Abstract
The fibroblast growth factor 1 (FGF1) gene is expressed primarily in the kidney and may contribute to hypertension. However, the biologic mechanisms underlying the association between FGF1 and BP regulation remain unknown. We report that the major allele of FGF1 single nucleotide polymorphism rs152524 was associated in a dose-dependent manner with systolic BP (P=9.65×10−5) and diastolic BP (P=7.61×10−3) in a meta-analysis of 14,364 individuals and with renal expression of FGF1 mRNA in 126 human kidneys (P=9.0×10−3). Next-generation RNA sequencing revealed that upregulated renal expression of FGF1 or of each of the three FGF1 mRNA isoforms individually was associated with higher BP. FGF1-stratified coexpression analysis in two separate collections of human kidneys identified 126 FGF1 partner mRNAs, of which 71 and 63 showed at least nominal association with systolic and diastolic BP, respectively. Of those mRNAs, seven mRNAs in five genes (MME, PTPRO, REN, SLC12A3, and WNK1) had strong prior annotation to BP or hypertension. MME, which encodes an enzyme that degrades circulating natriuretic peptides, showed the strongest differential coexpression with FGF1 between hypertensive and normotensive kidneys. Furthermore, higher level of renal FGF1 expression was associated with lower circulating levels of atrial and brain natriuretic peptides. These findings indicate that FGF1 expression in the kidney is at least under partial genetic control and that renal expression of several FGF1 partner genes involved in the natriuretic peptide catabolism pathway, renin-angiotensin cascade, and sodium handling network may explain the association between FGF1 and BP.
Keywords: gene expression, gene transcription, hypertension, blood pressure, glomerulus, kidney
Essential hypertension is a net product of genetic factors and environmental exposure acting together on regulatory systems in key organs for BP homeostasis. The kidney is central to BP regulation and drives the development of hypertension through numerous mechanisms including glomerular hemodynamics, tubular reabsorption of sodium, actions of the renin-angiotensin system, and natriuretic peptides.1 Rare genetic variants that affect expression of molecules and pathways operating within the kidney lead to elevated BP in monogenic forms of hypertension.2 Several common variants in genes associated with BP and/or susceptibility to hypertension are also believed to act through the renal mechanisms.3–7 Two of these variants map to the same fibroblast growth factor 1 signaling cascade—a pathway increasingly recognized as an important player in the cardiovascular system.6,7 Indeed, common alleles of fibroblast growth factor 1 (FGF1) gene and its chaperone molecule gene (fibroblast growth factor binding molecule, FGFBP1) cosegregated with familial susceptibility to hypertension in our previous studies.6,7 The central gene of this pathway—FGF1—was upregulated at both mRNA and protein level in the hypertensive kidney.6,7 The recent large-scale genetic analysis showed that genetic score composed of FGF1 signaling pathway single nucleotide polymorphisms (SNPs) provides a better explanation for variance in hypertension risk than the score calculated using a similar number of top variants identified by a genome-wide association study.8 Taken together, these data clearly suggest that FGF1 and its partner molecules play a role in genetic susceptibility to hypertension, possibly through kidney-related mechanisms. However, the exact biologic cause of the association between FGF1 and BP regulation remains unknown.
Here we examined the association between a common SNP of FGF1 and BP in a meta-analysis of approximately 15,000 individuals from five populations of white European ancestry. We then explored an effect of this SNP on expression of FGF1 mRNA in the largest to date analysis of 126 human kidneys collected in the TRANScriptome of renaL humAn TissuE (TRANSLATE) Study. Through next-generation RNA sequencing of human renal tissue we have investigated the network of most likely renal partner genes and transcripts of FGF1. Through further clinical association studies, in silico analyses and the investigations of biochemical read-outs of the most relevant partner molecules we have identified a biologically plausible network of transcripts that can mediate renal FGF1-related BP effect.
Results
A Common SNP of FGF1 (rs152524) Is Associated with Blood Pressure in the Meta-Analysis of Five Populations
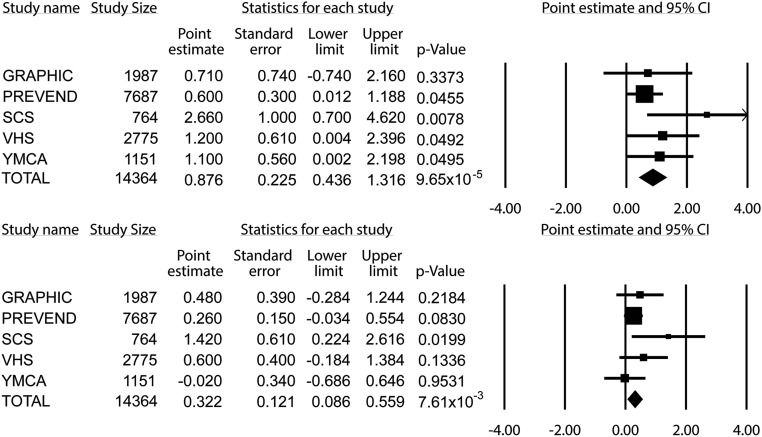
The major demographic and clinical characteristics of 14,364 individuals from five populations are given in Table 1. Distribution of rs152524 genotypes did not violate Hardy–Weinberg equilibrium6 in either of the cohorts and the minor allele frequency of rs152524 in all studies was typical for a white European population (Supplemental Table 1). There was at least nominally significant association between rs152524 and systolic BP in four out of five studies. The meta-analysis of all individuals with available genotypic and phenotypic information revealed a significant association between clinic systolic BP and rs152524—each major allele copy increased systolic BP by approximately 0.9 (±0.2) mmHg (P=9.65×10−5) (Figure 1). The association between rs152524 and clinic diastolic BP was directionally similar but the magnitude of the phenotypic effect of the SNP was smaller (P=7.61×10−3) (Figure 1).
Table 1.
