Abstract
Primary mediastinal B-Cell lymphoma (PMBL) is a recently defined entity comprising ~2–10% non-Hodgkin lymphomas (NHL). Unlike most NHL subtypes, PMBL lacks recurrent gene rearrangements to serve as biomarkers or betray target genes. While druggable, late chemotherapeutic complications warrant the search for new targets and models. Well characterized tumor cell lines provide unlimited material to serve as preclinical resources for verifiable analyses directed at the discovery of new biomarkers and pathological targets using high throughput microarray technologies. The same cells may then be used to seek intelligent therapies directed at clinically validated targets. Four cell lines have emerged as potential PMBL models: FARAGE, KARPAS-1106P, MEDB-1 and U-2940. Transcriptionally, PMBL cell lines cluster near c(lassical)-HL and B-NHL examples showing they are related but separate entities. Here we document genomic alterations therein, by cytogenetics and high density oligonucleotide/SNP microarrays and parse their impact by integrated global expression profiling. PMBL cell lines were distinguished by moderate chromosome rearrangement levels undercutting cHL, while lacking oncogene translocations seen in B-NHL. In total 61 deletions were shared by two or more cell lines, together with 12 amplifications (≥4x) and 72 homozygous regions. Integrated genomic and transcriptional profiling showed deletions to be the most important class of chromosome rearrangement. Lesions were mapped to several loci associated with PMBL, e.g. 2p15 (REL/COMMD1), 9p24 (JAK2, CD274), 16p13 (SOCS1, LITAF, CIITA); plus new or tenuously associated loci: 2p16 (MSH6), 6q23 (TNFAIP3), 9p22 (CDKN2A/B), 20p12 (PTPN1). Discrete homozygous regions sometimes substituted focal deletions accompanied by gene silencing implying a role for epigenetic or mutational inactivation. Genomic amplifications increasing gene expression or gene-activating rearrangements were respectively rare or absent. Our findings highlight biallelic deletions as a major class of chromosomal lesion in PMBL cell lines, while endorsing the latter as preclinical models for hunting and testing new biomarkers and actionable targets.
Introduction
Primary mediastinal B-Cell lymphoma arises in the mediastinum from transformed thymic B-cells and comprises 2–10% NHL. According to microarray profiling, PMBL is distinct from both germinal center and activated diffuse large B-cell lymphomas (DLBCL) bearing the closest pathological resemblance to classical Hodgkin lymphoma (cHL) nodular sclerosing subtype and mediastinal grey zone lymphoma.
Although PMBL responds initially to chemotherapy subsequent poor prognostic outcomes warrant the search for new targets and disease models [1, 2]. Like cHL, but unlike most NHL subtypes, PMBL lacks recurrent gene rearrangements to serve as diagnostic or prognostic biomarkers or portals to oncogenic drivers, and hence, potential therapeutic targets. PMBL and cHL show alterations at three loci, 2p16 (~50%), 9p24 (~75%), and 16p13 (~45%) [3–5]. Doubt has been cast on the clinical significance of SOCS1 the mooted target at 16p13 [6], while genomic neighbors of JAK2 the preferred candidate at 9p24, namely CD274/PDL1, PDCD1LG2/PDL2 which serve to fatigue reactive T-cells have emerged as alternative targets [7]. Recently, inactivating mutations of PTPN1 have been reported in both PMBL and cHL [8] compounding the list of targets shared by these entities.Low incidence has impeded ascertainment of oncogenomic changes in PMBL [2]. Should key changes be indeed found these may turn out to be rare or cryptic. By permitting in depth studies well characterized tumor cell lines have helped unravel the pathology of such rare or pathologically intractable cancers [9]. In the light of revised PMBL diagnostic criteria four well characterized PMBL cell lines have recently emerged [10]. The advent of forensic DNA profiling promises to dispel the threat of cross contamination widely perceived as a major hindrance [11].
In the quest for PMBL biomarkers and pathological targets we have assembled a panel of PMBL cell lines and documented genomic alterations therein using high density arrays offering circa 40–80x improvements over earlier studies. Candidacies of gene targets were evaluated by parallel expression array profiling and reference clinical data. Several new or unfamiliar potential oncogenomic targets were thus identified. Concordance with clinical data thus observed strengthens the validity of PMBL cell lines as useful models and resources.
Materials and Methods
Cell lines
FARAGE was established before 1992 from the lymph node of a 70-year old female at diagnosis of DLBCL sited parasternally [12]. The close similarity of its DNA methylation profile to primary PMBL cells warrants reassignment to that entity [13]. In 1984 KARPAS-1106P and its phenotypically indistinguishable sibling KARPAS-1106A were respectively established from a pleural effusion (at diagnosis) and ascites (during disease progression) of a 23-year old female with “mediastinal lymphoblastic B-NHL” [14]. MEDB-1 was derived in 1981 from the mediastinal mass of a 27-year old male with PMBL (mediastinal B-cell non Hodgkin lymphoma [B-NHL] stage IIb) during relapse [15]. U-2940 was established in 1990 from an 18-year old female diagnosed with B-NHL with mediastinal features subsequent to treatment for cHL [16], but recently reassigned to PMBL [17, 18]. The particulars and culture of these and remaining cell lines are detailled elsewhere [10, 19].
DNA profiling
To eliminate the risk of cross contamination STR DNA profiling was carried out using fluorescent PCR in combination with capillary electrophoresis as described previously [20]. Briefly, the PowerPlex VR 1.2 system (Promega, Mannheim, Germany) was set to run two-color DNA profiling allowing the simultaneous single-tube amplification of eight polymorphic STR loci plus amelogenin (gender). Loci were amplified by primers labeled with the Beckman/Coulter dye D3 (green; Sigma-Aldrich, Munich/Germany), while the STR loci D16S539, D7S820, D13S317 and D5S818 were amplified using primers labeled with D2 (black). Data (Table A in S1 File) were analyzed with the CEQ 8000 software (Beckman-Coulter, Krefeld, Germany), which enables an automatic assignment of genotypes and automatic export of resulting numeric allele codes into the reference DNA database of the DSMZ.
Cytogenetic analysis
Cytogenetic analyses were conducted as described previously [21]. Imaging was performed using a Zeiss Axioplan (Oberkochen/Germany) microscope configured to a Spectral Karyotyping (SKY) system (ASI Ltd, Migdal Haemek/Israel). Briefly, bacterial artificial chromosome (BAC) and fosmid clones were obtained from BACPAC Resources, Children's Hospital, Oakland, CA/USA and DNA labeled by nick translation with d-UTP fluors (Dyomics (Jena/Germany). Cell lines were investigated by fluorescence in situ hybridization (FISH) using BAC/fosmid tilepath clones which flanked or straddled loci consistently rearranged in lymphoid neoplasms including, BCL2, BCL6, BCL11A, CCND1, CCND3, IG-H/K/L, JAK2, LMO2, MYC, PAX5, REL.
Genomic Array Data
CytoScan High Density Arrays which combine oligonucleotide and SNP probes (Affymetrix, High Wycombe/UK) were used to detect genomic copy number alterations/gains/losses (CNA/G/L), losses of heterozygosity (LOH) and unbalanced chromosome translocation breakpoints at high resolution. DNA was prepared using the Qiagen Gentra Puregene Kit (Hilden/Germany). Labeling, hybridization and washing were performed using the recommended kits and CytoScan HD arrays according to the manufacturers protocols. Quality control criteria were those set by the manufacturer. Data were subsequently analyzed using the Chromosome Analysis Suite software version 2.0.1.2 (Affymetrix). This allows access to the Database of Genomic Variants (DGV) (http://dgv.tcag.ca/dgv/app/home) for immediate identification of polymorphic CNV.
Expression profiling
Total RNA (500 ng) were labelled with biotin according to the 3´ IVT Express Kit (Affymetrix). Circa 7.5 μg of biotinylated cDNA were fragmented and placed in a cocktail containing four biotinylated hybridization controls (BioB, BioC, BioD, and Cre) as recommended by the manufacturer. Samples were hybridized to Affymetrix GeneChip HG-U133 2.0 Plus for 16 h at 45°C. Washing and staining were performed with the fluidics station 450 according to the recommended FS450 protocol. Image analysis was performed on GCS3000 Scanner and GCOS1.2 Software Suite (Affymetrix). Analyses of microarray data were performed using GeneSpring 11.5.1 (Agilent, Santa Clara/CA/USA). Signal intensities (raw data) were log2 transformed and normalized. Comparison datasets were generously provided by Prof. Andreas Rosenwald (Institute of Pathology, University of Würzburg, Germany) or obtained from the BROAD Institute (www.broadinstitute.org). For creation of heat maps we used the software CLUSTER version 2.11 and TREEVIEW version 1.60 (http://rana.lbl.gov/EisenSoftware.htm). For clustering the algorithm was set to Euclidean distance based on p-values confirmed by multiscale bootstrap resampling [22]. For microRNA profiling the Affymetric GeneChip miRNA 2.0 system was used together with samples drawn from the DSMZ cell bank as comparison data sets. For data analysis Affymetrix cel-files were loaded into R/Bioconductor (3.1.0/2.14) using package oligo. External SET2 data are publicly available on GEO (GSM836163, GSM836164, GSM836165). Before principal component analysis (PCA), data were preprocessed, normalized and 10% of the most variable miRNA taken for calculating principal components.
qPCR
Reverse transcriptase and qPCR used total RNA extracted using the RNEasy Kit (Qiagen). cDNA was subsequently synthesized from 3 μg RNA by random priming, using Superscript II (Invitrogen, Darmstadt/Germany). qPCR was performed by the 7500 Fast Real-time PCR System (Applied Biosystems, Darmstadt/Germany), using SsoFast EvaGreen Real Mastermix (BIORAD, Hercules, CA/USA) and custom designed oligonucleotides (MWG Eurofins, Martinsried/Germany–for sequences see Table B in S1 File). For normalization of expression levels we used TATA box binding protein (TBP). Quantitative analyses were performed in triplicate and repeated twice. The analysis of relative quantitative expression was performed using the Applied Biosystems software (2^ΔΔCt), followed by statistical analysis and data visualization using the R-based ggplot2 package [23].
