig. 1). Among these altered miRNAs, miR-1260b was overexpressed while miR-23a-5p and miR-423-3p were underexpressed in the LN metastasis group (Table 1). Gene Ontology (GO) analysis was applied to explore the significant biological processes related to cancer development, proliferation, and metastasis. As shown in Table 2, the analyses of cancer related pathways and the predicted targets of miRNAs revealed that miR-1260b showed a deepest connection with cancer development and metastasis.
Figure 1.
Change of miRNAs in the LN metastasis group. (A) Volcano plot shows the fold change of miRNAs in the LN metastasis group compared to NLN group. The vertical lines correspond to 2.0-fold up and down respectively, and the horizontal line represents a P value of 0.05. The colored points in the plot represent the differentially expressed miRNAs with statistical significance. (B) Hierarchical clustering for differentially expressed miRNAs in the LN metastasis group versus NLN group using a volcano plot (fold change ≥ 0.5). Red indicates high relative expression and green indicates low relative expression.
Table 1.
Expression level of miRNAs in LN metastasis and NLN groups
| MiRNA | Fold -change (LN/NLN) | P value |
|---|---|---|
| Hsa-miR-1260b | 2.684 | 0.012 |
| Hsa-miR-23a-5p | 0.049 | 0.021 |
| Hsa-miR-423-3p | 0.057 | 0.010 |
LN: lymph node metastasis; NLN: lymph node free
Table 2.
Predicated gene targets of the differentially expressed miRNAs with Gene Ontology (GO) analysis
| miRNA | Pathway ID | KEGG name | Gene count | Gene list |
|---|---|---|---|---|
| miR-1260b | ||||
| hsa04310 | Wnt signaling pathway | 8 | GPC4,CCND2,CSNK2B,NKD1,FZD5,SOST,TCF7,SFRP1 | |
| hsa04350 | TGF-beta signaling pathway | 3 | SP1,INHBB,ZFYVE9 | |
| hsa05223 | Non-small cell lung cancer | 2 | SOS1,RXRA | |
| hsa04115 | p53 signaling pathway | 2 | CCND2,CHEK1 | |
| hsa04514 | Cell adhesion molecules (CAMs) | 1 | NRXN2 | |
| hsa04010 | MAPK signaling pathway | 7 | SOS1,MKNK2,DUSP3,ELK1,TAOK2,DUSP16,ELK4 | |
| hsa05200 | Pathways in cancer | 8 | SOS1,RXRA,FLT3,FZD5,TCF7,TRAF4,ABL1,ETS1 | |
| hsa04668 | TNF signaling pathway | 2 | ATF6B,NOD2 | |
| miR-423-3p | ||||
| hsa04010 | MAPK signaling pathway | 1 | FGFR2 | |
| hsa05200 | Pathways in cancer | 1 | FGFR2 | |
| miR-23a-5p | ||||
| hsa04514 | Cell adhesion molecules (CAMs) | 3 | CNTN1,PDCD1,JAM3 | |
| hsa04668 | TNF signaling pathway | 1 | CSF1 | |
| hsa04310 | Wnt signaling pathway | 1 | NKD1 | |
| hsa05200 | Pathways in cancer | 1 | ARNT | |
| hsa04010 | MAPK signaling pathway | 1 | STK3 | |
Clinical characteristics
A total of 85 NSCLC patients were recruited in a validation study with their clinical characteristics presented in Table 3. Tumor types can be classified as the two most common NSCLC type (adenocarcinoma and squamous carcinoma) and other types (like sarcomatoid carcinoma, large cell carcinoma, adenosquamous carcinoma, etc.) [17]. The analyses showed that there was no significant difference between LN metastasis and NLN groups with respect to age, gender, and tumor type (Table 3). Nevertheless, the LN metastasis cases were accompanied by a higher TNM stage and poorly differentiated degree (Table 3; P = 1.772E-11, 0.007, respectively).
Table 3.