Demographic and clinical characteristics of populations
| Variable | SCS | VFHS | PREVEND | GRAPHIC | YMCA | TRANSLATE |
|---|---|---|---|---|---|---|
| Number of subjects | 764 | 2755 | 7687 | 1987 | 1151 | 126 |
| Male/Female | 439/325 | 1334/1421 | 3784/3903 | 1005/982 | 1151/0 | 71/55 |
| Age (years) | 55.2±11.8 | 39.7±15.8 | 49.5±12.8 | 39.3±14.5 | 19.1±3.6 | 61.1±10.5 |
| Body mass index (kg/m2) | 27.5±4.2 | 25.1±4.1 | 26.1±4.2 | 26.1±4.6 | 22.8±3.0 | 27.6±4.4 |
| Clinic systolic BP (mmHg) | 131.3±19.1 | 123.3±15.2 | 129.3±20.3 | 127.1±17.8 | 118.0±13.1 | 136.4±13.5 |
| Clinic diastolic BP (mmHg) | 74.9±11.0 | 75.7±10.7 | 75.6±11.2 | 79.2±11.0 | 74.2±7.9 | 83.2±8.0 |
| Hypertension (%) | 549 (71.9) | 515 (18.6) | 2647 (34.4) | 571 (28.7) | 120 (10.4) | 85 (67.5) |
| Antihypertensive treatment (%) | 451 (59.0) | 222 (8.1) | 1219 (15.9) | 134 (6.7) | 18 (1.6) | 74 (58.7) |
Data are counts and percentages or means and SD. SCS, Silesian Cardiovascular Study; VFHS, Victorian Family Heart Study; PREVEND, Prevention of Renal and Vascular End-stage Disease Study; YMCA, Young Men Cardiovascular Association Study.
Figure 1.
Association between single nucleotide polymorphism of FGF1 (rs152524) and blood pressure in 14,344 individuals from five populations. Data are expressed as β-coefficients (point estimate), with SEM and lower/upper limits of the confidence intervals together with respective levels of statistical significance (P value); the data come from regression analysis of systolic BP (upper panel) or diastolic BP (lower panel) as independent variables together with rs152524 genotype and demographic phenotypes as dependent parameters; major allele or rs152524 (A) is a reference allele.
Major Allele of rs152524 Is Associated with Upregulation of FGF1 mRNA in the Human Kidney
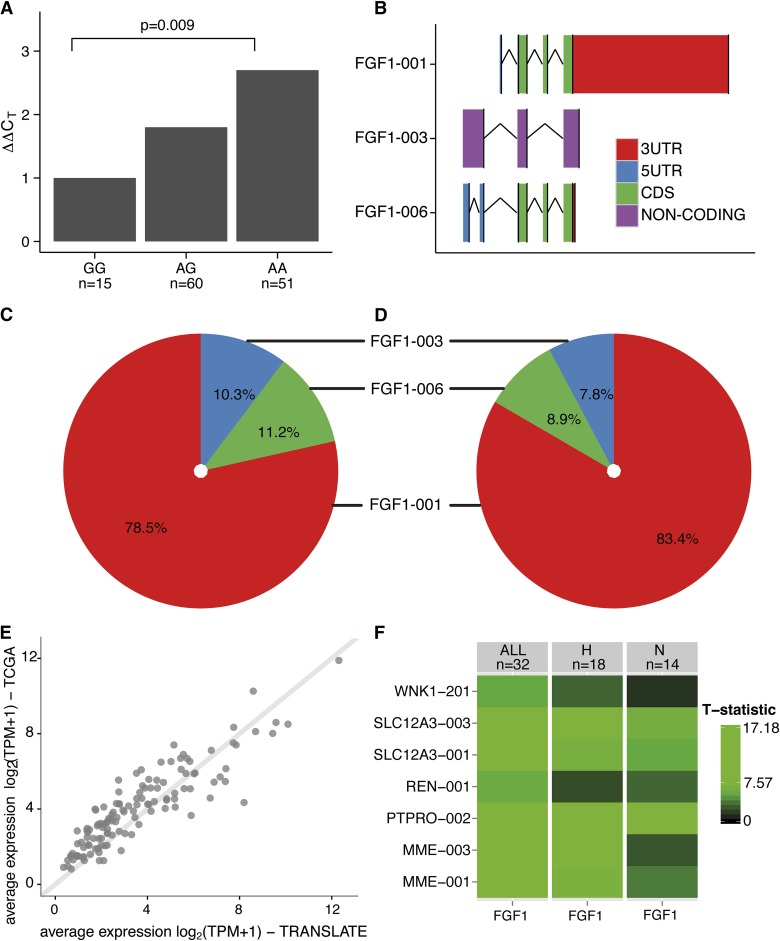
Clinical characteristics of 126 TRANSLATE Study individuals with informative genotype included in quantitative real-time PCR analysis of renal FGF1 are given in Table 1. rs152524 was associated with renal expression of total FGF1—compared with the reference genotype (rare homozygous), carriers of one and two copies of the major allele of rs152524 had 1.8- and 2.7-fold higher (respectively) levels of FGF1 mRNA in the kidney (P=0.009) (Figure 2).
Figure 2.
Renal expression of FGF1, its main mRNA isoforms and partner mRNAs in the human kidney. (A) Relative fold differences in expression of total FGF1 mRNA between rs152524 genotypes—quantitative real-time PCR analysis of 126 human kidneys, P, level of statistical significance; n, number of individuals in each genotype group. (B) The schematic structure of three renal FGF1 mRNA isoforms, 3UTR, 3′ untranslated region; 5UTR, 5′ untranslated region; CDS, coding sequence; FGF1–001 and FGF1–006 contain the same set of translated exons but differ in the structure of both 5′ and 3′ regions, FGF1–003 is a noncoding mRNA with a retained intron. (C, D) Percentage abundance of FGF1 mRNA isoforms in relation to total renal FGF1 mRNA in TRANScriptome of renaL humAn TissuE (TRANSLATE) Study (C) and The Cancer Genome Atlas (TCGA) resource (D). (E) A total of 126 mRNAs co-expressed with FGF1 in the human kidney—consistency in the average expression between the discovery population (TRANSLATE Study) and the replication resource (TCGA), log2 transcripts per million +1 is the unit of expression from next-generation RNA-sequencing. (F) Renal coexpression between FGF1 and seven transcripts in five genes with highest relevance to BP regulation; comparison of hypertensive (H) and normotensive (N) kidneys from the next-generation RNA-sequencing experiment; T-statistic, the magnitude of pair-wise coexpression calculated from a linear regression and expressed by color intensity—from black (least coexpressed) to green (most coexpressed).