Results
Authentication
STR profiles of PMBL cell lines are given in Table A in S1 File, all testing unique among circa 3000 known cell line profiles provided by the major cell banks (https://www.dsmz.de/services.html). Positive authentication was subsequently provided by cytogenetics (see below).
Transcriptional clustering and principal component analysis
Unsupervised clustering of microarray expression data were displayed as a dendrogram (S1A Fig) showing that PMBL and cHL cell lines jointly diverge from DLBCL, reflecting their supposed pathologic relationships (replotted from ref. 10). PCA of global microRNA expression arrays again showed the coherence of the PMBL group and their proximity to DLBCL, while segregated from T-cell lymphoma and myelomonocytic neoplasms (S1B Fig). Thus, PMBL cell lines form a coherent and separate entity neighboring cHL, and then DLBLCL.
Cytogenetics
Consensus ISCN karyotypes [24] of the four PMBL cell lines recorded at the DSMZ and previously by their originators were as follows:
FARAGE: 46(41–46)<2n>XX; no consistent abnormality detected. The originator karyotype was: idem +11. Loss of a chromosome 11 homolog is probably attributable to clonal divergence.
KARPAS-1106P: 49(45–51)<2n>XXX,+X,i(X)(p10),der(X)dup(X)t(X;13)(q25;q11), del(2)(p16p22), der(3)t(2;3)(p25;p14), +9,i(9)(p10),dup(12)(q11q14.1),del(14)(q12q21), del(15)(q11q15),+17, del(18)(q21q22), dup(18)(q12q21),+20,del(20)(q13.1q13.3)x2,add(21)(p13); and that of the originator: 49,XX,der(X)t(X;13;18)(q28;q12;q21),i(Xp),del(2)(p11.2p13), der(3)t(2;3)(p13;p25), +i(9p), ins(12;?)(q13q13),del(14)(q11.2q13),del(15)(q11q15), der(18)t(X;13;18)(q28;q12.;q21),-20, del(20)(q13q13)x2. Thus, both karyotypes are closely similar.
MEDB-1: 47(41–47)<2n>XY,inv(X)(p21;q12),+1,der(1)t(1;14)(p11;q11),t(2;12)(p24;p12),+9, der(10)t(10;20)(q25)(q11),-14,i(21)(q10). The originator karyotype was: 47,XY,inv(X)(p22q13), +der(1)t(1;14)(q10;q10),+9,-14,-21,i(21q), again closely resembling that observed.
U-2940: 45(43–45)X,-X,del(3)(p14p21),del(6)(q13q15),der(7)t(2;7)(q22;p22), dup(12)(q13q22), der(14)t(X;14)(q12;p11),t(16;16)(p12;p13),del(17)(p13); and that of the originator: 45–46,-X,del(3)(p13p21),del(6)(q14q16),del(7)(p11),+i(7)t(2;7)(q23;q31), dup(12)(q12q21), der(14)del(X)(q21.1q21.3)t(X;14)(q11;p11),t(16;16)(p12;p13.3), again resembling that observed.
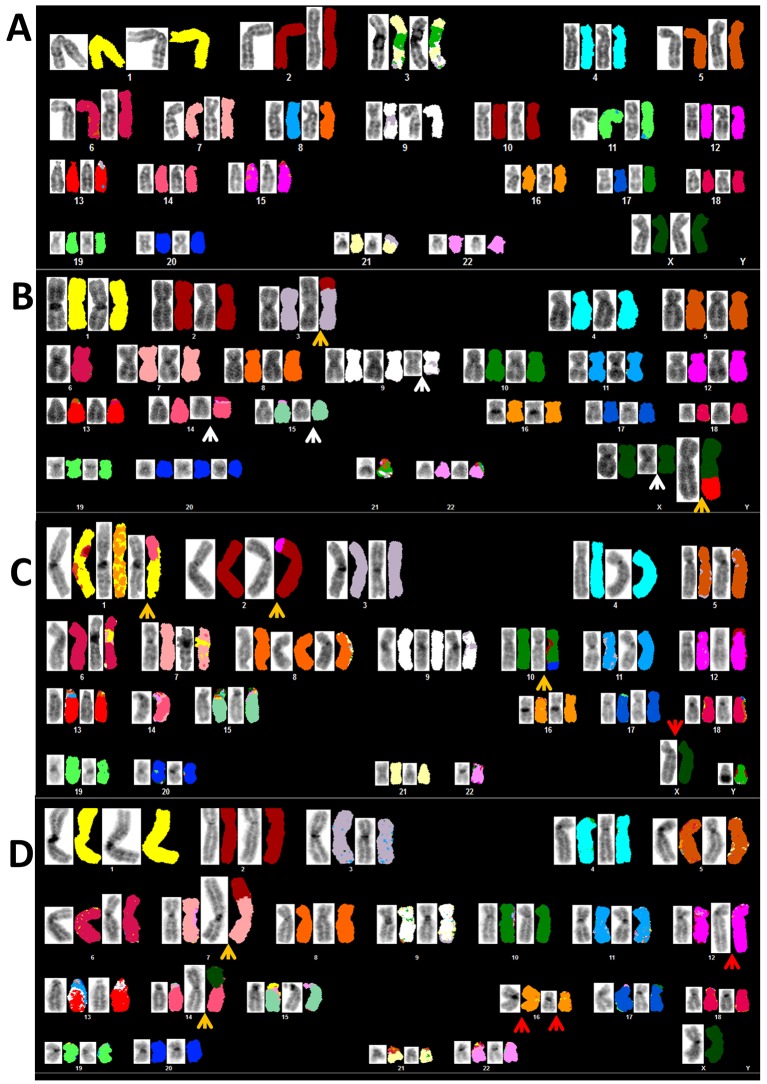
For representative SKY images see Fig 1A–1D.
Fig 1. Spectral Karyotyping (SKY): FARAGE (A), KARPAS-1106P (B), MEDB-1 (C) and U-2940 (D).
Inverse DAPI G-banding shown left of corresponding pseudocolored SKY. Arrows show deletions (white), duplications (red), and translocations (ochre). After trypsin G-banding (not shown) analyses were extended to prepare consensus karyotypes. Individual metaphases sometimes departed from consensus karyotypes, e.g. note loss of Y-chromosome in MEDB-1. Note absence of key cytogenetic rearrangements and relative lack of rearrangement when compared to cHL cell lines. Thus the salient cytogenetic features of PMBL are essentially negative with respect to neighboring entities, cHL and PMBL.
Thus, karyotypes of all four cell lines matched the originators’ data, both confirming their stability in vitro and authenticity suggested by their unique STR profiles (Table A in S1 File).
FISH analyses using BAC clones covering lymphoma breakpoints (BCL2, BCL6, BCL11A, CCND1, CCND3, IG-H/K/L, JAK2, LMO2, MYC, PAX5, REL) all tested normal, excluding rearrangements at these loci. Thus, although most PMBL cell line karyotypes bore chromosome translocations, none target known common lymphoma breakpoints.
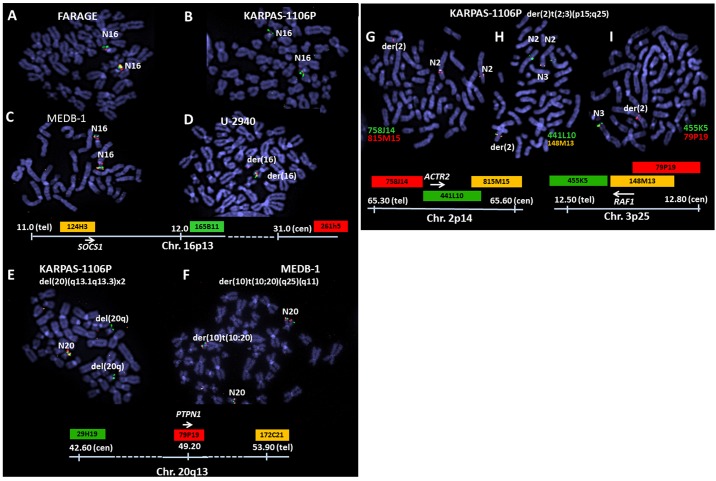
Cytogenetically visible deletions at 16p13 hosting SOCS1 were present in 3/4 cell lines: monoallelic in FARAGE, biallelic in Karpas-1106P and U-2940, but absent from MEDB-1 (Fig 2A–2D). Only in U-2940 was this rearrangement microscopically visible without FISH. Deletions of 20q13 were present in KARPAS-1106P (Fig 2E) but not in MEDB-1 (Fig 2F). This rearrangement effected PTPN1 monosomy in KARPAS-1106P, while in MEDB-1 where it is mutated [8], PTPN1 escaped deletion.
Fig 2. Fluorescence in situ hybridization (FISH) Analysis.
A-D: Shows the cytogenetic configuration of focal deletions affecting 16p13 including the SOCS1 locus (arrows) in three PMBL cell lines: monoallelic in FARAGE, biallelic in KARPAS1106P and U-2940 effected by t(16;16) rearrangement, but absent in MEDB-1. E/F: Shows deletions affecting the PTPN1 locus in KARPAS-1106P (E) and MEDB-1 (F). Twin partial chromosome 20 long-arm deletions in KARPAS-1106P effect PTPN1 monosomy, while in MEDB-1 where the gene is mutated this locus escapes deletion despite proximity to a translocation breakpoint therein. G-I: Shows analysis of der(3)t(2;3)(p14;p25) in KARPAS-1106P. The respective breakpoints at 2p14 and 3p25 were placed close to ACTR2 (within clone RP11-441L10) and RAF1 (within RP11-148M13). Coordinates and labelling scheme are shown below. Coordinates (MBp) are from HG19. FISH was performed using tilepath BAC clones.
