Abstract
Traditionally, genetic testing has been too slow or perceived to be impractical to initial management of the critically ill neonate. Technological advances have led to the ability to sequence and interpret the entire genome of a neonate in less than 50 hours. As the cost and speed of testing decreases, the utility of whole genome sequencing (WGS) of neonates for acute and latent genetic illness increases. Analyzing the entire genome allows for concomitant evaluation of the currently identified 5,430 single gene diseases. When applied to a select population of ill infants in a level IV neonatal intensive care unit, WGS yielded a diagnosis of a causative genetic disease in 57% of patients. These diagnoses may lead to clinical management changes ranging from transition to palliative care for uniformly lethal conditions to alteration or initiation of medical or surgical therapy to improve outcomes in others. Thus, institution of 2-day WGS at time of acute presentation opens the possibility of early implementation of precision medicine. This implementation may create opportunities for early interventional therapies, which would frequently be novel or off-label, that may alter disease trajectory in infants with what would otherwise be fatal disease. Widespread deployment of rapid WGS and precision medicine will raise ethical issues pertaining to interpretation of variants of unknown significance, discovery of incidental findings related to adult onset conditions and carrier status, and implementation of medical therapies for which little is known in terms of risks and benefits. Despite these challenges, precision neonatology has significant potential both to decrease infant mortality related to genetic diseases with onset in newborns and to facilitate parental decision-making regarding transition to palliative care.
Introduction
The completion of the first composite human genome sequence in April, 2003 marked the dawn of the promise of precision medicine – a new approach to medicine wherein diagnosis, treatment, and risk factor modification would be informed by an individual's unique genetic make-up. While mature models of precision medicine remain to be defined, changes in the speed and cost of whole genome sequencing (WGS) are bringing the details of initial applications into focus. NIH Director, Francis Collins, foresees a society in which every baby will have access to their sequenced genome in order to modify their strategies for disease prevention, detection and treatment[1]. In the 2015 State of the Union Address, President Barack Obama announced the creation of a precision medicine initiative, ultimately to provide each individual with personalized information to drive expedient diagnoses and individualized, more effective treatments. The transformation of healthcare through the use of personal WGS information has already begun in Neonatal Intensive Care Units (NICUs). Since 2011, neonatologists at our institution have, through research protocols, used research-based rapid WGS in acutely ill infants and their parents to diagnose the underlying genetic cause of the neonates’ conditions[2–4]. Furthermore, in a research setting, it is now possible to sequence human genomes at a cost of less than $1000 per individual. At this early stage in its evolution, we review the premise, practicality, and potential of rapid WGS for neonatal precision medicine.
Monogenic Diseases: Neonatal Impact and Incidence
Monogenic diseases are conditions causally related to genomic change(s), or variant(s), in a single gene. This collection of diseases is currently most amenable to diagnosis through WGS because the causative variants frequently involve one or a few contiguous DNA nucleotides in one or a handful of genes. These variants interfere with the efficient functioning of a gene product through disruption of transcription, translation, protein modification, complex assembly or function. They may be inherited from a parent or occur de novo as a mutation in the germ cell of one of the parents. It is estimated that each individual's germline genome harbors about 74 de novo single nucleotide variants [5–7]. When these de novo variants are associated with dominantly expressed phenotypes, they tend to present in the newborn period because they are often more deleterious than inherited variants due to the absence of evolutionary selection [8, 9].
As a proportion of overall disease burden, monogenic diseases decrease in importance with age, and their impact is highest in fetal, perinatal, and neonatal care respectively. The incidence of each individual monogenic disease is rare, but in toto, they are common. It is estimated that 60 million people in the United States and Europe have rare genetic diseases, of which 75% are children. Of these 45 million children, an estimated 30% will die before the age of 5 years[10]. Genetic diseases and birth defects are the leading cause of infant death in the United States with many of these being monogenic[11]. While the proportion of newborns admitted to the NICU with genetic disorders is unknown, 76% of NICU patients are admitted for reasons other than prematurity[12, 13]. A 1991 study from Scotland determined, in a cohort of 821 consecutive admissions to the NICU, that 5.7% of the admissions were for chromosomal or monogenic disorders[14]. This is likely a considerable underestimate given lack of NGS at that time. Infants with recognizable genetic disorders have disproportionately longer hospitalizations and more frequent neonatal death[15–20].
At present, newborns and infants with congenital malformations, syndromes, and inherited disorders typically undergo an extensive diagnostic process, with relatively low rates of etiologic diagnosis[4]. It is suspected that 3% of babies born in the US and Europe will have a major birth defect, with only 10–20% of these having an identifiable syndrome[21]. Acute management decisions are therefore typically made in the absence of a definitive diagnosis, which leads to delays in initiation of effectual treatments or to the use of empiric treatments that are ineffective, have adverse effects, or exacerbate symptoms. Thus, the timely return of definitive diagnoses of monogenic diseases during a NICU stay can potentially result in substantive changes in practice for neonatologists and consulting subspecialists[4]. In addition to having the potential to modify medical treatment in amenable cases, rapid genetic diagnosis allows for rational refocusing of care to diminish neonatal suffering and to support familial grieving in futile situations. These end-of-life decisions are common in neonatal genetic diseases, with most deaths resulting from withholding or withdrawing care[22]. Given the limitations to parental bonding and contact with the baby in the NICU setting, earlier holistic, end-of-life care decisions shifts focus from invasive medical management to the alleviation of suffering, allowing the family to bond, say “goodbye,” baptize or give last rites, and facilitate the grieving process. Thus, early definitive diagnosis may actually increase neonatal (28-day) mortality in patients with genetic diseases, whilst having the potential to decrease infant (1 year) mortality.