Potential Transcriptional Activity of rs152524 and its Statistical Proxies—Roadmap Epigenomics and ENCODE Analysis In Silico
rs152524 maps to the segment of FGF1 intron 1 showing histone modifications in cells from adipose tissue, brain, and skin indicating that the region may act as a transcriptional enhancer (Supplemental Table 2). In addition, the rs152524-containing part of FGF1 intron 1 is a DNase I hypersensitivity site, contains a HOXA5 transcription factor binding site and both alleles of rs152524 are predicted to show differential effect on binding of HOXA5. The rs152524 has at least five statistically similar (r2>0.8) SNPs, of which three lie within regulatory enhancer regions identified in several human tissues (Supplemental Table 2). The strength and quality of regulatory annotations for all proxy SNPs was less than that for rs152524 (Supplemental Table 2). All detected proxies are located within the FGF1 gene but none of them map to any of four previously reported promoters in the 5′ region of FGF1.9–11 No other known SNPs within a 1-Mb distance of rs152524 can account for the detected associations through linkage disequilibrium with rs152524 (Supplemental Figure 1).
Increased Expression of FGF1 and its Three Renal mRNA Isoforms Is Associated with Hypertension and Higher BP—Next-Generation RNA Sequencing of Human Kidney Transcriptome
The available characteristics of individuals whose renal samples underwent next-generation RNA sequencing are shown in Supplemental Table 3. A total of 54,043 mRNAs mapping to 18,677 genes were expressed in 32 human kidneys from the TRANSLATE Study. The RNA sequencing identified three mRNA isoforms of FGF1 and revealed that FGF1–001 was the major transcript accounting for 78.5% of detectable renal FGF1 (Figure 2). The abundance of two other FGF1 transcripts (FGF1–003 and FGF-006) was lower at 10.3% and 11.2% (respectively) (Figure 2). The expression of all FGF1 transcripts showed a high level of linear intercorrelation (Supplemental Table 4). The percentage abundance of FGF1 mRNA isoforms and their co-expression patterns were largely replicated in an independent collection of 70 apparently healthy renal tissues from The Cancer Genome Atlas (TCGA) project (Figure 2, Supplemental Table 5).
Both FGF1 globally and its three mRNAs separately were approximately 31%–37% more abundant in hypertensive than normotensive kidneys from the TRANSLATE Study (where appropriate phenotypic information was available) (Supplemental Table 6). Consistently, the expression of total FGF1 and its three renal mRNA isoforms was associated with clinic BP in the expected direction (Table 2).
Table 2.
Association between FGF1, its renal mRNAs and blood pressure in TRANSLATE Study—next-generation RNA-sequencing analysis of human kidneys
| mRNA | Systolic BP | Diastolic BP | ||
|---|---|---|---|---|
| β-coefficient (SEM) | P value | β-coefficient (SEM) | P value | |
| FGF1 total | 0.85±0.35 | 0.021 | 1.15±0.57 | 0.053 |
| FGF1–001 | 0.91±0.36 | 0.016 | 1.21±0.58 | 0.046 |
| FGF1–003 | 0.091±0.034 | 0.0083 | 0.13±0.06 | 0.028 |
| FGF1–006 | 0.13±0.05 | 0.016 | 0.14±0.08 | 0.092 |
Data are β-coefficients, SEM and level of statistical significance (P value) from linear regression models whereby clinic BP (adjusted for treatment) was a dependent variable, and mRNA expression level (in transcripts per million units) of total FGF1 or its individual mRNAs, age, sex and body mass index were independent parameters.
Renal FGF1 Shows a Very High Level of Coexpression with Over 100 mRNAs in the Human Kidney
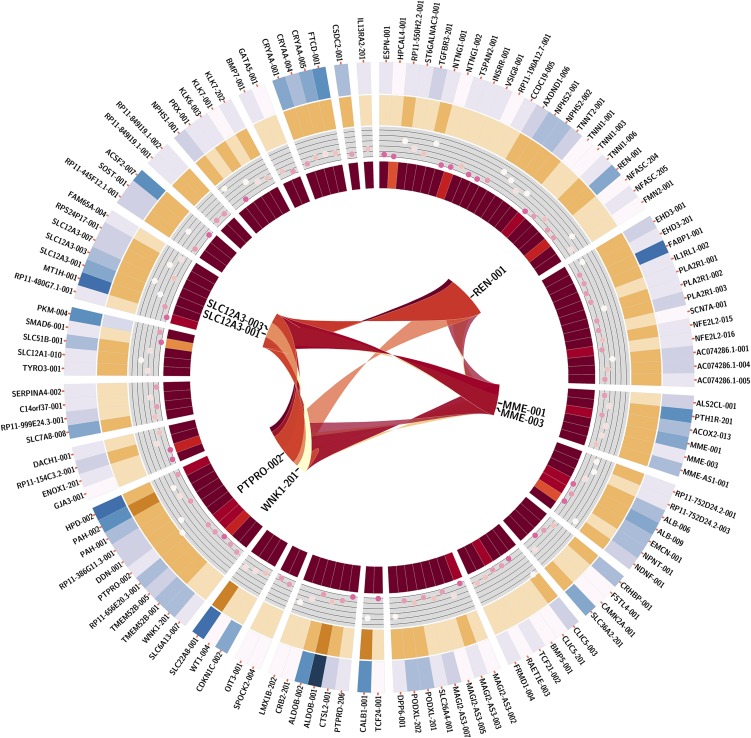
After correction for multiple testing (false discovery rate q<0.1%) a total of 747 mRNAs collapsed to 506 genes showed association with renal expression of FGF1 in the TRANSLATE Study (Supplemental Figure 2). A total of 126 non-FGF1 transcripts in 101 genes associated with FGF1 in the TRANSLATE population replicated at a conservative false discovery rate <0.1% in the TCGA (Figure 3, Supplemental Figure 3, Supplemental Table 7). There was excellent consistency in both average expression and the magnitude of association with FGF1 for those transcripts between the discovery and the replication population (Figure 2, Supplemental Figure 4).
Figure 3.