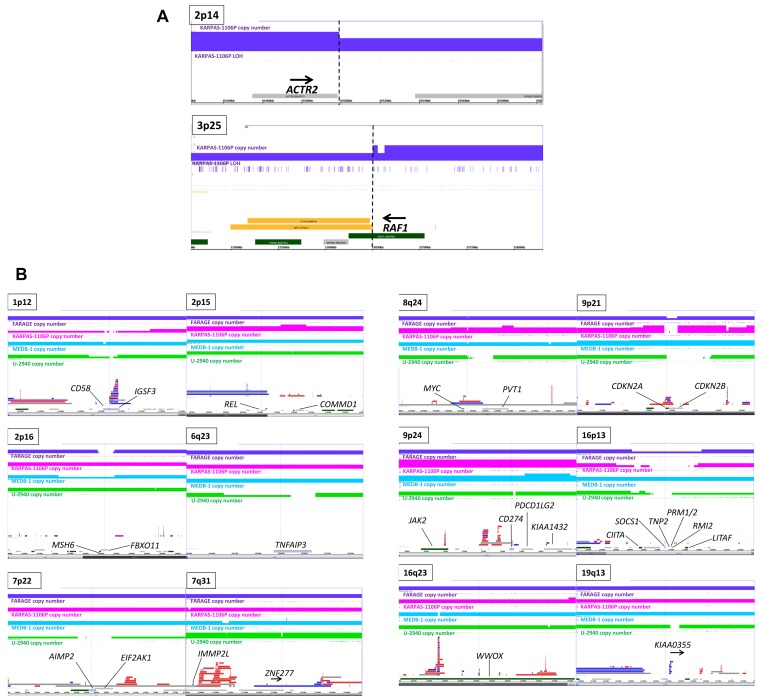
FISH mapped breakpoints of unbalanced der(3)t(2;3)(p14;p25) present in KARPAS-1106P to ACTR2 at 2p25 and RAF1 at 3p14 (Fig 2G–2I). Refinement by genomic array (Fig 3A) placed the 2p14 breakpoint just upstream of the ACTR2 coding region and that at 3p25 inside RAF1 consistent with FISH data. Although ACTR2-RAF1 mRNA fusion has been reported by RNAseq [25], we were unable to detect hybrid mRNA by RT-PCR using published coordinates (S2 Fig). Moreover, formation of the reported chimeric ACTR2-RAF1 mRNA demands an additional cryptic inversion to generate the required ORF.
Fig 3. Genomic array data.
A: The respective breakpoints and FISH clones at ACTR2 and inside RAF1 of der(2)t(2;3)(p14;p25) in KARPAS-1106P as revealed by genomic arrays expands the FISH data. B: Color coded plots of FARAGE (purple), KARPAS-1106P (pink), MEDB-1 (blue), U-2940 (green)—show genomic copy number (solid) and LOH (barred) at 12 loci (1p12, 2p15, 2p16, 6q23, 7p22, 7q31, 8q24, 9p21, 9p24, 16p13, 15q23, 19q13, together with OMIM genes below. Copy number polymorphic regions (http://dgv.tcag.ca/dgv/app/home) are shown between, listing gains (blue), losses (red), and copy number neutral alterations, such as inversions (gray).
Array comparative genomic hybridization (aCGH) and single nucleotide polymorphism (SNP)
Copy number variations/amplifications/deletions (CNV/A/D) and zygosity were investigated with high density combined oligonucleotide SNP arrays. Images depicting each chromosome are shown in S3 Fig Candidate targets of focal CNV, specifically protein coding and RNA genes located within higher level CNA (≥4x) and shared biallelic CND, are respectively listed in Table 1A and 1B, and those of shared monoallelic CND given in Table 1C. Twelve fourfold-or more amplified regions were present (Table 1A) of which four were shared with two or more cell lines, together with a further 12 shared biallelic/monoallelic deletions (Table 1B); these are discussed below. In addition, 49 monoallelic shared deletions were documented (Table 1C).
Table 1. Higher-Level Copy Number Alterations and recurrent deletions in PMBL Cell Lines.
| Chr. band | Coordinates (KBp) | Cell lines | Genes in region of interest | Co-incident previous studies | ||||
|---|---|---|---|---|---|---|---|---|
| FARAGE | KARPAS-1106P | MEDB-1 | U-2940 | Protein coding | Noncoding RNA | |||
| A: Gains (≥4x) | ||||||||
| 1p31 | 72309–72321 | 2 | 2 | 4 | 2 | NEGR1 (part) | - | |
| 1q31 | 193155–193158 | 2 | 2 | 4 | 4 | B3GALT2 (upstream), CDC73 (part) | - | |
| 2p15 | 61679–62435 | 2 | 4 | 2 | 2 | XPO1, FAM161A, CCT4, COMMD1, B3GNT2 (part) | - | [6, 26] |
| 2p14 | 66213–66220 | 4 | 3 | 4 | 2 | SLC1A4 (part) | - | |
| 2q33 | 207864–208167 | 2 | 2 | 2 | 5x | KLF7 | hsa-mir-2355, hsa-mir-1302-4 | [6 9] |
| 3q27 | 188046–188075 | 2 | 2 | 2 | 4x | LPP (part), BCL6 (regulatory region) | - | [6, 26] |
| 6q22 | 125228–126628 | 2 | 2 | 2 | 4–4.5 | NKAIN2 (part), STL, RNF217, TPD52L1, HDDC2, LOC643623,HEY2, NCOA7, TRMT11 | LOC643623 MIR5695 | |
| 9p24 | 3087–7795 | 2 | 4 | 3 | 2 | RFX3, GLIS3, PPAPDC2, SLC1A1, CDC37L1, AK3, RCL1, JAK2, INSL4, RLN2, RLN1, CD274, (INSERT), ERMP1, MLANA, IL33, GLDC, KDM4C | hsa-mir-101-2 | [6] |
| 9p24 (peak) | 5527–5766 | 3 | 4 | 4 | 2 | PDCD1lG2 (part), KIAA1432 (part) | - | |
| 8q24 | 129061–129153 | 2 | 6.5 | 2 | 0 | PVT1 (part) | [6, 26] | |
| 21q11 | 15639–16173 | 2 | 2 | 4 | 2 | ABCC13,HSPA13, SAMSN1, SAMSN1-AS1, LOC388813 | - | |
| 22q12 | 36679–36694 | 2 | 4 | 4 | 2 | MYH9 (part) | - | |
| B: Deletions (bilateral) | ||||||||
| 1p12 | 117068–117118 | 2 | 0 | 0 | 2 | CD58 | hsa-mir-320b | |
| 2p16 | 48006–48193 | 0 | 2 | 1 | 2 | MSH6, FBXO11 | - | |
| 7p22 | 60267–6095 | 2 | 2 | 2 | 0 | PMS2 (part), AIMP2, EIF2AK1 (part) | - | [26] |
| 8q24 | 128829–128990 | 2 | 3 | 2 | 0 | PVT1 | hsa miR-1204-1208 | |
| 9p21 | 21969–22067 | 1 | 0 | 3 | 0 | CDKN2A, CDKN2B | - | |
| 12q24 | 133272–133289 | 2 | 0 | 2 | 2 | PXMP2 | - | |
| 15q26 | 100148–100208 | 2 | 0 | 2 | 2 | MEF2A | - | |
| 16p13 | 10897–1099711169–11682 | 1 | 0 | 2 | 0 | FAM18A (part),CLEC16A (part), SOCS1, TNP2, PRM2, PRM1, RMI2, LITAF | hsa-mir-548h-2 | |
| 17q21 | 42570–42584 | 1 | 2 | 2 | 0 | GPATCH8 (part) | - | |
| 17q21 | 42596–42599 | 1 | 2 | 2 | 0 | GPATCH8 (5´) | - | |