Clinical characteristics of NSCLC patients in the validation cohort (n = 44)
| LN (n = 46) | NLN (n = 39) | P | |
|---|---|---|---|
| Age | 62.826 ± 7.159 | 62.231 ± 7.478 | 0.709 |
|
| |||
| Gender | 0.417 | ||
|
| |||
| Male | 28 | 27 | |
|
| |||
| Female | 18 | 12 | |
|
| |||
| TNM stage | 1.772E-11 | ||
|
| |||
| I | 0 | 17 | |
|
| |||
| II | 18 | 20 | |
|
| |||
| III | 28 | 2 | |
|
| |||
| Tumor type | 0.481 | ||
|
| |||
| Adenocarcinoma | 31 | 22 | |
|
| |||
| Squamous carcinoma | 11 | 14 | |
|
| |||
| Other type of NSCLC | 4 | 3 | |
|
| |||
| Differentiation | 0.007 | ||
|
| |||
| Poorly | 12 | 7 | |
|
| |||
| Moderately and poorly | 21 | 8 | |
|
| |||
| Moderately | 13 | 24 | |
|
| |||
NSCLC: non-small cell lung cancer; TNM: tumor-node-metastasis; LN: lymph node metastasis; NLN: lymph node free
Validation of the miRNA-1260b by qRT-PCR
To validate the results from the miRNA array, miR-1260b was assayed by qRT-PCR in the validation cohort. The selection of this miRNA was according to the overexpression in the miRNA array and the potential function analyzed by GO. The result obtained by qRT-PCR showed that the mean relative expression fold change (2-ΔΔCt) in the LN metastasis group (3.942) was significantly higher than that in the NLN group (1.743), which was in agreement with the data from microarray (Fig. 2, P = 1.179E-04). The value of fold-change between the LN metastasis and NLN groups was 2.262. These results suggest that overexpression of miR-1260b is associated with a higher risk of LN metastasis. Furthermore, we also discovered the association of miR-1260b expression with different clinical stages (P = 1.147E-06). Patients with TNM stage III had a higher relative expression fold change than patients in TNM stage I and II (Fig. 3, I:1.145; II: 2.159; III: 5.126; I vs. III: P = 1.347E-06; II vs. III: 1.662E-05, respectively). No significant difference was observed between patients in TNM stagesIandII (P = 0.159).
Figure 2.
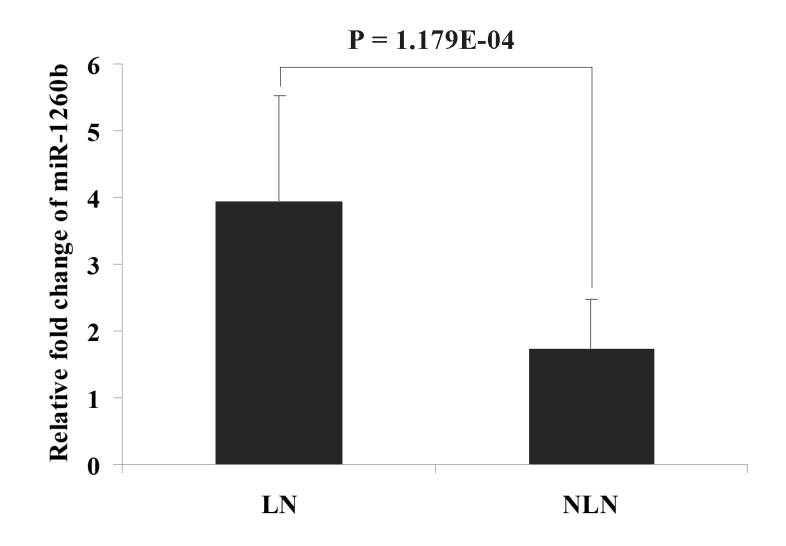
The miRNA expression level of has-miR-1260b detected by qRT-PCR in the LN metastasis and NLN groups. Error bars in the figure represent Mean ± SD. LN: lymph node metastasis; NLN: lymph node free.
Figure 3.