Replicated partner mRNAs of FGF1 in the kidney-next-generation RNA-sequencing in TRANScriptome of renaL humAn TissuE (TRANSLATE) Study. Replicated mRNAs, mRNAs associated with expression of FGF1 in both the TRANSLATE Study and The Cancer Genome Atlas resource. Outermost circle, symbols of 126 mRNAs ordered in circular manner; first circle in, level of renal expression for each replicated partner mRNA (in log2 transcripts per million +1 values) whereby white is lowest expression and navy blue is highest expression; second circle in, level of coexpression (measured as β-coefficient from linear regression) between each partner mRNA and FGF1 mRNA whereby dark brown represents strong positive coexpression and beige represents weak positive coexpression; third circle in, level of statistical significance (measured as –log10 P value from linear regression) for coexpression between each partner mRNA and FGF1 mRNA, whereby white represents strong statistical significance and pink represents weak statistical significance; innermost circle, level of connectivity of partner mRNAs whereby dark red represents highly connective mRNAs, orange represents mRNAs with low connectivity; center, coexpression between selected mRNAs relevant to BP regulation.
Renal Transcripts Coexpressed with FGF1 mRNA Are Associated with BP and Have a Strong Relevance to Human Hypertension
Of 126 non-FGF1 mRNAs correlated with expression of FGF1 in kidneys, 71 and 63 showed at least nominal association with systolic BP and diastolic BP, respectively, in the TRANSLATE Study (Supplemental Table 8). This translates into 57 and 51 individual genes whose mRNAs are associated with systolic BP and diastolic BP. Five genes (membrane metallo-endopeptidase gene, MME; protein tyrosine phosphatase, receptor type, O gene, PTPRO; renin gene, REN; solute carrier family 12 [sodium/chloride transporters], member 3 gene, SLC12A3; and WNK lysine-deficient protein kinase 1 gene, WNK1) associated with BP in the study and tightly coexpressed with FGF1 had direct prior annotation to BP regulation either through gene ontology or by manual data mining (Supplemental Table 9). The direction of association between the renal expression of these five genes and FGF1 abundance as well as BP was positive—the higher their expression, the higher renal abundance of FGF1 and the higher BP. In silico exploration of the existing data sets of renal expression profiling revealed that similar to FGF1, MME and PTPRO have the strongest enrichment for expression in the glomeruli (Supplemental Table 10). Further comparative co-expression analysis revealed that hypertensive kidneys exhibit much stronger co-regulation between MME transcripts and FGF1 mRNA than normotensive ones (Figure 2).
Renal Upregulation of FGF1 mRNA Is Associated with Lower Circulating Levels of Natriuretic Peptides
In 32 patients whose kidney samples underwent next-generation RNA sequencing, renal expression of FGF1 showed negative correlation with circulating levels of brain natriuretic peptide (BNP) (r=−0.359, P=0.044). This association retained its statistical significance after adjustment for other clinical variables (β=−0.022, SEM=0.009, P=0.023). Circulating levels of pro-atrial natriuretic peptide (pro-ANP) showed the same direction of correlation (r=−0.287, P=0.111). Upon specified criteria, renal FGF1 mRNA showed a negative association with circulating pro-ANP in multiple regression analysis (β=−0.018, SEM=0.009, P=0.057). The direction of associations between serum concentrations of both natriuretic peptides and renal expression of MME mRNA was similar to that of FGF1 mRNA but the level of statistical significance of these associations was somewhat weaker in multiple regression analysis (P=0.083 and P=0.138 for pro-ANP and BNP, respectively). FGF1 was also positively associated with NPR3 (natriuretic peptide receptor 3) gene at the renal mRNA expression level (P=3.4×10−8).
Discussion
Our study has provided several important new insights into the association between FGF1 and BP. First, we revealed that the common allelic variant of FGF1 previously associated with familial susceptibility to hypertension is also related to BP as a continuous quantitative trait in a large sample of individuals recruited primarily from the general population. Second, we provided data suggestive of an effect of this variant on FGF1 mRNA in the kidney. Third, through next-generation RNA sequencing we have characterized the diversity of renal FGF1 mRNAs and quantified the extent of their overexpression in the hypertensive kidney. Most importantly, through coexpression analysis we have identified the network of FGF1 partner transcripts and genes that may explain how upregulation of FGF1 mRNA in the kidney could relate to BP elevation.
Previous studies combining linkage analysis with fine mapping, rat-human synteny, and direct sequencing identified FGF1 as a strong positional and potentially functional candidate gene for hypertension.6,12 Subsequent family-based association analysis revealed that a major allele of common FGF1 SNP (rs152524) was transmitted from heterozygous parents to essentially hypertensive offspring more frequently than would be expected by chance.6 The data presented here clearly demonstrate that rs152524 is also associated with both systolic BP and diastolic BP as continuous traits and that the direction of allelic association with BP is concordant with that observed for hypertension. As expected from a common SNP, the magnitude of the allelic effect on BP is small (approximately 0.9 mmHg increase in systolic BP per major allele copy), consistent with the findings on common variants associated with BP in large genetic meta-analyses.3 However, in contrast to many previously identified genetic associations, the relationship between rs152524 and BP has a biologically meaningful functional context—a major allele of rs152524 not only correlated with BP elevation but also with an increase in renal expression of FGF1 mRNA. Further studies will be needed to confirm that rs152524 acts as a cis-acting gene expression Quantitative Trait Locus for FGF1 in the kidney. Our previous direct sequencing studies excluded common SNP within the coding sequences of FGF1 that could account for the detected associations by linkage disequilibrium with rs152524.6 The rs152524 maps to a putatively regulatory segment of FGF1 intron 1 in several tissues; the absence of kidneys in ENCODE and Roadmap Epigenomics did not permit us to explore a potential regulatory role of rs152524 in tissue relevant to our study. It is tempting to surmise that the characterized region in intron 1 may operate as an enhancer and possibly, in concert with previously identified alternative promoters,9–11 contribute to transcriptional regulation of FGF1. Indeed, four different promoters in the 5′ region of FGF1 were reported to control its transcription into four different mRNA isoforms in a cell/tissue-specific manner.9–11 5′ regions of two FGF1 mRNA isoforms identified in our next-generation RNA-sequencing experiment as FGF1–003 and FGF1–006 best overlap with sequences regulated by promoter 1A while FGF1–001—with promoter 1D.9–11 FGF1 promoter 1A was reported as operational in the kidney and promoter 1D was identified as active in vascular smooth muscle cells and fibroblasts.11 Both fibroblasts and vascular smooth muscle cells are resident cells of the human kidney.13 However, we should acknowledge the inherent limitation of selection of poly-adenylated RNA molecules in sample preparation—the use of this biochemistry is known to lead to 3′ bias in RNA-sequencing experiments, so fine-scale 5′ promoter usage may be more challenging to resolve.14,15
The kidney is a key organ for BP regulation and the site of highest FGF1 expression (http://www.proteinatlas.org/ENSG00000113578/tissue). FGF1 is one of the glomerulus-enriched genes16–19 and previous immunochemistry confirmed the exclusive expression of FGF1 within apparently normal human glomeruli.6,20 A number of mRNAs (i.e., Nephrosis 1, congenital, Finnish type [nephrin] gene, NPHS1; Nephrosis 2, idiopathic, steroid-resistant [podocin] gene, NPHS2) most tightly coexpressed with FGF1 in kidneys examined here are known as key structural constituents of the glomerulus.21 Together these data suggest that the main direct biologic actions of FGF1 in the kidney occur in glomeruli. Given a diversity of biologic processes that FGF1-coexpressed mRNAs map onto, we suspect that FGF1 may be involved in the regulation of structural integrity of glomeruli and renal maintenance of metabolic homeostasis.