| 19q13 | 34737–34809 | 2 | 2 | 3 | 0 | KIAA0355 | - | |
| 22q11 | 183978–18488 | 2 | 0 | 2 | 2 | MICAL3, | hsa-mir-648 | |
| C: Deletions (unilateral) | ||||||||
| 1p35 | 25686–25762 | 2 | 2 | 1 | 1 | RHCE | - | [26] |
| 1p13 | 110479–110484 | 1 | 1 | 2 | 2 | CSF1 (downstream) | - | [26] |
| 1q43 | 236900–236905 | 1 | 2 | 3 | 1 | ACTN2 (Part; partially overlaps deletion polymorphism) | - | |
| 1q44 | 241161–241667 | 1 | 2 | 1 | 2 | FH (part) | - | |
| 2p16 | 50546–50559 | 2 | 1 | 2 | 1 | NRXN1 (part) | - | |
| 2q11 | 100928–100951 | 2 | 2 | 1 | 1 | LONRF2 (part) | - | |
| 2q12 | 103099–103104 | 2 | 2 | 1 | 1 | SLC9M (part) | - | |
| 2q12 | 106384–106390 | 1 | 2 | 3 | 1 | NCK2 (part)17q21 | - | |
| 2q14 | 123282–123303 | 2 | 3 | 1 | 1 | - | - | |
| 2q34 | 211953–211971 | 1 | 1 | 2 | 3 | - | - | |
| 2q36 | 223660–223675 | 2 | 2 | 1 | 1 | - | - | |
| 3p21 | 50627–50638 | 2 | 1 | 2 | 1 | CISH (downstream) | - | [26] |
| 3p14 | 69532–69554 | 2 | 1 | 1 | 2 | - | - | |
| 3q13 | 117961–117972 | 1 | 1 | 2 | 1 | - | - | |
| 4p11 | 48765–48777 | 2 | 1 | 2 | 1 | FRYL (part) | - | |
| 4q13 | 62870–62877 | 2 | 2 | 1 | 1 | LPHN3 (part) | - | [26] |
| 4q32 | - | 2 | 1 | 1 | 2 | PALLD (non-overlapping losses within same gene) | - | [26] |
| 5q34 | 167062–167071 | 2 | 1 | 1 | 2 | TENM2 (part) | - | |
| 6q25 | 149377–149382 | 2 | 1 | 1 | 2 | UST (part) | - | [26] |
| 6q26 | 164446–164467 | 1 | 1 | 2 | 2 | - | - | |
| 7p21 | 11162–11167 | 2 | 2 | 1 | 1 | PHF14 (part) | - | [26] |
| 7p15 | 21579–21584 | 2 | 2 | 1 | 1 | DNAH11 (part) | - | |
| 7p15 | 27224–27236 | 1 | 1 | 2 | 2 | HOXA11 (part), HOXA11-AS | - | |
| 7p14 | 30000–30003 | 2 | 2 | 1 | 1 | SCRN1 (part) | - | |
| 7p14 | 42267–42268 | 1 | 2 | 1 | 1 | GLI3 (part) | - | |
| 7q21 | 95043–95053 | 1 | 2 | 2 | 1 | PON2 | - | |
| 8q22 | 95398–95399 | 1 | 2 | 1 | 2 | RAD54B(part) | - | |
| 8q22 | 102670–102674 | 2 | 1 | 1 | 2 | GRHL2 (part) | - | |
| 9p13 | 37997–37999 | 2 | 2 | 1 | 1 | SHB (part) | - | |
| 10q12 | 49731–49739 | 1 | 1 | 1 | 1 | ARHGAP22 (part)—adjacent to loss DGV48558) | - | |
| 10q12 | 51818–51850 | 2 | 1 | 1 | 2 | FAM21B (part) | - | |
| 10q22 | 73532–73538 | 1 | 1 | 1 | 2 | C10orf54 (5´), CDH23 (part)–adjacent to loss DGV29865 | - | |
| 10q22 | 108236–108253 | 2 | 1 | 1 | 2 | - | - | |
| 11q22 | 102212–102214 | 2 | 2 | 1 | 1 | BIRC3 (3´), BIRC2 (5´) | - | |
| 11q23 | 118140–118154 | 1 | 2 | 1 | 2 | MPZL2 (5´) | - | |
| 11q23 | - | 2 | 2 | 1 | 1 | SORL1—non-overlapping losses within same gene | - | |
| 12q23 | 101601–101611 | 1 | 2 | 1 | 2 | SLC5A8 (part) | - | |
| 13q31 | 93488–93497 | 2 | 3 | 1 | 1 | GPC5 (part)—adjacent to loss DGV86974 | - | |
| 14q22 | 52271–52276 | 2 | 1 | 2 | 1 | GNG2 (5´)–adjacent to loss DGV87223 | - | |
| 14q23 | 68248–68251 | 1 | 1 | 2 | 2 | ZFYVE26 (part) | - | |
| 14q24 | 76430–76438 | 1 | 1 | 2 | 2 | TGFB3 (part) | - | |
| 14q31 | 80199–80210 | 2 | 2 | 1 | 1 | NRXN3 (part) | - | |
| 15q26 | 101032–101037 | 1 | 1 | 2 | 2 | CERS3 (part) | - | |
| 16p13 | 7553–7561 | 1 | 2 | 2 | 1 | RBFOX1 (part) | - | |
| 16p13 | - | 1 | 2 | 1 | 2 | LOC283856 (part)—non-overlapping losses within same gene | - | |
| 18q21 | 5840–358444 | 1 | 1 | 2 | 2 | Gene desert: nearest gene CDH20 | - | |
| 19p13 | 6483–6496 | 1 | 1 | 2 | 2 | TUBB4A (part) | - | |
| 19p11 | 23557–23564 | 2 | 1 | 2 | 1 | CST9L (5´) | - | |
| 19q13 | 56076–56085 | 2 | 1 | 1 | 2 | CTCFL (part) | - | |
Table lists coordinates (HG19) and hosted loci in PMBL cell lines bearing (A) significant CNV, both gains (≥4x) and (B) losses (null) and both protein coding and noncoding RNA genes located within. Part C lists shared unilateral deletions including non-overlapping deletions in the same genes. Where multiple cell lines are involved coordinates of common affected regions are shown. DGV polymorphisms are excluded.
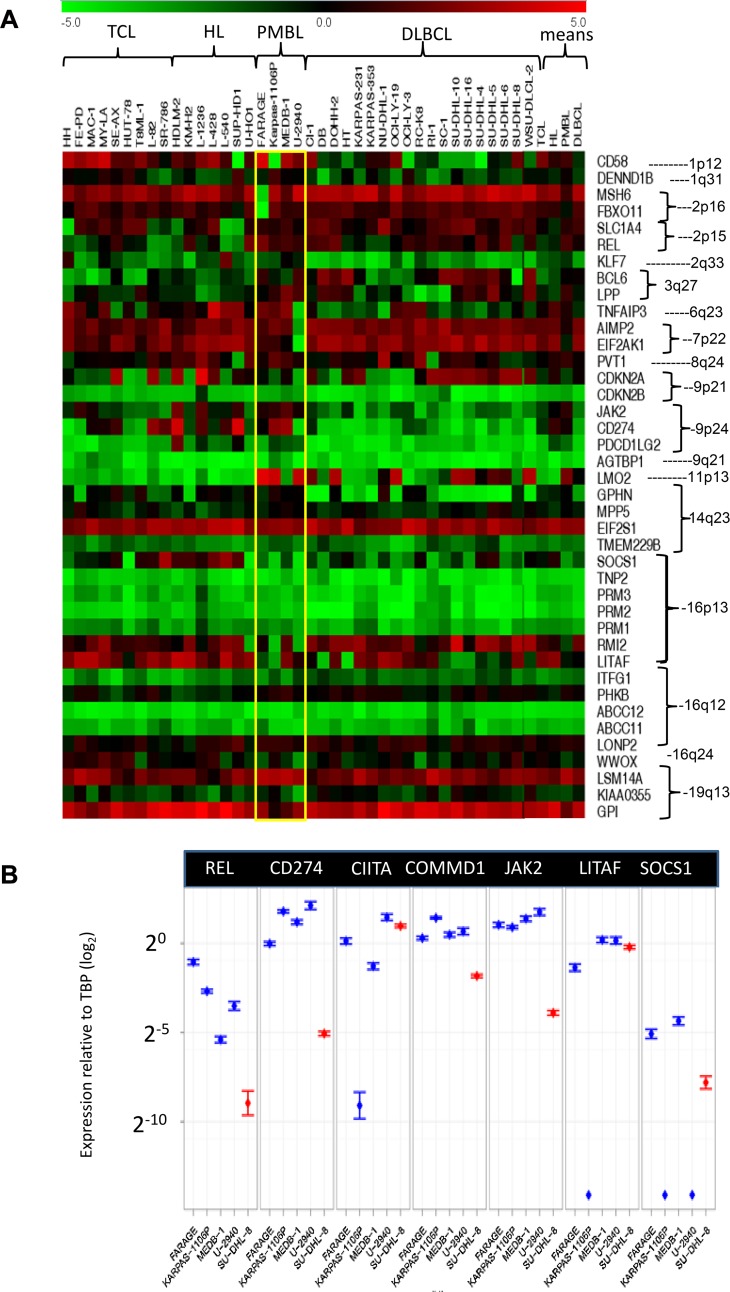
Table 2 lists 72 LOH shared overlapping regions present in two or more PMBL cell lines and genes encoded within. Detailed genomic plots of 12 conspicuously altered loci at 1p12, 2p15, 2p16, 6q23, 6q27, 7p22, 7q31, 8q24, 9p21, 9p24, 16p13, 16q23 and 19q13 are presented in Fig 3B. A corresponding microarray gene expression heatmap covering candidate genes mainly within genomically altered loci is shown (Fig 4A) with select validation by qPCR (Fig 4B).