Genetic diseases also have significant societal costs associated with profound emotional, financial, social, and physical stress within families[23, 24]. The impact of newborn genetic diseases and birth defects on family structure is profound with studies identifying increased maternal depression and anxiety. The presence of maternal anxiety and depression are associated with childhood behavioral, developmental, and persistent health complications[25]. In a 1997 report, parental divorce occurred in 50 % of families with a child with a genetic disease[26]. Rapid, precise diagnosis coupled with robust treatment and support teams may offset not just direct medical expense but larger familial and societal costs of genetic disease in infancy.
Rapid Whole Genome Sequencing Methods
While the specifics of rapid WGS will differ from institution to institution, we have reported on our three year experience of sequencing selected neonates and infants for diagnosis of likely genetic diseases[2–4] described briefly as follows. Enrollment of parental and proband trios is preferred, and every effort is made to sequence both parents. After informed consent is obtained, the presenting clinical features are ascertained by review of electronic health records and translated into structured Human Phenotype Ontology terms[2, 27]. These terms are then mapped to the approximately 5430 known monogenic disorders and 3353 genes using either an in-house clinicopathologic correlation tool, Symptom & Sign Assisted Genome Analysis (SSAGA), or publically available software, such as Phenomizer,[2, 3, 28, 29] generating a rank ordered differential diagnosis of diseases and assocatied genes. DNA is isolated from participants and sequenced using an Illumina HiSeq 2500 in rapid mode. Short reads are computationailly aligned to the GRCh37 human reference genome, and variants identified with software, including short nucleotide substitutions, deletions, and insertions.[2–4, 30, 31].
Each individual sample sequence yields 4 to 5 million nucleotide variants that differ from the human reference genome. Through a variety of commercially available and in-house computer programs, each of these variants is genotyped and annotated[2]. The annotation process incorporates data from ENSEMBL Variant Effect Predictor software[32] comparing variants from the NCBI Single Nucleotide Polymorphism Database, Human Gene Mutation Database disease-causing variants[33, 34], and performing additional in silico prediction of variant consequences using RefSeq and ENSEMBL gene interpretations[35, 36]. Variants are categorized according to ACMG recommendations for reporting sequence variation[34, 37] along with a minor allele frequency from our in-house database[2]. Variants are filtered using a minor allele frequency of <1% and ACMG categories 1–5 (With a focus on Cat 1–3 known pathogenic, likely pathogenic, or unknown significance respectively). Analysis is further limited to variants in genes that ranked high in correspondence to the phenotype of the affected infant or child. If a single, likely-causative variant is identified for an autosomal recessive condition, the entire coding region is manually inspected using the Integrated Genomics Viewer[38]. Expert interpretation and literature curation are performed for all likely-causative variants with regard to evidence for pathogenicity[37]. Rapid WGS, from sample procurement to test result, can be completed in less than 50 hours[2]. Currently, all causative variants identified by WGS are confirmed by Sanger sequencing prior to clinical reporting. If the subject's phenotype differs from those previously reported for mutations in the suspected disease gene, additional expert consultation and functional confirmation is performed. We do not currently report variants of unknown significance, carrier status, or predisposition for adult onset diseases. Reports in the health record are limited to confirmed variants that explain the presenting phenotype of the infant.
Experience with Whole Genome Sequencing in Neonates
Our early experience with rapid WGS involved 35 acutely ill infants whose genomes were sequenced with their families as parent-child trios[4]. All infants were less than 4 months of age at time of enrollment, had a suspected genetic cause of their symptoms, and lacked a molecular or genetic diagnosis. The infants enrolled for sequencing had diverse presentations, with symptoms typically apparent at birth (Table 1) and received multiple standard genetic tests in addition to WGS. In this highly selected group of NICU infants, rapid WGS provided a genetic diagnosis in 20 patients (57%), in contrast to only 9% diagnostic rate with standard genetic testing in the same individuals. Sanger sequencing confirmed 100% of WGS diagnostic findings. In all cases examined, WGS analysis also identified variants of unknown significance that did not explain the etiology of illness or lacked sufficient evidence of pathogenicity and were not reported. The significance of these variants may change as more information is acquired on the role and functions of these genes and variants.
Table 1.
Clinical findings in 20 infants who received genetic disease diagnoses by rapid WGS.
| Demographics | Value |
|---|---|
| Symptom onset (Average, range, days) | 0.5 (0–7) |
| Multisystem Congenital Anomalies | 5 (25%) |
| Neurologic findings | 4 (20%) |
| Cardiac findings/Heterotaxy | 3 (15%) |
| Hydrops/Pleural Effusion | 2 (10%) |
| Metabolic findings (inc. Hypoglycemia) | 2 (10%) |
| Renal findings | - |
| Arthrogryposis | 2 (10%) |
| Respiratory findings | - |
| Hepatic findings | 1 (5%) |
| Dermatologic findings | 1 (5%) |
No presenting symptoms seemed to confer a higher diagnostic rate with WGS (Table 1). Recurrent genes with causative variants were PTPN11 (Noonan/LEPOARD syndrome), CHD7 (CHARGE syndrome), and SCN2A (Early infantile epileptic encephalopathy). Dominant de novo mutations were the most commonly found mechanism of disease variant accrual (65%). The recognition of de novo and somatic mutations as common causes of neonatal genetic diseases was important since family history is negative in such situations and the disease appears to be sporadic in origin. WGS of parent – infant trios is critical for recognition of de novo variants as the absense of the variant in the unaffected parents lends strong indirect support to the pathogenicity of the variant identified in the patient. WGS also provides good coverage of the mitochondrial genome and yielded one maternally-inherited diagnosis in the 35 cases. Of five patients with autosomal recessive inheritance, four had compound heterozygous variants, and one, from a genetically isolated population, had a homozygous causative variant. This mix of inheritance patterns is similar to that seen in recently published large case series of exome and genome sequencing (Table 2).
Table 2.