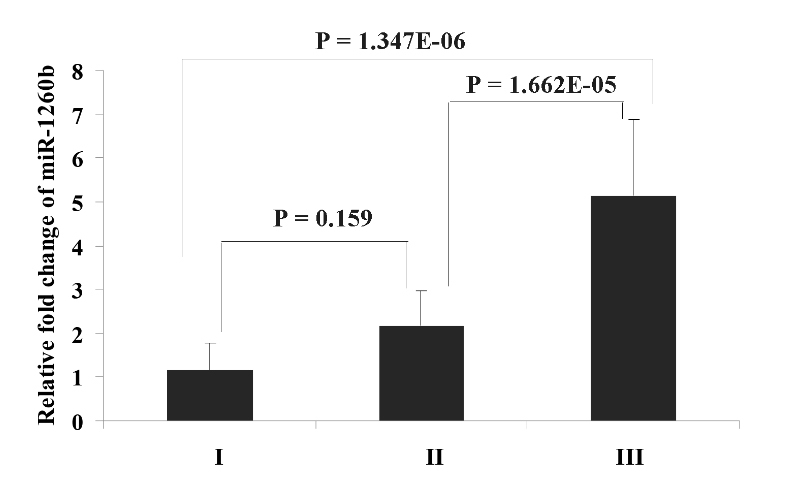
The miRNA expression level of has-miR-1260b in different TNM stages. Error bars in the figure represent Mean ± SD.I: TNM stage I; II: TNM stage II; III: TNM stage III.
DISCUSSION
Non-small cell lung cancer is regarded as a heterogeneous disease with multiple histopathological and clinical features. Patients with metastasis that frequently occurs in NSCLC often have poor prognosis and high mortality. Previous high-throughput array analyses have demonstrated that the expression of miRNAs present substantial variances in tumors [18, 19]. Overexpression of miRNAs may contribute to its oncogenesis by downregulating tumor suppressors, whereas underexpression of miRNAs may negatively regulate oncogenes or factors related to tumorigenesis and progression [20]. Thus, we explore comprehensive miRNAs profile in NSCLC with LN metastasis.
In this study, we discovered three abnormally expressed miRNAs (miR-1260b, miR-23a-5p and miR-423-3p). Through further qRT-PCR validation, overexpressed miR-1260b was demonstrated in LN metastasis tissue of NSCLC patients compared with LN free NSCLC patients. A variety of studies have identified certain miRNAs that suppress the metastasis in lung cancer [9,10,21]. Let-7 is one of the earliest identified miRNAs with significantly reduced expression in a large number of malignant tumors and is known to attenuate the development of lung cancer [22,23]. As for miR-10b and miR-155, the upregulated expression level was significantly positively correlated with TNM stage, lymph node involvement, and poor survival [11,12]. The higher expression of miR-31 was observed in lung adenocarcinoma tissues from patients with LN metastases rather than those without LN metastases [24]. In line with our verification, miR-1260b was highly expressed in prostate cancer (PC) tissues and renal cell carcinoma (RCC) tissues [25,26]. On the other hand, miR-1260b was underexpressed in periodontitis tissues compared with healthy tissues [27]. Interestingly, miRNAs were not only tissue specific but also body fluid specific. MiR-1260b was first discovered to be specifically expressed in vaginal secretions [28]. The discrepancy of these studies might be derived from different tumor types and heterogeneity. Two miRNAs were newly detected in NSCLC tissue i.e., miR-423-3p and miR-23a-5p, which were underexpressed in the LN group. MiR-423-3p can promote tumor progression through similar signaling pathways as in laryngeal hepatocellular carcinomas [29,30]. It could also be a signature to differentiate hereditary and non-hereditary breast cancers [31]. The miR-23a-5p was only identified as a participant in the pathogenesis of traumatic injury [32].
No study has explored the association of miR-1260b expression and metastasis. One study revealed that the higher expression of miR-1260b was observed in stage II, III, and IV of PC patients rather than stage I patients [25]. In addition, in vitro experiments showed that miR-1260b promoted renal cell carcinoma cells proliferation and invasion [26]. Consistent with the results, the expression of miR-1260b was significantly higher in stage III patients than stage I and II in our study. Clinically, TNM stage III patients almost always present with metastasis. Therefore, we can infer that the miR-1260b may play an important role in lung cancer metastasis.