Table 2. Genomic Regions with Shared LOH.
| Chr. band | Coordinates (KBp) | PMBL cell lines | Genes in region of interest | ||||
|---|---|---|---|---|---|---|---|
| FARAGE | KARPAS-1106P | MEDB-1 | U-2940 | Protein coding | RNA/miR | ||
| 1p36 | 16321–18176 | + | + | - | |||
| 1p34 | 31583–31823 | - | + | + | + | SDC3, PUM1, NKAIN1, SNRP40, | - |
| 1p34 | 32266–32755 | + | + | + | + | SPOCD1 (part), PTP4A2, KDSRBH1, TMEM39B, KPNA6 | - |
| 1p32 | 51871–52884 | - | + | + | + | EPS15, OSBPL9, NRD1, RAB3B, TXNDC12, BTF3L4,ZFY3E9, CC2D1B, ORC1, | MIR761 |
| 1p32 | 52884–53444 | - | + | + | - | ZCCHC11 | - |
| 1p22 | 92031–93395 | + | + | - | - | HSP90B3P, TGFBR3, BRDT, EPHX4, BTBD8, KIAA1107, C1orf146, GLMN, RPAPq2, GFI1, EVI5, RPL5, FAM69A, | U21, U66 |
| 1p21 | 103783–104829 | + | + | + | - | RNPGC3, ACTG1P4, AMY2B, AMY2A, AMY1A, | - |
| 1q44 | 248127–249177 | + | + | + | + | ORL2L13,ORL2M-cluster, ORL2T-cluster,SH3BP5L, ZNF672, PGBD2 | hsa-mir-3124 |
| 2p16 | 47460–48198 | + | - | + | - | EPCAM, MSH2, KCKN12,MSH6, FBXO11 | hsa-mir-559 |
| 2p15 | 61675–62428 | - | + | - | - | XPO1, FAM161A, CCT4, COMMD1B3GN2 | - |
| 2p14 | 65230–65231 | + | + | + | - | SLC1A4 | - |
| 2p11 | 83872–84993 | + | + | + | - | SUCLG1, DNAH6, | - |
| 2q32 | 195244–196180 | + | + | + | + | - | - |
| 3p21 | 50664–52208 | + | + | + | + | MAPKAPK3, DOCK3, VPRBP, RAD54L2,GRM2, IQCF-cluster, RRP9, PARP3, PCBP4, RRP9, ABHD14B, DUSP7, POC1A, ALAS1, | hsa-let-7G, hsa-mir-1 |
| 3p11 | 89163–90486 | + | - | + | - | EPHA3 | - |
| 4p15 | 33305–33646 | + | - | + | + | - | - |
| 4q31 | 151969–152275 | + | - | + | + | DCLK2, LRBA, RPS3A, SH3D19 | U73b |
| 5q12 | 61391–62553 | - | - | + | + | KIF2A, IPO11 | - |
| 5q31 | 130539–130631 | + | - | + | + | LYRM7, CDC42SE2 | - |
| 6p22 | 26860–26944 | + | + | - | + | - | GUSBP2, LINC00240, LOC100270746 |
| 6p22 | 27212–28454 | + | + | - | + | PRSS16, POM121L2, VN1R10P, ZNF204P, ZNF391, ZNF184, HIST1H-cluster, OR2B2, OR2B6, ZSCAN12P1, ZSCAN16, ZNF192, TOB2P1, ZNF193, ZSCAN4, NKAPL, ZNF187, PGBD1, ZNF323, ZSCAN3, ZSCAN12, ZSCAN23 | LOC100507173 |
| 6q12 | 64251–64611 | + | + | + | - | PTP4A1, PHF3, EYS | - |
| 6q22 | 125776–126616 | - | + | + | + | HEY2,NCOA7,HINT3, TRMT11, CENPW | LOC643623, MIR5695 |
| 6q27 | 138167–138216 | - | + | - | + | TNFAIP3 | LOC100130476 |
| 7p11 | 56771–57261 | + | - | + | + | LOC100130849, ZNF479, GUSBP10, | MIR4283-1, MIR3147 |
| 7q36 | 156187–157470 | + | + | - | - | RNF32, LMBR1, NOM1, MNX1, UBE3C, DNAJB6, PTPRN2 | hsa-mir-153-2LOC285889LINC00244 |
| 8p11 | 42264–43779 | + | + | + | - | VDAC3, SLC20A2, CHRNB3, CHRNA6, THAP1, RNF170, HOOK3, FNTA, HGSNAT, POTEA | - |
| 9p24 | 2011–2025 | + | + | + | + | SMARCA2 | - |
| 9p24 | 5466–5765 | - | + | + | + | CD274, PDCD1GL2 | KIAA1432 |
| 9p24 | 6655–6712 | - | + | + | + | GLDC, KDM4C | - |
| 9p23 | 10481–10532 | + | + | + | + | PTPRD | - |
| 9p21 | 21435–23074 | + | + | + | + | IFNA1, MTAP, CDKN2A, CDKN2B, DMRTA1, FLJ35282 | MIR31HG |
| 9q22 | 94424–95917 | - | - | + | + | ROR2, SPTLC1, IARS, NOL8, CENPP, OGN,OMD, ASPN, ECM2, IPPK, BICD2,ANKRD19P, ZNF484, FGD3,SUSD3, C9orf89, NINJ1 | - |
| 9q22 | 97056–98129 | + | - | + | - | ZNF169, HIATL1, FBP2, FBP1, C9orf3, FANCC, | hsa-mir-23b, hsa-mir-24-1,hsa-let7-1/7dMIR23BLOC100132077 |
| 9q33 | 125867–126692 | + | + | + | + | OR1-cluster, PDCL, RC3H2, ZBTB6, ZBTB26, RABGAP1, GPR21,STRBP, CRB2, DENND1A, | MIR600 |
| 10q21 | 69833–69912 | - | + | + | + | MYPN | - |
| 10q22 | 74721–75779 | + | + | - | + | P4HA1, NUDT13, ECD, DNAJC9, TTC18, ANXA7,MSS51, PPP3CB, USP54, MYOZ1, SYNPO2L, BMS1P4, SEC24C, KIAA0913, CAMK2G, PLAU, C10orf55, VCL | - |
| 10q24 | 104198–104246 | - | - | + | + | GBF1, FBXL, CUEDC2, | hsa-mir-146b |
| 10q26 | 124319–124341 | + | + | + | - | DMBT1 | - |
| 11p11 | 46232–47689 | + | + | + | + | CREB3L1, DGKZ, AMBRA1, ATG13, ZNF408, F2, CKAP5, LRP4, C11orf49, ARFGAP2, PACSIN3, DDB2, NR1H3, MADDMYBPC3, SPI1, SLC39A13, RAPSN, CELF1,PTPMT1,NDUFS3,NUP160 | hsa-mir-3160-1/2, HBII-166hsa-mir-3161 |
| 11q13, | 71561–72385 | - | + | + | + | LOC100133315, RNF121, NUMA1, LRTOMT, LAMTOR1, FOLR3, FOLR1, IL18BP, ANAPC15, FOLR2, INPPL1, PHOX2A, CLPB, PDE2A, | hsa-mir-3165, hsa-mir-139 |
| 12p13 | 313216–315336 | - | - | + | + | TEAD4 | - |
| 13q12 | 25725–25758 | + | + | - | - | FAM123A | |
| 13q12 | 30015–31233 | - | + | + | - | MTUS2, UBL3, KATNAL1, HMGB1, USPL1 | LINC00297, LOC440131LINC00426 |
| 14q13 | 35870–37786 | - | + | - | + | NFKBIA, RALGAPA1, BRMS1L, PTSC3, MBIP, SFTA3, NKX2-1, PAX9, SLC25A21, MIPOL1 | - |
| 14q23 | 66556–67954 | - | + | + | + | GPHN, MPP5, EIF2S1, TMEM229B | - |
| 14q23 | 68248–68251 | + | + | - | + | ZFYVE26 | - |
| 15q13 | 28297–28560 | - | + | + | + | OCAC2, HERC2 | - |
| 15q15 | 41781–41868 | - | + | + | + | LTK, ITPKA,RPAP1, TYRO3 | - |
| 15q15-21 | 43928–45972 | + | + | - | + | CKMT1A, PDIA3, ELL3, SERF2, SERINC4, WDR76, PIN4P1, FRMD5, CASC4, CTDSPL2, LOC645212,MFAP1, C15orf63, EIF3J, SPG11, PATL2, B2M, TRIM69 | hsa-mir-1282 |
| 16p13 | 10901–10971 | + | + | - | + | FAM18A,CIITA | - |
| 16p13 | 11129–11826 | + | + | - | + | CLEC16A, SOCS1, TNP2, PRM2, PRM1, RMI2, KITAF, SNN, TXNDC11 | hsa-mir-548h-2 |
| 16p11 | 34198–35221 | + | - | + | + | UBE2MP1, RN5S411, | hsa-mir-1826RNU6-76, LOC283914LOC146481, LOC100130700, FLJ26245 |
| 16q12 | 47308–48308 | + | - | + | + | ITFG1,PHKB, ABCC12, ABCC11, LONP2 | MIR648AE2, |
| 16q22 | 66448–68214 | + | - | + | + | BEAN1, TK2, CKLF, CMTM1, CMTM2, CMTM3, CMTM4, DYNC1LI2, CCDC79, NAE1, PDP2, CDH16, FAM96B, CES3, CES4A, CBFB, C16orf70, HSF4, NOL3EXOC3L1, E2F4, LRRC29, SLC9A5, PLEKHG4, LRRC36, TPPP3, HSD11B2, ATP6VOD1, FAM65A, CTCF, RLTPR, ACD, GFPD2, RANBP10, THAP11, NUTF2, EDC4, PSKH1, PSMB10, CTRL, SLC12A4, DPEP3, DDX28, DUS2L, NFATC3 | hsa-mir-328 |
| 17q21 | 40514–41964 | - | + | + | + | STAT3, PTRF, ATP6VOA1, NAGLU, HSD17B1, COASY, PSMC3IP, FAM134C, MLX, TUBG1, TUBG2, PLEKHH3, CNTNAP1, EZH1, RAMP2, VPS25, CNTD1,BECN1, pSME3, AOC3, AOC4, G6PC, AARSD1, RPL27, RUNDC1, IFI35, RND2, BRCA1, NBR1, DHX8, ETV4, MEOX1, SOST, DUSP3, MPP3, CD300LG, MPP2 | LOC100190938,TMEM106A-AS1, LOC100130581, ARL4D, MIR2117, |
| 17q21 | 43704–45703 | + | + | - | + | CRHR1, SPPL2C, MAPT, STH, KANSL1, ARL17A, LRRC37A, NSFP1, LRRC37A2, NSF, WNT3, WNT9B, GOSR2, RPRML, CDC27, MYL4, ITGB3, C17orf57, MRPL45P2, NPEPPS | MGC57346, MAPT-AS1, MIR5089, |
| 17q22 | 46608–48139 | + | + | - | + | HOXB1,HOXB2, HOXB3, HOXB4, HOXB5, HOXB6, HOXB7, HOXB8, HOXB9, PRAC, HOXB13, TTLL6, ATP5G1, UBE2Z, SNF8, GIP, IGF2BP1, B4GALNT2, GNGT2, ABI3, FLJ40194, ZNF652,PHB, NGFR, NXPH3, SPOP, SLC35B1, FAM117A, KAT7, TAC4, DLX4, DLX3, ITGA3, | MIR10A, HOXB-AS3, MIR196A1, MIR3185, LOC294080, |
| 17q23 | 57929–59172 | + | + | - | + | RPS6KB1, TUBD1, HEATR6, CA4, USP32, C17orf64, APPBP2, PPMID, BCAS3 | TBC1D3P1-DHX40P1, LOC653653, |
| 19q13 | 42006–42422 | - | + | + | + | CEACAM21, CEACAM4, CEACAM7, CEACAM5, CEACAM6, CEACAM3, DMRTC2, LYPD4, RPS19, CD79A, ARHGEF1 | LOC100505495 |