FGF1 is a strong mitogenic signaling molecule22 but the mechanisms underlying its association with BP are not clear. Given that levels of mRNA/gene co-expression are good measures for activity of networks that control complex genetic mechanisms,23 we used renal next-generation RNA-sequencing data to identify FGF1 partner molecules and elucidate how increased expression of FGF1 in the kidney may link to BP regulation. Of FGF1-coexpressed mRNAs, those associated with systolic and/or diastolic BP in the TRANSLATE Study and those with direct prior annotation to BP regulation are the strongest putative mediators of FGF1-related BP effect. Indeed, four of these genes (REN, MME, WNK1, and SLC12A3) belong to key BP regulatory cascades and three (REN, MME, and SLC12As) are direct targets for antihypertensive medications. MME encodes membrane metallo-endopeptidase (neutral endopeptidase or neprilysin)—an enzyme responsible for biochemical degradation of circulating peptides, many of which promote vasodilation and/or natriuresis, i.e., bradykinin, ANP, and BNP.24 Pharmacologic inhibition of neprilysin leads to increased levels of natriuretic peptides and was proposed as a BP lowering therapy.24 Indeed, hypertension is increasingly recognized as a state of ANP and BNP deficiency.24 Similar to FGF1, MME is highly expressed within renal glomeruli.25 Renin is responsible for cleavage of angiotensin I from angiotensinogen—a rate-limiting step in activation of a fundamental pathway of BP regulation.26 The products of WNK1 and SLC12A3 belong to a key cellular mechanism of sodium reabsorption in the distal convoluted tubule of the nephron and their upregulation is a recognized renal mechanism of hypertension.27 Unlike FGF1 and MME, REN is expressed primarily within the juxto-glomerular apparatus and collecting duct whereas WNK1 and SLC12A3 are expressed in the distal nephron.26,27 Taken together these data indicate that increased expression of FGF1 in the kidney correlated with upregulation of MME, REN, SLC12A3, and WNK1 transcripts may contribute to BP elevation, potentially through heightened catabolism of natriuretic peptides clearance/degradation (MME), activation of the renin-angiotensin cascade (REN), and increased sodium reabsorption (SLC12A3 and WNK1). Furthermore, the direction of association between renal abundance of MME, REN, SLC12A3, and WNK1 mRNAs and BP in the TRANSLATE Study (increased expression equals higher BP) fits into their potential role as mediators of the BP-elevating effect of FGF1 overexpression. The correlation between decreased circulating levels of key substrates (natriuretic peptides) for MME and upregulation of renal FGF1 further suggests that at least one of its identified renal coexpressions translates into a biologically active mechanism measurable in the systemic circulation. One of the potential explanations for the somewhat weaker associations between natriuretic peptides and MME mRNA than those identified between both peptides and FGF1 mRNA is that increased renal expression of FGF1 mRNA correlates with up-regulation of another, MME-independent mechanism of natriuretic peptide catabolism in the kidney. Our data support this hypothesis—renal expression of FGF1 showed a positive association with NPR3 mRNA—an important clearance receptor for natriuretic peptides. In this context, increased expression of glomerular FGF1 may lead to a drop in levels of natriuretic peptides, potentially through renal activation of their clearance/degradation pathway, whereas increased expression of some other FGF1-coexpressed mRNAs in the kidney (i.e., REN, SLC12A3, WNK4) may simply indicate the activation of mechanisms secondary to the reduction in ANP/BNP circulating levels. To this end, lower levels of natriuretic peptides are synonymous with the reduction of their inhibitory effect on tubular sodium reabsorption and their weaker opposing effect on suppression of renin.28 Indeed, our data showed that expression of MME mRNA correlates positively with abundance of REN, WNK1, and SLC12A3 mRNAs in kidneys from both TRANSLATE and TCGA cohorts (Figure 3, Supplemental Figure 3—the innermost parts).
We should acknowledge that given the inherent limitations of our RNA-sequencing experiments, we cannot assign the direction to the identified co-expression patterns/associations and/or assess the extent to which they are primary (causal) or secondary (reactive). The association between FGF1 mRNA and its common SNP suggests that at least a proportion of renal increase of FGF1 mRNA is genetically regulated. It is also fair to point out that other FGF1-corelated transcripts and genes, including several non-coding RNAs without prior linkage to BP regulation in ontology terms, may mediate/contribute to the association between FGF1 and BP. We appreciate the limitations of RNA profiling of the kidney as the whole organ.17 Indeed, cellular diversity of the kidney may lead to variation in transcriptome composition between different segments of the nephron.17 Single-cell transcriptomics will be helpful to refine the identified coexpression patterns. Finally, we appreciate that our study is based primarily on expression analysis of FGF1 mRNA. Further experiments at the protein level will be necessary to confirm our findings.