Results of five large case studies that to date have retrospectively examined the diagnostic rate of genome or exome sequencing in children with suspected genetic diseases.
| Citation | Publication Date |
Site | Number of subjects |
Disease | Age (mean or median) |
Diagnosis Rate |
De novo mutation |
Management changed by Dx |
|
|---|---|---|---|---|---|---|---|---|---|
| Soden | Sci Trans Med | Oct-14 | CMH | 100 | NDD | 7 | 47% | 51% | 49% |
| Srivastava | Ann Neurol | Oct-14 | JHU | 78 | NDD | 9 | 41% | 56% | 100% |
| Yang | JAMA | Nov-14 | Baylor | 1756 | any | 6 | 27% | 49% | n.d. |
| Lee | JAMA | Nov-14 | UCLA | 520 | any | <18 | 26% | 50% | n.d. |
| Wright | Lancet | Dec-14 | UK | 1133 | NDD | 6 | 27% | 62% | n.d. |
Abbreviations: Dx: diagnosis; NDD: neurodevelopmental disabilities; n.d.: not deterimined.
Among infants receiving WGS diagnoses, the degree of overlap between the classical clinical features of the disease and the presenting symptoms of the infants was frequently modest. Of the 20 infants receiving a diagnosis by WGS, 9 (45%) were conditions that had not been considered in the differential diagnosis at the time of enrollment. These infants either had yet to develop the classical disease presentations (i.e. to “grow into their phenotype”) or represented unappreciated disease pleiotropy. Prior to WGS, there has not been a generalizable method for genetic disease diagnosis in newborns, and it is anticipated that our current knowledge of newborn presentations of genetic diseases may represent the tip of the phenotypic iceberg. In two of 35 cases, the genetic disease was novel and previously unpublished. It was encouraging in these situations that rapid WGS, nevertheless, yielded diagnoses. However, as noted above, there is probably yet an under-diagnosis of genetic diseases in infants with variants of unknown significance with our methods that will be clarified with time. There likely are many more novel genetic diseases that present as stillbirths or as extreme presentations of otherwise normal neonatal illness.
For this preliminary data, the average age at enrollment for WGS was 26 days, with the median time to confirmed, reported diagnosis of 23 days. Median time from enrollment to WGS analysis was 5 days, with interpretation and Sanger confirmation taking the remaining time to report clinically. Of the 35 infants in this initial experience, the median NICU or PICU stay was 42 days with a range of 3 – 387 days. 120-day mortality was 40% overall (14 of 35) but higher in those with a genetic diagnosis identified by WGS (55%, 11 of 20). These data indicate limitations of rapid WGS at present. First, the window for possible intervention is small when one accounts for the delay in enrollment coupled with the significant early mortality in the patients with genetic diagnoses. Additionally, if an infant is acutely ill due to an early presentation of an inborn error of metabolism, the turn-around time for basic biochemical testing is still more rapid than rapid WGS. WGS complements but does not replace conventional tandem mass spectrometry (MS/MS) newborn screening. Where a singular genetic disease diagnosis is likely, and if conventional molecular testing can be performed in-house, rapid WGS is unlikely to be superior due to current research restrictions and required Sanger confirmation which currently adds nearly a week to return of results.
Clinical Outcomes and Impact of Genomic Diagnoses
The clinical impact of WGS testing was positive in 65% of diagnoses according to clinician report. Specific services enabled by these rapid genetic diagnoses included institution of palliative care, initiation of new subspecialist consultant, or change in medication, diet, imaging study, surgical procedure, or specific genetic counseling. Of the 13 diagnoses made prior to discharge or death, 11 (85%) were considered to have acute clinical utility. Palliative care was instituted more often in infants receiving a genetic diagnoses than those who did not (6 of 20, 30%, versus 0 of 15, respectively).
Two Illustrative Cases of Clinical Impact
Of the previously published cases, two are presented as illustrations of potential clinical impact[4]. The first, CMH487, was admitted to the NICU at birth with multiple congenital anomalies. He developed acute hepatic failure on day of life (DOL) 56. Intravenous corticosteroids and immunoglobulin were started empirically on DOL 67 and 69, respectively. The infant-parent trio was enrolled on DOL 71. Rapid WGS gave a provisional molecular diagnosis of hemophagocytic lymphohistiocytosis. Since this diagnosis was actionable and the infant was at imminent risk of death, the provisional molecular diagnosis was reported verbally on DOL 74, before confirmatory testing. Subsequently, this diagnosis was confirmed by Sanger sequencing, and formally reported on DOL 77. The diagnosis was further solidified by functional studies on NK cells. On DOL 81, the knowledge of the genetic diagnosis allowed institution of published treatment protocols for this patient with discontinuation of extraneous and potentially harmful empiric therapies. The patient had resolution of coagulopathy by DOL 88. At 24 months of age, the child has normal liver function.
CMH569 was admitted to the PICU on DOL 34 with a blood glucose of 18 mg/dL. Hypoglycemia was refractory to glucose infusion and diazoxide. Hyperinsulinemia was detected. The infant-parent trio received rapid WGS on day of life 41. A provisional molecular diagnosis of type 1 familial hyperinsulinism was reported on DOL 45. Furthermore, rapid WGS suggested the disease to be focal (adenomatous hyperplasia that involved only part of the pancreas). The Sanger-sequence confirmed diagnosis was reported on DOL 50. Functional imaging confirmed focal pancreatic lesions, allowing targeted partial pancreatectomy instead of the previously planned total pancreatectomy which would have led to life long brittle diabetes mellitus. Rapid WGS shortened the PICU stay by approximately three weeks, as well as the morbidity associated with breakthrough hypoglycemia. At 17 months of age, the patient is euglycemic without need for insulin therapy.
Ongoing Research
Using our baseline three year experience with rapid WGS of selected neonates and infants for diagnosis of likely genetic diseases, we developed a prospective study of the diagnostic, clinical, and psychosocial utility of rapid WGS in the NICU (ClinicalTrials.gov Identifier: NCT02225522). This study is part of a multicenter investigation funded by the NIH under the Newborn Sequencing In Genomic medicine and public HealTh (NSIGHT) collaborative which seeks to explore the implications, challenges, and opportunities associated with the use of genomic sequence information in the newborn period.