| 20q13 | 46104–58874 | - | + | - | + | NCOA3, PREX1, ARFGEF2,STAU1, CSE1L, KCNB1, B4GALT5, PTGIS, SLC9A8, RNF114, SNAI1, SPATA2, UBEV1, CEBPB, LOC284751, PTPN1, PARD6B, BCAS4, ADNP, DPM1, KCNG1, NFATC2, ATP9A, SALL4, ZPF64, TSHZ2, ZNF217, SUMP1P1, BCAS1, CYP24A1, PFDN4, DOK5, CBLN4, MC3R, AURKA, TFAP2C, BMP7, SPO11, RAE1, RBM38, CTCFL, PCK1, ZBP1, PMEPA1, C20orf85, PPP4R1L, RAB22A, VAPB, LOC149773, STX16, GNAS, TH1L, CTSZ, TUBB1, EDN3, PHACTR3, PP1R3D, CDH26, C20orf197, LOC284757 | LINC00494, ZNFX1-AS1, U106,HBII-99B, hsa-mir-645, hsa-mir-1302-5, LOC100887755, hsa-mir-3194, hsa-mir-4325, MIR4532, STX16-NPEPL1, hsa-mir-296, |
| 22q13 | 41303–42236 | + | + | + | + | XPNPEP3, RBX1, EP300, L3MBTL2, CHADL, RANGAP1, ZC3H7B, TEF, TOB2, ACO2, POLR3H, PMM1, CSDC2, DESI1, XRCC6, NHP2L1, MEI1, SREBF2, | hsa-mir-1281, hsa-mir-33a, |
| 22q13 | 50294–50297 | + | + | + | - | ALG12 | - |
| Xp22 | 19115–20973 | - | + | + | + | PDHA!, MAP3K15, SH3KBP1, Cxorf23, MAP7D2, EIF1AX, RPS6KA3, | - |
| Xp11 | 36152–37511 | + | + | + | + | Cxorf59, Cxorf30, FAM47C, PRRG1, LANCL3 | - |
| Xp11 | 42327–43823 | + | + | + | + | MAOA, MAOB, NDP | - |
| Xq13 | 46298–46607 | + | + | + | + | ZNF673, ZNF674, CHST7, SLC9AS7, RP2, PHF16, RGN, UBA1, INE1, RBM10, CDK16, USP11, ZNF157, ZNF41, ARAF, TIMP1, CFP, ELK1, SYN1, CXXC1P11 | SNORA11C |
| Xq13 | 74360–75444 | + | + | + | + | ABCB7UPRT, ZDHHC15, TTC3P1, MAGEE2 | - |
| Xq22 | 98574–99573 | + | + | + | + | PCDH19, TNMD, TSPAN6 | LOC442459 |
| Xq23 | 109983–111118 | + | + | + | + | CHRDL1, PAK3, CAPN6, DCX1, DKFZp686DO853, ALG13, TRPC5, TRPC5OS | |
| Xq26 | 128870–132178 | + | + | + | + | XPNPEP2, SASH3, ZDHHC9, UTP14A, BCORL1, ELF4, AIPM1, RAB33A, ZNF280C, SLC25A14, GPR119, RBMX2, FAM45B, ENOX2, ARHGAP36, IGSF1, OR13H1, MST4, FRMD7, RAP2C, MBNL3, | LOC286467 |
| Xq28 | 147885–148078 | + | + | + | + | AFF2 | - |
Lists recurrent loci in PMBL cell lines bearing LOH and/or mild coincident CNV with both protein coding and RNA genes located within.
Fig 4. Gene expression at genomically rearranged loci.
A: Microarray expression data for genes implicated at CNV (mainly deletions). Note, for example gene silencing accompanying focal biallelic deletions at multiple loci: including, CD58 at 1p12 in KARPAS-1106P; MSH6 and FBXO11 at 2p26 in FARAGE; TNFAIP3, AIMP2 at 6q23 in U2940; EIF2AK1 at 7p22 in U-2940; CDKN2A at 9p21 in KARPAS-1106P and U-2940; CD274 at 9p24 in U-2940; SOCS1 at 16p13 in KARPAS-1106P and U-2940; and KIAA0355 at 19q13 in U-2940. B: Shows qPCR expression of select target genes at recurrent PMBL amplicons (2p15, 9p24) and a deletion (16p13) set against TBP reference in cell lines FARAGE, KARPAS-1106P, MEDB-1, and U-2940 (PMBL) shown blue, alongside reference SU-DHL-8 (DLBCL) shown red. Diamonds indicate undetectable expression. Quantitative data were verified by twofold or more biological replication.
Classical CNA which promotes oncogene activity by increasing mRNA levels occurred only sporadically in PMBL cell lines: singular amplifications, moderate in level (3.5–6.5 fold), were detected in three cell lines (Table 1A). Of 12 CNA seven were focal housing at most one or two genes but were restrained in level and frequency when compared to cHL and DLBCL cell lines. Focal CNA were present in KARPAS-1106P (8q24, Xq11), MEDB-1 (1q31, 9p21) and U-2940 (2q33, 3q28) together with a slightly larger CNA region (ca. 1.2 Mbp) at 6q22. Correlating genomic with microarray expression data (Fig 4A) showed that of four protein coding genes mapping within focal CNA loci, one showed above average (KLF7/2q3), and three near average expression (AGTBP1/9q21, DENN1B/1q31, LPP/3q27), discounting a major role for CNA in gene activation in PMBL cells.
Focal deletions abolish gene expression whether acting alone or in concert with inactivating mutations or epigenetic modifications of residual alleles. Focal null deletions were observed in three cell lines: namely, FARAGE (2p16), KARPAS-1106P (1p12, 1p31, 1p36, 9p21, 16p13, 20q11; plus subclonally at 12q26); and U-2940 (7p22, 8q24, 9p21, 16p11, 19q13) together with 1p12. Of these, three loci bore overlapping null deletions, at 1p12, 9p21 and 16p13. Validation of deletions covering the SOCS1 locus was obtained by FISH for three cell lines, FARAGE, KARPAS-1106P and MEDB-1 (Fig 2A–2C), and for the PTPN1 locus for KARPAS-1106P and MED-B1 (Fig 2E and 2F).
Parallel microarray expression data (Fig 4A) were also used to assess the candidacies of CNV as potential targets and selectively validated by qPCR (Fig 4B and 4C). Independent PMBL patient expression data compared to cHL were extracted from online expression data GSE40160 (extracted from ref. 27) and shown in S4 Fig This exercise highlighted CNV correlated with gene expression, mostly deletions accompanied by gene silencing. These included:
1p12: The CD58 locus was targeted by microdeletion, biallelic in KARPAS-1106P, monoallelic in MEDB-1 and U-2940 (Fig 3B). Expression levels reflected CNV status (Fig 4A). CD58 is also conspicuously silent in PMBL patients (S4A Fig), while neighboring IGSF3 expression remained inconspicuous reflecting that in cell lines (not shown). Although this region carries a polymorphic CNV (Fig 3B), only IGSF3 was affected.
1q31: The DENN/MADD Domain I (DENND1B) gene which is upregulated in PMBL cell lines is amplified in MEDB-1 only (not shown).
2p15: Although genomic array data confirmed the reported CNA in KARPAS-1106P at 2p15, its peak plateau (4n) excluded REL, the mooted target in PMBL (Fig 3B). REL expression, although consistently high in PMBL cells–exceeding that in both cHL and DLBCL–was thus but weakly correlated with CNV. The sole gene inside the amplicon peak region which was consistently upregulated was COMMD1 (Fig 4B), also the most conspicuously upregulated in patients S4A Fig).
2p16: Both MSH6 and the adjacent FBXO11 were deleted in FARAGE (biallelic) and MEDB-1 (monoallelic) (Fig 3) accompanied by dose-related transcriptional downregulation, markedly so in FARAGE (Fig 4A). Both genes are conspicuously silent in PMBL patients and may be deemed candidate tumor suppressor genes (S4A Fig).
3q27: Expression of BCL6 and LPP like DLBCL surpassed that in cHL cells placing PMBL closer to the former entity (Fig 4A). BCL6 copy numbers remained diploid (not shown) although expression was raised in FARAGE and U-2940 and lowered in KARPAS-1106P and MEDB-1. Interestingly, BCL6 was conspicuously well expressed in PMBL patients (S4A Fig).
6q23: Biallelic deletion affecting TNFAIP3/A20 (Fig 3B) accompanied conspicuous silencing of this gene in U-2940 cells (Fig 4A). Silencing also affected some DLBCL and TCL cell lines (Fig 4A). LOH at this locus was present in KARPAS-1106P (not shown) where expression levels were intermediate (Fig 4A). The same cell line reportedly carries an inactivating TNFAIP3 mutation [28]. TNAFAIP3 silencing was also apparent among PMBL patients (S4A Fig).