Our study is the first to reveal the role of miR-1260b in NSCLC metastasis. The overexpression of miR-1260b can be regarded as a specific signature for early progression and prognosis of NSCLC lymph node metastasis. However, formal verification for these findings is required in a larger cohort of tissue samples. Furthermore, the information of smoking history and the 5-year survival rate ought to be followed up. The predicted target genes and pathways responsible for cancer metastasis and development also need to be elucidated in future studies. We plan to dissect the mechanism for metastasis and find more miRNAs that might intervene with the metastatic process. We also hope that some of the miRNA strategies will be further developed and used, alone or in combination with chemotherapy, to improve the specificity of diagnosis and treatment for NSCLC patients.
Acknowledgement
This work was supported by a grant from the Key Program of the Natural Science Foundation of Zhejiang Province, China (No. Z2101431).
References
- [1].Siegel R,Ma J,Zou Z,Jemal A (2014). Cancer statistics, 2014. CA: a cancer journal for clinicians, 64: 9-29 [DOI] [PubMed] [Google Scholar]
- [2].Ricciuti B,Mecca C,Crino L,Baglivo S,Cenci M,Metro G (2014). Non-coding RNAs in lung cancer. Oncoscience, 1: 674-705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Siegel R,Naishadham D,Jemal A (2013). Cancer statistics, 2013. CA: a cancer journal for clinicians, 63: 11-30 [DOI] [PubMed] [Google Scholar]
- [4].Wang H,Wu S,Zhao L,Zhao J,Liu J,Wang Z (2015). Clinical use of microRNAs as potential non-invasive biomarkers for detecting non-small cell lung cancer: a meta-analysis. Respirology, 20: 56-65 [DOI] [PubMed] [Google Scholar]
- [5].Li J,Yi H,Liu Z,Zhang H,Zhang D,Yue W,et al. (2015). Association between VEGFR-3 expression and lymph node metastasis in non-small-cell lung cancer. Experimental and therapeutic medicine, 9: 389-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Krol J,Loedige I,Filipowicz W (2010). The widespread regulation of microRNA biogenesis, function and decay. Nature reviews. Genetics, 11: 597-610 [DOI] [PubMed] [Google Scholar]
- [7].Iorio MV,Croce CM (2009). MicroRNAs in cancer: small molecules with a huge impact. Journal of clinical oncology: official journal of the American Society of Clinical Oncology, 27: 5848-5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Calin GA,Sevignani C,Dumitru CD,Hyslop T,Noch E,Yendamuri S,et al. (2004). Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America, 101: 2999-3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ling DJ,Chen ZS,Zhang YD,Liao QD,Feng JX,Zhang XY,et al. (2015). MicroRNA-145 inhibits lung cancer cell metastasis. Molecular medicine reports, 11: 3108-3114 [DOI] [PubMed] [Google Scholar]
- [10].Cui G,Cui M,Li Y,Liang Y,Li W,Guo H,et al. (2014). MiR-186 targets ROCK1 to suppress the growth and metastasis of NSCLC cells. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine, 35: 8933-8937 [DOI] [PubMed] [Google Scholar]
- [11].Zhang J,Xu L,Yang Z,Lu H,Hu D,Li W,et al. (2014). MicroRNA-10b indicates a poor prognosis of non-small cell lung cancer and targets E-cadherin. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico; [DOI] [PubMed] [Google Scholar]
- [12].Yanaihara N,Caplen N,Bowman E,Seike M,Kumamoto K,Yi M,et al. (2006). Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer cell, 9: 189-198 [DOI] [PubMed] [Google Scholar]
- [13].Sobin LH,Gospodarowicz MK,Wittekind C (2010) TNM Classification of Malignant Tumours (Uicc International Union Against Cancer), Wiley-Blackwell, New York [Google Scholar]
- [14].Sato F,Tsuchiya S,Terasawa K,Tsujimoto G (2009). Intra-platform repeatability and inter-platform comparability of microRNA microarray technology. PloS one, 4: e5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang Z,Zhang H,Zhang P,Li J,Shan Z,Teng W (2013). Upregulation of miR-2861 and miR-451 expression in papillary thyroid carcinoma with lymph node metastasis. Med Oncol, 30: 577. [DOI] [PubMed] [Google Scholar]