At our site, we are understaking a prospective, random blinded study of the utility of rapid WGS in the care of ill neonates. Potentially eligible newborns are nominated for the study by neonatologists in our level IV NICU, which has an annual census of approximately 900 neonates. Inclusion criteria for enrollment require that either a genetic test or genetic subspecialty consult has been ordered, a major congenital anomaly or multiple minor defects is present, or a poor response to routine care for a condition is identified (raising the suspicion of an underlying genetic etiology). Exclusion criteria include infants > 4 months of age, features pathognomonic for a known chromosomal anomaly, or a confirmed molecular genetic diagnosis. Upon acceptance of nominations, informed consent from both parents is obtained prior to participation as required by the IRB. Efforts are made to enroll and sequence the proband and both parents when possible. To date, timely nomination (i.e. within days of life 0–5) has proven difficult; the rates of nomination vary widely by neonatologist (from 0 – 10%) and consent is obtained in ~50% of accepted nominations. Major reasons for failure to obtain consent are the unavailability of a second parent, underage parents, mothers who do not wish the father contacted, and unwillingness to undergo WGS (primary stated reason is related to perceived limitations of the Genetic Information Nondiscrimination Act of 2008). Enrolled infants and family members then undergo sequencing and variant identification as described above.
Current Limitations
Rapid WGS is a quickly evolving technology that still has multiple limitations. Causative larger, structural variants that affect single loci are sought using computational tools, but these methods currently lack sufficient sensitivity and specificity for clinical use. The short sequences generated in rapid WGS preclude their use for diagnosis of triplet repeat expansion disorders and in some disease genes with nonfunctional but highly homologous intronic regions called pseudogenes. Sequencing advances will likely be able to address these limitations in the near future. A much more difficult hurdle in rapid WGS application involves interpretation of variant pathogenicity. That is, determining if the variant both effects the gene function, and if that effect reasonably may be causing the patient’s symptoms. Each patient has many variants that are unique or private to them and determination of whether the variant may be pathogenic may be inferred from various sources, including similar changes that have been reported or in silico analysis of the importance of the variant. However, determining variant causality in the absence of prior literature involves functional testing. This method of testing for each variant, while compelling, is expensive and labor intensive.
Outside of variants of unknown significance, there are technologic limitations of rapid WGS that need to be understood. False positive results are variants labeled in the literature as pathogenic or predicted to be pathogenic by software tools that are actually not disease causing. The “over-interpretation” of variant pathogenicity was an even greater problem prior to WGS or exome sequencing, when only a few candidate genes were sequenced. At this early phase of precision medicine, some false positive errors due to erroneous medical literature are unavoidable and may become more common as testing moves to wider groups of newborns with lower pre-test probability of genetic disease. In one sense, even with Sanger technical confirmation, these diagnoses remain provisional until either enough literature is accumulated to confirm that association, the child develops a full phenotypic manifestation of disease, or orthogonal functional testing is performed. The ACMG has recently issued detailed guidelines for structured methods for evaluating the evidence in support of a variant being disease causing[37]; however, the adoption of these new guidelines is time consuming and necessitates highly expert laboratory directors.
There will also inevitably be false negative results with expanded use of WGS. These can occur from missing variant calls, miscategorization of variants in introns, untranslated regions and regulatory elements as “silent”, or from inherent insensitivity of WGS for detection of structural variants. The most common form of false negative, though, likely comes from undiscovered disease genes, changes in deep intronic regions that affect gene expression, and incomplete knowledge of the spectrum of clinical presentations of known disease genes. At present there are more than twenty novel disease gene discoveries or substantive phenotype expansions reported each month. As our knowledge of gene function, network function and regulatory mechanisms grow, these gaps in diagnosis will diminish. An additional source of error is identification of the wrong pattern of inheritance, which can lead to erroneous genetic counseling. One example is under-reporting of de novo variants, which have extremely low likelihood of recurrence. These difficulties with variant interpretation are the basis from which many ethical arguments arise.
Ethics of Widespread Genome Sequencing
The prospect of WGS of infants is forcing society to grapple with ethical issues such as the child’s right to an open future, a family's right to know about health predispositions, the nature of informed consent, and returning of results related to adult onset diseases or risk factors of conditions not manifesting during infancy and/or childhood. Much literature exists related to the issue of return of secondary (or incidental) genetic variant findings, but without coalescence to a consensus. Initially, the ACMG had suggested that all genomic sequencing tests should report incidental findings on 56 disease genes for which treatments are available, irrespective of the reason for the sequencing. This has now been modified with a position statement allowing parents and patients to opt out of receiving this information. The original position statement raised concerns about engendering anxiety within the families of our patients and impinging on the right of pediatric patients to be free from the burden of predisposition to adult onset diseases. This "Right to an Open Future" informed our current practice of restricting our search for disease-causing variants to changes in genes with some reported relationship to the presenting symptoms. As mentioned above, many of our patients with rapid WGS diagnoses had not yet fully manifested the classical symptoms of their diagnosis, so this approach may be overly restrictive. However, through discussion with our pediatric bioethics center, filtering our examination of variants by the patient's presenting symptoms was deemed to be the best compromise between increasing sensitivity of diagnosis and protecting the child's rights. Specifically, if we do not review the variants for breast cancer or Alzheimer disease, then we cannot report this information. Even in the analysis of relevant variants, though, finding some variants with impact on adult onset conditions is inevitable. Our solution to addressing these issues is to not to return any results clinically that do not directly impact on the nominating symptoms. Parents are informed of this policy prior to obtaining consent. These measures provide some guidance for returning results, but ethical issues associated with testing are still present. For these unforeseen cases, we turn to advice from our colleagues in pediatric bioethics to help guide decision making.