7p22: Focal biallelic deletion of AIMP2 and EIF2AK1 (Fig 3B) accompanied conspicuous silencing of both genes in U-2940 (Fig 4A). Of these, EIF2AK1 evidenced the greater silencing in PMBL patients (S4A Fig).
7q31: A complex pattern of deletion and amplification was observed. ZNF277 was downregulated in U-2940, and IMMP2L in MEDB-1 (not shown) which respectively bore focal monoallelic deletions at these loci (Fig 3B). In PMBL patients both genes are downregulated in patient subsets (S4A Fig).
8q24: A complex CNV pattern was observed around PVT1 and the associated miR cluster while excluding MYC (Fig 3B). The centromeric microdeletion present in all four cell lines is apparently non-polymorphic. Biallelic deletion in U-2940 and four-fold amplification in KARPAS-1106P affecting PVT1 were reflected transcriptionally (Fig 4A), while the embedded miR cluster (miR-1204-1208) remained inconspicuously silent throughout in miR arrays (not shown). Likewise, PMBL patients expressed widely fluctuating PVT1 levels (S4B Fig).
9p21: In KARPAS-1106P and U-2940 biallelic, and in FARAGE monoallelic, focal deletions affected the adjacent CDKN2A/B loci (Fig 3B). Although trisomic, the locus in MEDB-1 also evidenced LOH (not shown). Gene expression data showed silencing of CDKN2B in all cell lines irrespective of ploidy, and of CDKN2A except in MEDB-1, but conspicuously so in U-2940 and KARPAS-1106P (Fig 4A). qPCR confirmed CDKN2A/B silencing (not shown). In patients as in cell lines CDKN2A was conspicuously silenced, and CDKN2B unevenly so (S4B Fig). Collectively, these findings raise the spectre of epigenetic controls at this locus, such as silencing by DNA methylation.
9p24: At this site of recurrent CNV ploidy gains were ubiquitous in both PMBL and cHL, sparing only FARAGE. Tetraploidy in KARPAS-1106P was extended plateau-like, but focal in MEDB-1 cresting a wide triploid region, while a microdeletion in U-2940 impinged CD274. Consensus gains included PDCG1LG2 and KIA1432 (Fig 3B). There was no clear correlation between CNA and transcriptional upregulation, as both JAK2 and CD274 which showed unambiguous upregulation (Fig 4A and 4B) shunned the amplicon peak. Thus, while endorsing the existence of a 9p24 amplicon in PMBL its relationship to JAK2 remains enigmatic, the combined CNA and expression data marginally favoring CD274 while failing to exclude multiple targeting.
16p13: Genomic deletions, monoallelic in FARAGE, biallelic in KARPAS-1106P/MEDB-1/U-2940, covered 6 transcriptionally silenced genes at 16p13: namely, CIITA, LITAF, SOCS1, RM1/2 and TNP2, (Fig 3B). On expression arrays the flanking candidates CIITA and LITAF undercut cHL, DLBCL or T-ALL cell lines (Fig 4A). According to qPCR data, SOCS1 showed the most consistent silencing, just ahead of LITAF which lay outside the CDR (Fig 4B). Uniquely, in U-2940 a cytogenetic rearrangement t(16;16) accompanied deletion (Fig 1D). In patients, of these candidates only LITAF was conspicuously underexpressed, and RMI2 but moderately so (S4B Fig). CIITA/MHC2TA expression remained nondescript in both settings (Fig 3C and Fig 4B).
16q23: Biallelic and monoallelic deletions affecting the common fragile site FRA16D gene WWOX were present in MEDB-1 and U-2940 (Fig 3B) correlated with modest expression seen therein (Fig 4A). Interestingly, both deletions are intronic and perfectly correspond to known polymorphisms (Fig 4A). In PMBL patients WWOX is conspicuously downregulated (S4C Fig).
17p13: Although TP53 may be inactivated in PMBL, this locus displayed neither copy number losses nor LOH. Expression remained unperturbed (not shown).
17q21: GPATCH8 uniquely bore a singleton biallelic deletion in U-2940 which abolished transcription, while the monoallelic deletion in FARAGE accompanied depressed microarray expression therein (not shown).
19q13: A biallelic focal deletion was present in U-2940 (Fig 3B) accompanied by conspicuous silencing of KIAA0355 (Fig 4A) seen also in patients (S4C Fig).
21q12: A discrete fourfold amplicon of circa 15 Kbp was present in both KARPAS-1106P and MED-B1 (Table 1A) which although apparently identical in both cell lines is not hitherto recorded as a polymorphism. This region includes part of the coding region of MYH9 which was, however, inconspicuously expressed in both patients and cell lines (not shown).
In addition to biallelic deletions which ipso facto enforce gene silencing, 49 shared monoallelic deletions (SMD) were recorded (Table 1C). On average shorter than biallelics, these sometimes shared identical coordinates and may represent unrecorded polymorphisms. To assess their significance SMD were also checked against microarray expression. This exercise revealed two examples correlating with gene expression, one positively (2q12/NCK2) and one negatively at 10q12/FAM21B, i.e. within expected stochastic noise levels.
Losses of heterozygosity (LOH)
LOH are thought to promote cancer by exposing recessive gene mutations or epigenetic silencing modifications among tumor suppressor genes. Alternately, LOH may activate oncogenes by silencing their cognate micro-RNAs. In total 72 shared LOH were recorded the most frequent and widespread alterations present (Table 2). Most LOH were extensive covering multiple genes, hampering target identification. A minority were focal pinpointing candidate gene targets within a few loci, e.g. at 6q23 (TNFAIP3) and 9p24 (CD274)–both targeted separately by focal deletions in other cell lines (Figs 3B and 4A and 4B). LOH also occurred independently of deletions, e.g. at 16q12 FARAGE, MEDB-1 and U-2940 (Table 2) negatively correlated with gene expression at this locus (Fig 4A): of potentially inactivated genes inside the common LOH region (EIF2S1, GPHN, and MPP5) were also underexpressed in PMBL patients (S4B Fig). In the same three cell lines LOH was also observed at 16q12 where ITFG1 and PHKB were downregulated both in the affected cells (Fig 4A) and in PMBL patients in general (S4 Fig). Singleton LOH was accompanied by conspicuous LMO2 silencing in MEDB-1 covering the 3´ distal regulatory region (Fig 4A). In the PMBL patient series LMO2 was highly expressed throughout highlighting downregulation in MEDB-1 (S4B Fig). LOH at 14q23, in KARPAS-1106P, MED-B1 and U-2940 was seemingly accompanied by downregulation of GPHN and MPP5 (Fig 4B). In PMBL patients, however, the same genes were conspicuously silent as was the adjoining EIF2S1 (S4B Fig). While microRNA loci also fell within genomically altered regions offering a possible explanation for upregulation accompanying LOH—parallel miR expression arrays provided no evidence that these impacted transcription (not shown).
Discussion
By integrating genomic with parallel transcriptional data sets we both documented genomic alterations in PMBL cell lines and prioritized gene candidates for further assessment. Given the degree to which cell lines portray the emerging oncogenomic picture of PMBL we conclude that they provide suitable resources for pursuing further investigations into this enigmatic neoplasm. Shared impactful changes were shown to feature in two or more cell lines forming patterns of recurrence warranting further study. Although their rearrangement spectrum partially overlaps neighboring entities cHL and DLBCL, PMBL cell lines stand apart bearing far fewer rearrangements than the former, while lacking the recurrent B-cell oncogene translocations of the latter. These evidences plus its snug transcriptional niche close to both neigboring entities show that the PMBL cell lines fill a unique oncogenomic niche consistent with their attribution to PMBL.
The likely import of deletions in PMBL was first bestowed by low density array studies [6, 26]. While broadly consistent with these groundbreaking studies (Table 1), the increased sensitivity afforded by high density arrays allowed detection of an additional layer of CNV, mainly unilateral deletions invisible to BAC arrays. The CNV impacting gene expression the most were, however, bilateral deletions affecting protein coding genes. Top genes/loci verifiably impacted by deletions and/or LOH included MSH6/FBXO11 at 2p16 (in two cell lines), TNFAIP3 at 6q23 (three), CDKN2A at 9p21 (four), and SOCS1 (four) together with LITAF (three) at 16p13—all plausible oncogenic agents, including some already known in PMBL. Although additional CNV were detected, notably the unilateral short CND discussed above, their targets and impact, remain doubtful after assessment with parallel expression data. The significance of other hitherto uncharacterized deletion candidates, e.g. the ubiquitously expressed GPATCH8 at 17q21 and KIAA0355 at 19q13, also awaits further study.
Among validated candidates, SOCS1 deletion/mutation in KARPAS-1106P and MEDB-1 has been already described where it delays JAK2 degradation to allow pSTAT6 stockpiling [29, 30]. Our findings add FARAGE and U-2940 to the list of cell lines bearing 16p13 deletions and confirm concurrent SOCS1 inactivation throughout. Our data also indict neighboring LITAF (and possibly RMI2) as collateral targets. While SOCS1 remains the prime target of 16p13 deletions, LITAF (lipopolysaccharide-induced TNF-alpha factor)—a DNA binding protein which promotes cytokine production including TNF-α, seems to be affected by this lesion. Promoter methylation and biallelic deletion of LITAF has been reported in DLBCL [31]. Here we show that LITAF is both deleted and downregulated in PMBL cell lines while conspicuously silenced in PMBL patients (S4C Fig). LITAF has been identified as a BCL6 target in mature B-cell lymphomas where it regulates autophagy [32]. Together with STAT6, LITAF governs CCL2 expression via NFKB1, and its ancient genomic proximity to SOCS1 may reflect a need for co-regulation. A tumor suppressor role for LITAF has also been reported [33], and this role invites further study in PMBL. Nearby RMI2 is a component of the BLM complex via processing of Holiday junctions formed after DNA repair [34] and its role, if any, in PMBL requires to be established.