Within the interpretational limitations discussed above, our data suggest that the expression of FGF1 mRNA in the human kidney is at least partly genetically controlled and that FGF1 maps onto several major renal pathways of BP regulation. Our study also illustrates the potential of next-generation RNA sequencing in relevant human tissues to provide insights into mechanisms underlying associations between candidate genes and complex phenotypes.
Concise Methods
Populations
Five populations (Silesian Cardiovascular Study29; Victorian Family Heart Study30; Prevention of Renal and Vascular End-stage Disease Study31,32; Genetic Regulation of Arterial Pressure of Humans in the Community Study (GRAPHIC)5,33; and Young Men Cardiovascular Association Study34) with relevant genetic (directly genotyped rs152524 SNP) and phenotypic (clinic BP) information were included in the genetic association analysis. All individuals were of white European ethnicity.
The TRANSLATE Study provided human kidney samples for quantitative real-time PCR analysis of FGF1 (n=133) and the discovery phase of the next-generation RNA-sequencing analysis (n=32). In brief, the TRANSLATE Study recruited individuals of white European ancestry eligible for unilateral elective nephrectomy because of noninvasive renal cancer in three reference centers.7 Small fragments of kidney tissues were collected from the healthy (unaffected by cancer) pole of the kidney and immersed in RNAlater (Ambion, Austin, Texas) immediately after nephrectomy. Information on an additional set of 70 human kidneys characterized by RNA-sequencing used in the replication phase of next-generation RNA-sequencing analysis was obtained from the TCGA database.35 The tissue material was secured from tissue unaffected by cancer (healthy control) after renal nephrectomy for cancer.36
The Bioethical Committees have approved the studies and all subjects have given written informed consent for participation. The studies adhered to the Declaration of Helsinki. Information on the recruitment and phenotyping of each individual study is summarized in the Supplementary material.
DNA Analysis
DNA was extracted from peripheral blood leukocytes according to the previously described protocols.5,12,29,34 In all populations, the SNP of interest was genotyped directly—in GRAPHIC by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MassARRAY system; Sequenom Inc., San Diego, CA) and in other cohorts—by TaqMan (ABI PRISM 7900HT Sequence Detection System; Applied Biosystems, Foster City, CA).
RNA Analysis
RNA was extracted from kidneys using previously reported protocols.6 The measurement of total FGF1 mRNA in 126 TRANSLATE Study kidneys with informative rs152524 genotype was conducted by SYBR Green-based real-time quantitative PCR on Eppendorf Realplex PCR equipment (Eppendorf, Sydney, Australia). The discovery next-generation RNA-sequencing experiment in 32 TRANSLATE kidneys was conducted on an Illumina HiSeq 2000 sequencer (Illumina, San Diego, CA) using 100-bp paired-end reads. Seventy TCGA kidneys were also sequenced on an Illumina HiSeq 2000 and raw (level 1) 50-bp reads were downloaded from CGHub (https://cghub.ucsc.edu/).
Biochemical Studies
Serum levels of BNP and pro-ANP(1–98) were measured in TRANSLATE individuals who underwent renal RNA-sequencing by direct chemiluminescence on a Centaur XP platform (Siemens, Munich, Germany) and enzyme immunoassay (BioMedica, Vienna, Austria) on Asys Hitech Expert 96 (Biochrom Ltd., Cambridge, UK), respectively.
Statistical and Bioinformatic Analysis
The analysis of association between rs152524 and treatment-corrected clinic BPs was conducted by multiple linear regression after adjustment for additional clinical variables and under an additive model of inheritance in each cohort individually and then jointly in fixed-effects inverse variance meta-analysis. The effect of rs152524 on quantitative PCR expression measures of FGF1 mRNA was examined under the same model of inheritance and after adjustment for demographic and technical variables in linear regression. In silico analysis of biologic activity of rs152524 was conducted using data from Roadmap Epigenomics37 and ENCODE.38 Circulating levels of BNP and pro-ANP underwent log and inverse transformation before association studies. Their association with RNA-sequencing measurement of FGF1 expression in the kidney was examined using Pearson’s correlation and multiple linear regression after adjustment for clinical and demographic parameters.
The renal RNA-sequencing data were processed through the Tuxedo workflow and the ultimate abundance of each mRNA was calculated in log2 transcripts per million units. Baseline coexpression analysis with FGF1 was conducted for all renal mRNAs (individually and after collapsing to genes) by LIMMA with adjustment for available clinical and demographic variables in both the TRANSLATE Study and TCGA. The correction for multiple testing calculated by false discovery rate was set at 0.1% in both data sets.
The extended version of the statistical and bioinformatics analysis is available in the Supplementary material.
Disclosures
None.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Health & Medical Research Council of Australia, the Federation University Australia ‘Self-sustaining Regions Research and Innovation Initiative’, an Australian Government Collaborative Research Network grant (to F.J.C.) and British Heart Foundation (PG/12/9/29376 to M.T. and J.E.). AFD was a holder of a British Heart Foundation Chair in Cardiovascular Medicine. NJS holds a British Heart Foundation Chair in Cardiology and is an NIHR Senior Investigator. This work is part of the portfolio of research supported by NIHR Leicester Biomedical Research Unit in Cardiovascular Disease.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2014121211/-/DCSupplemental.