Future Implications for Precision Medicine
The evidence to date, while retrospective, strongly suggests that rapid WGS does have utility for timely genetic disease diagnosis for ill NICU infants, even before a fully developed symptom complex evolves. However, prospective evidence has not yet been published. A goal of the Children’s Mercy NSIGHT study is to prospectively assess the diagnostic yield of rapid WGS with that of standard genetic testing in a randomized, controlled study. We seek to also address important questions of which diseases and presentations rapid WGS does or does not have diagnostic effectiveness, those in which diagnoses change acute medical management, and whether there are potential harms of rapid WGS in the NICU. The provision of a diagnosis frequently holds power for a family regardless of the impact on clinical management. Families speak of ‘not having to search anymore,’ of ‘being able to give a name to the disease,’ and feel like they can ‘stop looking.’ With increased connectedness, this also allows families of children with rare diseases to find support from others with the same or similar diagnosis, even when separated by great distances. Thus, rapid WGS is also anticipated enable personalized genetic counseling and, in some cases, allow a more natural death with retraction of medical technology that separates the parents and child. This study will further allow the collection of multiple use cases, from which initial answers to these questions can be informed. In particular, there is a great need to define processes for clear communication of genetic disease diagnoses with counseling and support to help parents to process this information and navigate their options for their babies who will have a future of major morbidity from a genetic disease. While a diagnosis alone is impactful for families, the great hope for rapid WGS-based diagnoses is that earlier instigation of precise, effective care – before irreversible organ damage or disease progression – will result in change in care and improved clinical outcomes in a subset of infants (Box 1, Figure 1). While that subset may be small today, the ability to make a timely diagnosis may render some genetic diseases tractable from a pharmaceutical development standpoint[39]. Currently, for many genetic diseases of newborn onset there is not an evidenced-based therapeutic literature. These diseases are individually very rare, thus, precluding adequate power for standard randomized trial designs of investigational new treatments (INDs) that compare multiple strategies for optimal efficacy[39]. Newborn-onset genetic diseases also frequently have rapid progression – 120-day mortality was more than 50% in our case series[4]. This rapid progression, when combined with delayed molecular diagnosis, negates any window for consideration or implementation of INDs. Rapid WGS has the potential to alter this dynamic by efficiently identifying diagnoses that are so rare as to be without accepted treatments thereby creating the time interval needed to define and implement a modified N-of-1 therapeutic study guided by the effected biological pathway involving currently unproven treatments[39]. Such N-of-1 studies would identify novel therapeutic approaches by examination of the mechanism whereby a mutated gene causes pathophysiology through literature review, seeking molecular opportunities to intervene, and evaluating other genes for which drugs have been developed with which the disease gene product may interact to cause pathophysiology[39]. N-of-1 studies would also be based on the premise of case reports of prior novel therapeutic interventions in that specific disease, which, although not rising to the status of a proven treatment, merit additional study. Currently approved medications (which are likely to be off-label for these very rare conditions) and/or dietary supplements could then be repurposed in the setting of a structured, IRB-approved, N-of-1 study, with defined end-points, surrogate biochemical effect markers, and dose escalation[39].
Box 1. Precision Medical Management following rapid genetic disease diagnosis in the NICU.
-
1
Psychosocial benefits for parents (answers, knowledge of prognosis, planning, psychological and religious support).
-
2
Precision treatments for affected infants that prevent death, diminish disease severity, delay progression or improve quality of life.
-
4
Earlier avoidance of futile or painful treatments, unnecessary or invasive testing, and planning of withdrawal of care.
-
5
Time to plan and implement investigative new treatments.
-
6
Basis for increased coordination of care among providers.
-
7
Genetic counseling regarding recurrence risk.
-
8
Parental referral to specific support groups.
-
9
Reduced lifetime cost of care.
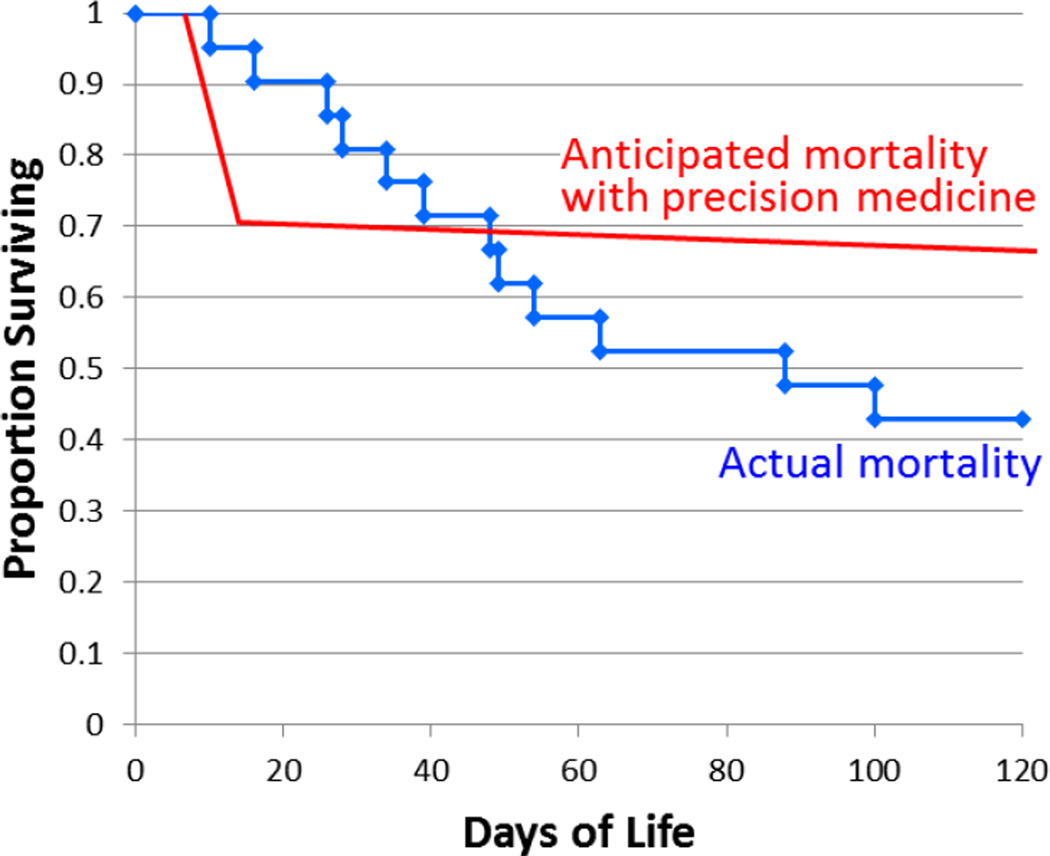
Figure 1.