- [16].Xu LM,Li LQ,Li J,Li HW,Shen QB,Ping JL,et al. (2015). Upregulation of miR-1280 expression in non-small cell lung cancer tissues. Chinese medical journal, 128: 670-673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Beasley MB,Brambilla E,Travis WD (2005). The 2004 World Health Organization classification of lung tumors. Seminars in roentgenology, 40: 90-97 [DOI] [PubMed] [Google Scholar]
- [18].Casalini P,Iorio MV (2009). MicroRNAs and future therapeutic applications in cancer. Journal of B.U.ON.: official journal of the Balkan Union of Oncology, 14 Suppl 1: S17-22 [PubMed] [Google Scholar]
- [19].Zhang B,Pan X,Cobb GP,Anderson TA (2007). microRNAs as oncogenes and tumor suppressors. Developmental biology, 302: 1-12 [DOI] [PubMed] [Google Scholar]
- [20].Zhao Q,Ping LI,Junrong MA,Xijie YU (2015). MicroRNAs in Lung Cancer and Lung Cancer Bone Metastases: Biomarkers for Early Diagnosis and Targets for Treatment. Recent patents on anti-cancer drug discovery [DOI] [PubMed] [Google Scholar]
- [21].Suh SS,Yoo JY,Cui R,Kaur B,Huebner K,Lee TK,et al. (2014). FHIT suppresses epithelial-mesenchymal transition (EMT) and metastasis in lung cancer through modulation of microRNAs. PLoS genetics, 10: e1004652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Trang P,Wiggins JF,Daige CL,Cho C,Omotola M,Brown D,et al. (2011). Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Molecular therapy: the journal of the American Society of Gene Therapy, 19: 1116-1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhao B,Han H,Chen J,Zhang Z,Li S,Fang F,et al. (2014). MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer letters, 342: 43-51 [DOI] [PubMed] [Google Scholar]
- [24].Meng W,Ye Z,Cui R,Perry J,Dedousi-Huebner V,Huebner A,et al. (2013). MicroRNA-31 predicts the presence of lymph node metastases and survival in patients with lung adenocarcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research, 19: 5423-5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hirata H,Hinoda Y,Shahryari V,Deng G,Tanaka Y,Tabatabai ZL,et al. (2014). Genistein downregulates onco-miR-1260b and upregulates sFRP1 and Smad4 via demethylation and histone modification in prostate cancer cells. British journal of cancer, 110: 1645-1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hirata H,Ueno K,Nakajima K,Tabatabai ZL,Hinoda Y,Ishii N,et al. (2013). Genistein downregulates onco-miR-1260b and inhibits Wnt-signalling in renal cancer cells. British journal of cancer, 108: 2070-2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Stoecklin-Wasmer C,Guarnieri P,Celenti R,Demmer RT,Kebschull M,Papapanou PN (2012). MicroRNAs and their target genes in gingival tissues. Journal of dental research, 91: 934-940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Park JL,Park SM,Kwon OH,Lee HC,Kim JY,Seok HH,et al. (2014). Microarray screening and qRT-PCR evaluation of microRNA markers for forensic body fluid identification. Electrophoresis, 35: 3062-3068 [DOI] [PubMed] [Google Scholar]
- [29].Guan G,Zhang D,Zheng Y,Wen L,Yu D,Lu Y,et al. (2014). microRNA-423-3p promotes tumor progression via modulation of AdipoR2 in laryngeal carcinoma. International journal of clinical and experimental pathology, 7: 5683-5691 [PMC free article] [PubMed] [Google Scholar]
- [30].Lin J,Huang S,Wu S,Ding J,Zhao Y,Liang L,et al. (2011). MicroRNA-423 promotes cell growth and regulates G(1)/S transition by targeting p21Cip1/Waf1 in hepatocellular carcinoma. Carcinogenesis, 32: 1641-1647 [DOI] [PubMed] [Google Scholar]
- [31].Murria Estal R,Palanca Suela S,de Juan Jimenez I,Egoavil Rojas C,Garcia-Casado Z,Juan Fita MJ,et al. (2013). MicroRNA signatures in hereditary breast cancer. Breast cancer research and treatment, 142: 19-30 [DOI] [PubMed] [Google Scholar]
- [32].Yang J,Liang Y,Han H,Qin H (2013). Identification of a miRNA signature in neutrophils after traumatic injury. Acta biochimica et biophysica Sinica, 45: 938-945 [DOI] [PubMed] [Google Scholar]