References
- 1.Hall JE, Granger JP, do Carmo JM, da Silva AA, Dubinion J, George E, Hamza S, Speed J, Hall ME, ME : Hypertension: physiology and pathophysiology. Compr Physiol 2: 2393–2442, 2012 [DOI] [PubMed] [Google Scholar]
- 2.Lifton RP, Gharavi AG, Geller DS: Molecular mechanisms of human hypertension. Cell 104: 545–556, 2001 [DOI] [PubMed] [Google Scholar]
- 3.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O’Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sõber S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjögren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimäki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND, Aspelund T, Garcia M, Chang YP, O’Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Hopewell JC, Ongen H, Dreisbach AW, Li Y, Young JH, Bis JC, Kähönen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Bolton JA, Köttgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grässler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Sun YV, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Soler Artigas M, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, McKenzie CA, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stančáková A, Raffel LJ, Yao J, Kathiresan S, O’Donnell CJ, Schwartz SM, Ikram MA, Longstreth WT, Jr, Mosley TH, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli CS, Mani KR, Yajnik CS, Hofman A, Mattace-Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, Ala-Korpela M, Kangas AJ, Lyytikäinen LP, Soininen P, Tukiainen T, Würtz P, Ong RT, Dörr M, Kroemer HK, Völker U, Völzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G, Meschia JF, Nalls MA, Sharma P, Terzic J, Kumar MV, Denniff M, Zukowska-Szczechowska E, Wagenknecht LE, Fowkes FG, Charchar FJ, Schwarz PE, Hayward C, Guo X, Rotimi C, Bots ML, Brand E, Samani NJ, Polasek O, Talmud PJ, Nyberg F, Kuh D, Laan M, Hveem K, Palmer LJ, van der Schouw YT, Casas JP, Mohlke KL, Vineis P, Raitakari O, Ganesh SK, Wong TY, Tai ES, Cooper RS, Laakso M, Rao DC, Harris TB, Morris RW, Dominiczak AF, Kivimaki M, Marmot MG, Miki T, Saleheen D, Chandak GR, Coresh J, Navis G, Salomaa V, Han BG, Zhu X, Kooner JS, Melander O, Ridker PM, Bandinelli S, Gyllensten UB, Wright AF, Wilson JF, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller PP, Elosua R, Soranzo N, Sijbrands EJ, Altshuler D, Loos RJ, Shuldiner AR, Gieger C, Meneton P, Uitterlinden AG, Wareham NJ, Gudnason V, Rotter JI, Rettig R, Uda M, Strachan DP, Witteman JC, Hartikainen AL, Beckmann JS, Boerwinkle E, Vasan RS, Boehnke M, Larson MG, Järvelin MR, Psaty BM, Abecasis GR, Chakravarti A, Elliott P, van Duijn CM, Newton-Cheh C, Levy D, Caulfield MJ, Johnson T, International Consortium for Blood Pressure Genome-Wide Association Studies. CARDIoGRAM consortium. CKDGen Consortium. KidneyGen Consortium. EchoGen consortium. CHARGE-HF consortium : Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478: 103–109, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trudu M, Janas S, Lanzani C, Debaix H, Schaeffer C, Ikehata M, Citterio L, Demaretz S, Trevisani F, Ristagno G, Glaudemans B, Laghmani K, Dell’Antonio G, Loffing J, Rastaldi MP, Manunta P, Devuyst O, Rampoldi L, Swiss Kidney Project on Genes in Hypertension (SKIPOGH) team : Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med 19: 1655–1660, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tobin MD, Tomaszewski M, Braund PS, Hajat C, Raleigh SM, Palmer TM, Caulfield M, Burton PR, Samani NJ: Common variants in genes underlying monogenic hypertension and hypotension and blood pressure in the general population. Hypertension 51: 1658–1664, 2008 [DOI] [PubMed] [Google Scholar]
- 6.Tomaszewski M, Charchar FJ, Lynch MD, Padmanabhan S, Wang WY, Miller WH, Grzeszczak W, Maric C, Zukowska-Szczechowska E, Dominiczak AF: Fibroblast growth factor 1 gene and hypertension: from the quantitative trait locus to positional analysis. Circulation 116: 1915–1924, 2007 [DOI] [PubMed] [Google Scholar]
- 7.Tomaszewski M, Charchar FJ, Nelson CP, Barnes T, Denniff M, Kaiser M, Debiec R, Christofidou P, Rafelt S, van der Harst P, Wang WY, Maric C, Zukowska-Szczechowska E, Samani NJ: Pathway analysis shows association between FGFBP1 and hypertension. J Am Soc Nephrol 22: 947–955, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taal HR, Verwoert GC, Demirkan A, Janssens AC, Rice K, Ehret G, Smith AV, Verhaaren BF, Witteman JC, Hofman A, Vernooij MW, Uitterlinden AG, Rivadeneira F, Ikram MA, Levy D, van der Heijden AJ, Jaddoe VW, van Duijn CM, Cohort for Heart and Aging Research in Genome Epidemiology and Early Genetics and Lifecourse Epidemiology consortia : Genome-wide profiling of blood pressure in adults and children. Hypertension 59: 241–247, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myers RL, Payson RA, Chotani MA, Deaven LL, Chiu IM: Gene structure and differential expression of acidic fibroblast growth factor mRNA: identification and distribution of four different transcripts. Oncogene 8: 341–349, 1993 [PubMed] [Google Scholar]
- 10.Madiai F, Hackshaw KV, Chiu IM: Characterization of the entire transcription unit of the mouse fibroblast growth factor 1 (FGF-1) gene. Tissue-specific expression of the FGF-1.A mRNA. J Biol Chem 274: 11937–11944, 1999 [DOI] [PubMed] [Google Scholar]
- 11.Hsu YC, Liao WC, Kao CY, Chiu IM: Regulation of FGF1 gene promoter through transcription factor RFX1. J Biol Chem 285: 13885–13895, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomaszewski M, Brain NJ, Charchar FJ, Wang WY, Lacka B, Padmanabahn S, Clark JS, Anderson NH, Edwards HV, Zukowska-Szczechowska E, Grzeszczak W, Dominiczak AF: Essential hypertension and β2-adrenergic receptor gene: linkage and association analysis. Hypertension 40: 286–291, 2002 [DOI] [PubMed] [Google Scholar]
- 13.Strutz F, Zeisberg M: Renal fibroblasts and myofibroblasts in chronic kidney disease. J Am Soc Nephrol 17: 2992–2998, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B: Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628, 2008 [DOI] [PubMed] [Google Scholar]