Actual and desired 120-day mortality of NICU infants receiving rapid genetic disease diagnoses. In suffering neonates with hopeless diagnoses, rapid diagnosis will allow planned withdrawal of support in a more timely manner. Precision medicine interventions in remaining infants are anticipated to reduce infant mortality.
For example, Kabuki syndrome is a rare genetic disease characterized by typical facial, minor skeletal anomalies, intellectual disability, and growth deficiency. Patients with Kabuki syndrome frequently have increased susceptibility to infections and autoimmune disorders, seizures, endocrine abnormalities, feeding problems, and deafness[40]. Kabuki syndrome is caused by heterozygous mutation in KMT2D or KDM6A. The FDA approved antibiotic, gentamicin, can induce read-through of nonsense codons that result in haploinsufficiency of KMT2D and KDM6A in cell lines derived from patients with Kabuki syndrome[41]. Thus, an N-of-1 study of gentamicin[42, 43] in Kabuki syndrome could potentially be designed, in which the expression of KMT2D target genes and post-natal growth were employed as a surrogate markers of treatment effect[41]. Markers of potential adverse effects of nephrotoxicity and ototoxicity would also require careful monitoring in the case of gentamicin use.
Through the development of a culture of N-of-1 studies of genetic diseases in the NICU, a knowledge base of treatments of rare diseases can eventually be built. It should be noted that many genes act through common biochemical pathways, so evidence generated in one genetic disease may support the use of a treatment in another, guided by an understanding of gene pathways.
In this protected population, precision neonatology must avoid heroic efforts that prolong suffering without the promise of significant therapeutic benefit. Guiding these measures must be respect for the infant and family, with benefits maximized, and risks must be reasonable and minimized. Parents must be informed of the experimental nature of such trials and only non-exploitative procedures should be used. The challenge going forward will be to develop teams and practices that can respond to rapid genetic diagnosis with specific interventions and treatments, including pharmacologic interventions, pulling from multispecialty teams that will realize the promise of precision medicine[39].
In cases where diagnosis leads to either palliation or effective treatments, rapid WGS is anticipated to shorten hospital stays, reduce empiric treatments, and simplify the diagnostic work-up increasing the cost-effectiveness of rapid genomic analysis. There are multiple current formats for WGS at present and an acuity guided strategy seems most appropriate at present, where rapid WGS is reserved for acutely ill infants in whom a diagnosis may be genetic. Exome sequencing, for example, costs about one third that of standard WGS but incurs about one day of additional turnaround time. Standard WGS takes approximately 5 days of additional turnaround time. Two day WGS is about three to four times more expensive than standard WGS. Even faster formats are now possible. We can now reproducibly perform WGS and analysis in ~30 hours, and strategies have been described that can reduce this to less than 20 hours[44]. From a practical standpoint, the greatest benefit is likely if rapid WGS is instituted within the first day of life and completed within 2 days (Figure 2). As noted above, average age at NICU enrollment was 23 days[4], somewhat blunting the current impact of diagnosis and certainly arguing that a standard WGS or exome protocol would provide comparable results in most cases.
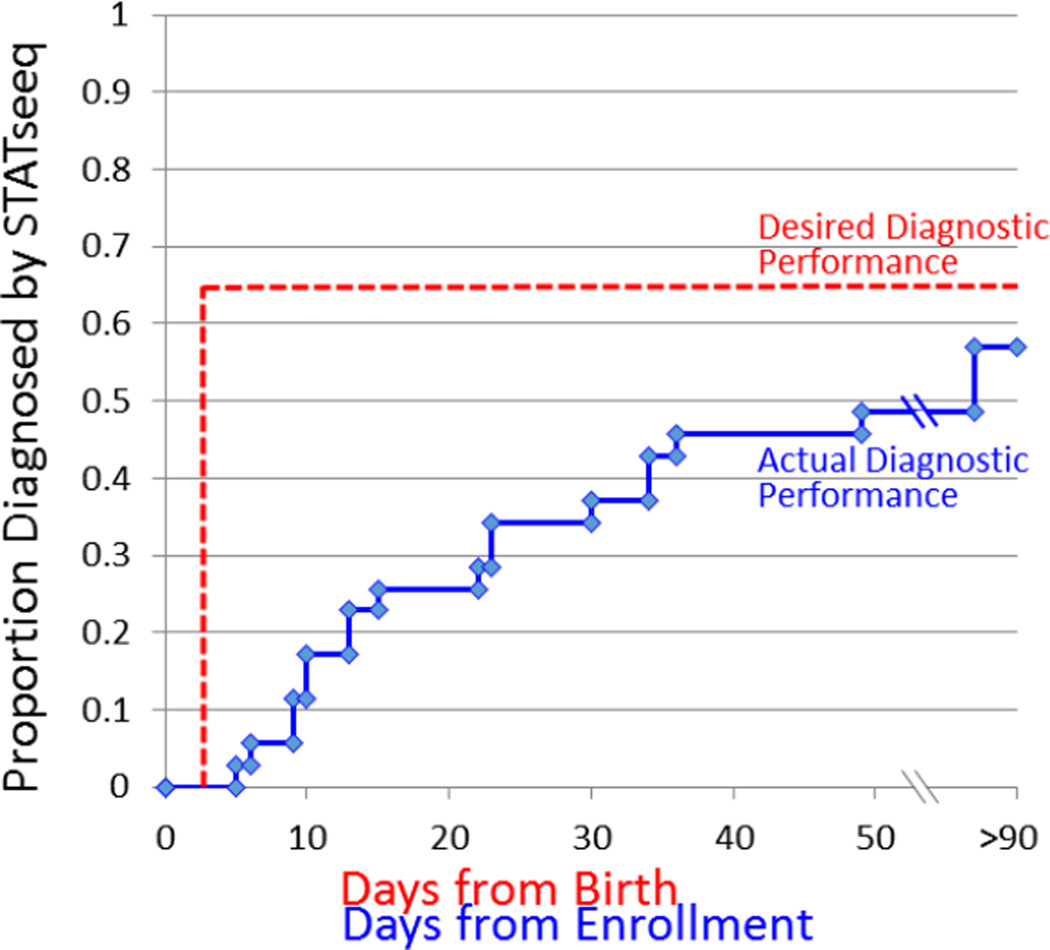
Figure 2.
Actual and desired time to genetic disease diagnosis by rapid WGS (STATseq). Ideally, blood samples would be obtained at birth and diagnoses would be returned by DOL 2 to optimize provision of precision medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Joshua E. Petrikin, The University of Missouri Kansas City School of Medicine, Department of Pediatrics, Division of Neonatal and Perinatal Medicine, Director of Neonatal Genomics, Center for Pediatric Genomic Medicine, Children's Mercy Hospital Kansas City, Kansas City, Missouri 64108, Phone: 816-701-4806, Fax: 816-802-1111, jepetrikin@cmh.edu.
Laurel K. Willig, The University of Missouri, Kansas City School of Medicine, Department of Pediatrics, Division of Pediatric Nephrology, Center for Pediatric Genomic Medicine, Children's Mercy Hospital Kansas City, Kansas City, Missouri 64108 USA, Phone: 816-701-4806, Fax: 816-802-1111, lkwillig@cmh.edu.
Laurie D. Smith, The University of Missouri Kansas City School of Medicine, Department of Pediatrics, Center for Pediatric Genomic Medicine, Children's Mercy Hospital Kansas City, Kansas City, Missouri 64108 USA, Phone: 816-701-4806, Fax: 816-802-111, ldsmith@cmh.edu.
Stephen F. Kingsmore, Dee Lyons/Missouri Endowed Chair in Pediatric Genomic Medicine, Department of Pediatrics, Department of Pathology and Laboratory Medicine, Director, Center for Pediatric Genomic Medicine, Children's Mercy Hospital Kansas City, Kansas City, Missouri, 64108 USA, Phone: 816-701-4806, Fax: 816-802-1111, sfkingsmore@cmh.edu.
References
- 1.F. C. Francis Collins Says Medicine in the Future Will Be Tailored to Your Genes. Wall Street Journal. 2014 [Google Scholar]
- 2.Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Science translational medicine. 2012;4(154):154ra35. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soden SE, Saunders CJ, Willig LK, Farrow EG, Smith LD, Petrikin JE, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Science translational medicine. 2014;6(265):265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willig L, Petrikin J, Smith L, Saunders C, Thiffault I, Miller N, et al. Retrospective analysis of diagnostic and clinical findings among critically ill infants receiving rapid whole genome sequencing for identification of mendenlian disorders. The Lancet Respiratory Medicine. 2015 doi: 10.1016/S2213-2600(15)00139-3. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genome of the Netherlands C. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nature genetics. 2014;46(8):818–825. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 6.Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Human mutation. 2003;21(1):12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- 7.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nature reviews Genetics. 2012;13(8):565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 8.Crow JF. The origins, patterns and implications of human spontaneous mutation. Nature reviews Genetics. 2000;1(1):40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 9.Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nature reviews Genetics. 2007;8(8):610–618. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]
- 10.Project. GG. 2012 Available from: https://globalgenes.org/wp_content/uploads/2013/06/Review2012_brochure_web.pdf.