- 15.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L: Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol 12: R22, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindenmeyer MT, Eichinger F, Sen K, Anders HJ, Edenhofer I, Mattinzoli D, Kretzler M, Rastaldi MP, Cohen CD: Systematic analysis of a novel human renal glomerulus-enriched gene expression dataset. PLoS ONE 5: e11545, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chabardès-Garonne D, Mejéan A, Aude JC, Cheval L, Di Stefano A, Gaillard MC, Imbert-Teboul M, Wittner M, Balian C, Anthouard V, Robert C, Ségurens B, Wincker P, Weissenbach J, Doucet A, Elalouf JM: A panoramic view of gene expression in the human kidney. Proc Natl Acad Sci U S A 100: 13710–13715, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He L, Sun Y, Takemoto M, Norlin J, Tryggvason K, Samuelsson T, Betsholtz C: The glomerular transcriptome and a predicted protein–protein interaction network. J Am Soc Nephrol 19: 260–268, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nyström J, Fierlbeck W, Granqvist A, Kulak SC, Ballermann BJ: A human glomerular SAGE transcriptome database. BMC Nephrol 10: 13, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossini M, Cheunsuchon B, Donnert E, Ma LJ, Thomas JW, Neilson EG, Fogo AB: Immunolocalization of fibroblast growth factor-1 (FGF-1), its receptor (FGFR-1), and fibroblast-specific protein-1 (FSP-1) in inflammatory renal disease. Kidney Int 68: 2621–2628, 2005 [DOI] [PubMed] [Google Scholar]
- 21.Saleem MA, Ni L, Witherden I, Tryggvason K, Ruotsalainen V, Mundel P, Mathieson PW: Co-localization of nephrin, podocin, and the actin cytoskeleton: evidence for a role in podocyte foot process formation. Am J Pathol 161: 1459–1466, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingwell K: Obesity and diabetes: FGF1 goes long to tackle diabetes. Nat Rev Drug Discov 13: 652–653, 2014 [DOI] [PubMed] [Google Scholar]
- 23.Rotival M, Petretto E: Leveraging gene co-expression networks to pinpoint the regulation of complex traits and disease, with a focus on cardiovascular traits. Brief Funct Genomics 13: 66–78, 2014 [DOI] [PubMed] [Google Scholar]
- 24.Mangiafico S, Costello-Boerrigter LC, Andersen IA, Cataliotti A, Burnett JC, Jr: Neutral endopeptidase inhibition and the natriuretic peptide system: an evolving strategy in cardiovascular therapeutics. Eur Heart J 34: 886–893, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubiak-Wlekły A, Perkowska-Ptasińska A, Olejniczak P, Rochowiak A, Kaczmarek E, Durlik M, Czekalski S, Niemir ZI: The comparison of the podocyte expression of synaptopodin, CR1 and neprilysin in human glomerulonephritis: Could the expression of CR1 be clinically relevant? Int J Biomed Sci 5: 28–36, 2009 [PMC free article] [PubMed] [Google Scholar]
- 26.Prieto-Carrasquero MC, Botros FT, Kobori H, Navar LG: Collecting duct renin: A major player in angiotensin II-dependent hypertension. J Am Soc Hypertens 3: 96–104, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossier BC, Staub O, Hummler E: Genetic dissection of sodium and potassium transport along the aldosterone-sensitive distal nephron: importance in the control of blood pressure and hypertension. FEBS Lett 587: 1929–1941, 2013 [DOI] [PubMed] [Google Scholar]
- 28.Volpe M: Natriuretic peptides and cardio-renal disease. Int J Cardiol 176: 630–639, 2014 [DOI] [PubMed] [Google Scholar]
- 29.Tomaszewski M, Charchar FJ, Barnes T, Gawron-Kiszka M, Sedkowska A, Podolecka E, Kowalczyk J, Rathbone W, Kalarus Z, Grzeszczak W, Goodall AH, Samani NJ, Zukowska-Szczechowska E: A common variant in low-density lipoprotein receptor-related protein 6 gene (LRP6) is associated with LDL-cholesterol. Arterioscler Thromb Vasc Biol 29: 1316–1321, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrap SB, Stebbing M, Hopper JL, Hoang HN, Giles GG: Familial patterns of covariation for cardiovascular risk factors in adults: The Victorian Family Heart Study. Am J Epidemiol 152: 704–715, 2000 [DOI] [PubMed] [Google Scholar]
- 31.Pinto-Sietsma SJ, Janssen WM, Hillege HL, Navis G, De Zeeuw D, De Jong PE: Urinary albumin excretion is associated with renal functional abnormalities in a nondiabetic population. J Am Soc Nephrol 11: 1882–1888, 2000 [DOI] [PubMed] [Google Scholar]
- 32.Hillege HL, Fidler V, Diercks GF, van Gilst WH, de Zeeuw D, van Veldhuisen DJ, Gans RO, Janssen WM, Grobbee DE, de Jong PE, Prevention of Renal and Vascular End Stage Disease (PREVEND) Study Group : Urinary albumin excretion predicts cardiovascular and noncardiovascular mortality in general population. Circulation 106: 1777–1782, 2002 [DOI] [PubMed] [Google Scholar]
- 33.Tomaszewski M, Debiec R, Braund PS, Nelson CP, Hardwick R, Christofidou P, Denniff M, Codd V, Rafelt S, van der Harst P, Waterworth D, Song K, Vollenweider P, Waeber G, Zukowska-Szczechowska E, Burton PR, Mooser V, Charchar FJ, Thompson JR, Tobin MD, Samani NJ: Genetic architecture of ambulatory blood pressure in the general population: insights from cardiovascular gene-centric array. Hypertension 56: 1069–1076, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charchar FJ, Tomaszewski M, Lacka B, Zakrzewski J, Zukowska-Szczechowska E, Grzeszczak W, Dominiczak AF: Association of the human Y chromosome with cholesterol levels in the general population. Arterioscler Thromb Vasc Biol 24: 308–312, 2004 [DOI] [PubMed] [Google Scholar]
- 35.The Cancer Genome Atlas (TCGA) data portal: Available at: https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm. Accessed April 4, 2014
- 36.Zhao Q, Caballero OL, Davis ID, Jonasch E, Tamboli P, Yung WK, Weinstein JN, Shaw K for TCGA research network. Strausberg RL, Yao J: Tumor-specific isoform switch of the fibroblast growth factor receptor 2 underlies the mesenchymal and malignant phenotypes of clear cell renal cell carcinomas. Clin Cancer Res 19: 2460–2472, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.http://www.roadmapepigenomics.org/
- 38.ENCODE Project Consortium : An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.