- 11.Martin JA, Hamilton BE, Ventura SJ, Osterman MJ, Mathews TJ. Births: final data for 2011. National vital statistics reports : from the Centers for Disease Control and Prevention, National Center for Health Statistics, National Vital Statistics System. 2013;62(1):1–69. 72. [PubMed] [Google Scholar]
- 12.Dimes. Mo. Special care nursery admissions. 2011 Available from: https://www.marchofdimes.org/peristats/pdfdocs/nicu_summary_final.pdf. [Google Scholar]
- 13.Hamilton BE, Hoyert DL, Martin JA, Strobino DM, Guyer B. Annual summary of vital statistics: 2010–2011. Pediatrics. 2013;131(3):548–558. doi: 10.1542/peds.2012-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FitzPatrick DR, Skeoch CH, Tolmie JL. Genetic aspects of admissions to a paediatric intensive care unit. Archives of disease in childhood. 1991;66(5):639–641. doi: 10.1136/adc.66.5.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Acikalin A, Bagir EK, Torun G, Ates BT, Erdogan S, Uguz A, et al. Perinatal autopsy evaluation of 2150 autopsies in the Cukurova region of Turkey. Turk patoloji dergisi. 2014;30(3):189–194. doi: 10.5146/tjpath.2014.01266. [DOI] [PubMed] [Google Scholar]
- 16.Cunniff C, Carmack JL, Kirby RS, Fiser DH. Contribution of heritable disorders to mortality in the pediatric intensive care unit. Pediatrics. 1995;95(5):678–681. [PubMed] [Google Scholar]
- 17.McCandless SE, Brunger JW, Cassidy SB. The burden of genetic disease on inpatient care in a children's hospital. American journal of human genetics. 2004;74(1):121–127. doi: 10.1086/381053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simpson CD, Ye XY, Hellmann J, Tomlinson C. Trends in cause-specific mortality at a Canadian outborn NICU. Pediatrics. 2010;126(6):e1538–e1544. doi: 10.1542/peds.2010-1167. [DOI] [PubMed] [Google Scholar]
- 19.Yoon PW, Olney RS, Khoury MJ, Sappenfield WM, Chavez GF, Taylor D. Contribution of birth defects and genetic diseases to pediatric hospitalizations. A population-based study. Archives of pediatrics & adolescent medicine. 1997;151(11):1096–1103. doi: 10.1001/archpedi.1997.02170480026004. [DOI] [PubMed] [Google Scholar]
- 20.Zlotogora J, Leventhal A, Amitai Y. The impact of congenital malformations and Mendelian diseases on infant mortality in Israel. The Israel Medical Association journal : IMAJ. 2003;5(6):416–418. [PubMed] [Google Scholar]
- 21.Carmichael SL. Birth defects epidemiology. European journal of medical genetics. 2014;57(8):355–358. doi: 10.1016/j.ejmg.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Weiner J, Sharma J, Lantos J, Kilbride H. How infants die in the neonatal intensive care unit: trends from 1999 through 2008. Archives of pediatrics & adolescent medicine. 2011;165(7):630–634. doi: 10.1001/archpediatrics.2011.102. [DOI] [PubMed] [Google Scholar]
- 23.Behrman R, Butler A. Societal costs of preterm birth. In: Behrman R, Butler A, editors. Preterm birth-causes, consequences and prevention. Washington, DC: National Academies; 2007. pp. 398–429. [PubMed] [Google Scholar]
- 24.Hack M, Taylor HG, Drotar D, Schluchter M, Cartar L, Andreias L, et al. Chronic conditions, functional limitations, and special health care needs of school-aged children born with extremely low-birth-weight in the 1990s. Jama. 2005;294(3):318–325. doi: 10.1001/jama.294.3.318. [DOI] [PubMed] [Google Scholar]
- 25.Wachs TD, Black MM, Engle PL. Maternal Depression: A global threat to children's health, development, and behavior and to human rights. Child Development Perspectives. 2009;3:51–59. [Google Scholar]
- 26.Hall JG. The impact of birth defects and genetic diseases. Archives of pediatrics & adolescent medicine. 1997;151(11):1082–1083. doi: 10.1001/archpedi.1997.02170480012002. [DOI] [PubMed] [Google Scholar]
- 27.Kingsmore SF, Saunders CJ. Deep sequencing of patient genomes for disease diagnosis: when will it become routine? Science translational medicine. 2011;3(87):87ps23. doi: 10.1126/scitranslmed.3002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohler S, Doelken SC, Rath A, Ayme S, Robinson PN. Ontological phenotype standards for neurogenetics. Human mutation. 2012;33(9):1333–1339. doi: 10.1002/humu.22112. [DOI] [PubMed] [Google Scholar]
- 29.Kohler S, Schulz MH, Krawitz P, Bauer S, Dolken S, Ott CE, et al. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. American journal of human genetics. 2009;85(4):457–464. doi: 10.1016/j.ajhg.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grada A, Weinbrecht K. Next-generation sequencing: methodology and application. The Journal of investigative dermatology. 2013;133(8):e11. doi: 10.1038/jid.2013.248. [DOI] [PubMed] [Google Scholar]
- 31.Ng PC, Kirkness EF. Whole genome sequencing. Methods in molecular biology. 2010;628:215–226. doi: 10.1007/978-1-60327-367-1_12. [DOI] [PubMed] [Google Scholar]
- 32.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26(16):2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maddalena A, Bale S, Das S, Grody W, Richards S Committee ALQA. Technical standards and guidelines: molecular genetic testing for ultra-rare disorders. Genetics in medicine : official journal of the American College of Medical Genetics. 2005;7(8):571–583. doi: 10.1097/01.gim.0000182738.95726.ca. [DOI] [PubMed] [Google Scholar]
- 34.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genetics in medicine : official journal of the American College of Medical Genetics. 2008;10(4):294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 35.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic acids research. 2012;40(Database issue):D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shashi V, McConkie-Rosell A, Rosell B, Schoch K, Vellore K, McDonald M, et al. The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genetics in medicine : official journal of the American College of Medical Genetics. 2014;16(2):176–182. doi: 10.1038/gim.2013.99. [DOI] [PubMed] [Google Scholar]
- 37.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine : official journal of the American College of Medical Genetics. 2015 doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith L, Kingsmore S. N-of-1 genomic medicine for the rare pediatric genetic diseases. Expert opinion on orphan drugs. 2014;12(2):1279–1290. [Google Scholar]
- 40.Adam MP, Hudgins L, M H. Seattle (WA): University of Washington, Seattle; 2011. Kabuki Syndrome. [updated Sep 1 [Updated 2013 May 16]. cited 1993–2105]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK62111/. [PubMed] [Google Scholar]
- 41.Micale L, Augello B, Maffeo C, Selicorni A, Zucchetti F, Fusco C, et al. Molecular analysis, pathogenic mechanisms, and readthrough therapy on a large cohort of Kabuki syndrome patients. Human mutation. 2014;35(7):841–850. doi: 10.1002/humu.22547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bushby K, Finkel R, Wong B, Barohn R, Campbell C, Comi GP, et al. Ataluren treatment of patients with nonsense mutation dystrophinopathy. Muscle & nerve. 2014;50(4):477–487. doi: 10.1002/mus.24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryan NJ. Ataluren: first global approval. Drugs. 2014;74(14):1709–1714. doi: 10.1007/s40265-014-0287-4. [DOI] [PubMed] [Google Scholar]
- 44.Kingsmore SF. personal communication. 2015 [Google Scholar]