In 21% DLBCL cases the immunosurveillance marker CD58 (LFA3) is inactivated [35], as reflected by our cell line (Fig 4) and patient data (S4A Fig) consistent with previous reports [4, 35]. Loss of CD58 expression as seen in KARPAS-1106P recalls an immune escape mechanism reported in DLBCL [36].
FBXO11 at 2p16 forms a complex which targets BCL6 for ubiquitylation and proteasomal degradation in DLBCL [37]. Biallelic deletion and silencing of FBXO11 among lymphoma cell lines occurred uniquely in FARAGE along with raised BCL6 expression. MSH6—co-deleted and silenced along with FBXO11—is subject to inactivation in DLBCL associated with microsatellite instability, increased structural rearrangement and altered mutation signatures [38]. PMBL cell lines also carry mutations in other genes affecting genome stability (Ehrentraut et al., in preparation). BCL6 and FBXO11 are also weakly expressed in PMBL cell lines and merit clinical evaluation.
Centromeric of FBXO11 lies REL at 2p15 which though highly expressed in PMBL cell lines (Fig 4A and 4B) falls outside the CNA region in KARPAS-1106P implying deregulation other than via CNV. Interestingly, genome wide association studies have shown that single polymorphic base changes, at 2p15 near REL and 8q24 near PVT1 where recurrent CNV were detected (Fig 3B), increase the risk of cHL [39]. Alhough COMMD1 activation is less emphatic, it lies within the amplicon peak and is also well expressed in PMBL patients. COMMD1 shortens survival in DLBCL [40], perhaps via deregulation of NFκB [41], and its candidacy merits consideration alongside REL whose overexpression is firmly linked to CNA at 2p15 [42].
TNFAIP3 inactivating mutations occur widely in DLBCL, cHL and in PMBL patients and have been reported in KARPAS-1106P [28, 43]. Mutations inactivating TNFAIP3 are also present in MEDB-1 (Ehrentraut et al., in preparation). These findings, together with the expression data pinpoint TNFAIP3 as a key tumor suppressor gene. Inactivation has been described in a variety of T/B-cell lymphomas and leukemias where it is also thought to promote activation of NFκB [44], a gene well expressed in all 4 PMBL cell lines.
Although deletions affecting 9p21 are widespread in aggressive leukemia and have been described in DLBCL, genes at this locus emerge relatively unperturbed in PMBL [45]. Here lie the coding regions for CDKN2A/B which yield several transcript variants, including p14/ARF from a different reading frame. CDKN2A/B foster self-renewal and, though epigenetically silenced throughout hematopoiesis, subsequently remain poised for reactivation by oncogenic stress. Oncogenomic losses posited to affect this locus are attended by special caveats concerning polymorphism (both gains and losses) and the possibility of immortalization artifact. However, clinical data show that CDKN2A inactivation is widespread in PMBL patients (S4B Fig) buttressing the cell line data.
Our data also show that the posited genomic upregulation of JAK2 which outlies the CNA peak also warrants further study. Although JAK2 is constitutively phosphorylated in MEDB-1 at least [46], “smoking gun” mutations or translocations remain unreported in PMBL and cHL alike [47]. Nevertheless, the hypersensitivity of PMBL cell lines to JAK2 inhibitors has been taken to imply oncogene addiction [48]. JAK-STAT signalling in PMBL is relayed via STAT6, itself constitutively activated in both KARPAS-1106P and MEDB-1 recalling cHL and DLBCL [46, 49]. BCL6 silencing has been identified as a downstream target of JAK-STAT signalling [50] further highlighting this pathway as an actionable target in PMBL. Again, our data highlighted an alternative target at 9p24, CD274/PDL1, which confers immune privilege by binding the PD1 receptor on reactive CD8+ T-cells and thus inhibits proliferation [51], offering a novel candidate for blockade therapy [52].
High density oligonucleotide/SNP arrays remove some bias inherent in BAC arrays while offering improved resolution, parallel LOH data, and—thanks now to the continually updated Database of Genomic Variants—the ability to sift out natural polymorphic CNV. Integrating genomic and transcriptional microarray data allows prioritizing of gene candidates targeted by genomic rearrangements at a first pass level. By delineating ever shorter regions of interest, high density arrays simplify candidate ranking. Finding little evidence that chromosomal translocations impact cancer gene alterations, our findings pinpoint bilateral microdeletions as the predominant class of impactful genomic lesion in PMBL cell lines. Thus our findings strengthen the case for applying high density arrays to clinical samples. And while LOH was in a few cases also correlated with gene silencing, its likely transcriptional—hence oncogenic—consequence remains doubtful. Parallel studies addressing the impact and role of gene mutations in PMBL cell lines are nearing completion in this lab, while those addressing the role of epigenetic modifications, such as DNA methylation, in gene regulation in PMBL are underway.
This study combined high density oligonucleotide/SNP arrays with cytogenetics to profile the genomic features of PMBL cell lines revealing subtle genomic changes therein, notably short genomic deletions. By integrating genomic lesions with global cell line and patient expression data we shortlisted potentially impactful gene targets therein, comprising known, suspected and hitherto inconspicuous candidates. The current investigation endorsed the validity of this panel to serve as an oncogenomic resource and scaffold in pursuit of novel biomarkers and actionable gene targets in PMBL and evaluate “intelligent” therapies directed against specific lesions in cells bearing them.
Supporting Information
A: Transcriptional profiling of PMBL and other hematopoietic cell lines was used to construct a cluster diagram–(updated from ref. 10)—of mRNA expression. The AU value (printed red) gives the "approximately unbiased" p-value, which is calculated by multiscale bootstrap resampling. The bootstrap probability value is less stringent than AU value when testing significance. Clusters (edges) with high AU values are strongly correlated. Results are shown as a dendrogram, showing different expression profiles as early dividing branches. B: PCA plot of microRNA expression. Note discrete clustering of both mRNA and miR expression showing that PMBL cell lines occupy a unique niche apart from other hematopoietic entities. Abbreviations: AML, acute myeloid leukemia; DLBCL, diffuse large B-cell lymphoma; erythro-megakaryocytic leukemia; ITL, immortalized T-cell; PMBL, primary mediastinal B-cell lymphoma; TCL, T-cell lymphoma.
(TIF)
Shows absence of product yielded for ACTR2-RAF1 fusion suggested by genomic breakpoints and reported recently [25]. Amplification of ETV6 served as positive control for confirmation of cDNA quality. NTC: no template control. Control cell line HL-60 is derived from a patient with acute myeloid leukemia.
(TIF)
Shows global microarray expression for select genes in PMBL patients compared to cHL. Data extracted from ref [27].
(PDF)
(DOCX)
Acknowledgments
The authors thank donors of cell lines used here, Andreas Rosenwald for access to microarray expression data and Marshall Kadin for reviewing the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no support or funding to report.
References
- 1. Boleti E, Johnson PW. Primary mediastinal B-cell lymphoma. Hematol Oncol 2007; 25: 157–163. [DOI] [PubMed] [Google Scholar]
- 2. Dunleavy K, Wilson WH. Primary mediastinal B-cell lymphoma and mediastinal gray zone lymphoma: do they require a unique therapeutic approach? Blood 2015; 125: 33–9. 10.1182/blood-2014-05-575092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Joos S, Otano-Joos MI, Ziegler S, Brüderlein S, Du Manoir S, Bentz M, et al. Primary mediastinal (thymic) B-cell lymphoma is characterized by gains of chromosomal material including 9p and amplification of the REL gene. Blood 1996; 87: 1571–8. [PubMed] [Google Scholar]
- 4. Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med 2003; 198: 851–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mestre C, Rubio-Moscardo F, Rosenwald A, Climent J, Dyer MJ, Staudt L, et al. Homozygous deletion of SOCS1 in primary mediastinal B-cell lymphoma detected by CGH to BAC microarrays. Leukemia 2005; 19: 1082–4. [DOI] [PubMed] [Google Scholar]
- 6. Wessendorf S, Barth TFE, Viardot A, Mueller A, Kestler HA, Kohlhammer H, et al. Further delineation of chromosomal consensus regions in primary mediastinal B-cell lymphomas: An analysis of 37 tumor samples using high-resolution genomic profiling (array-CGH). Leukemia 2007; 21: 2463–9. [DOI] [PubMed] [Google Scholar]
- 7. Twa DDW, Chan FC, Ben-Neriah S, Woolcock BW, Mottok A, Tan KL, et al. Genomic rearrangements involving programmed death ligands are recurrent in primary mediastinal large B-cell lymphoma. Blood 2014; 123: 2062–5. 10.1182/blood-2013-10-535443 [DOI] [PubMed] [Google Scholar]
- 8. Gunawardana J, Chan FC, Telenius A, Woolcock B, Kridel R, Tan KI, et al. Recurrent somatic mutations of PTPN1 in primary mediastinal B cell lymphoma and Hodgkin lymphoma. Nature Genet 2014; 46: 329–35. 10.1038/ng.2900 [DOI] [PubMed] [Google Scholar]
- 9. MacLeod RA, Nagel S, Scherr M, Schneider B, Dirks WG, Uphoff CC, et al. Human leukemia and lymphoma cell lines as models and resources. Curr Med Chem 2008; 15: 339–59. [DOI] [PubMed] [Google Scholar]
- 10. Drexler HG, Ehrentraut S, Nagel S, Eberth S, MacLeod RAF. Malignant hematopoietic cell lines: in vitro models for the study of primary mediastinal B-cell lymphomas. Leuk Res 2015; 39: 18–29. 10.1016/j.leukres.2014.11.002 [DOI] [PubMed] [Google Scholar]
- 11. Alston-Roberts C, Barallon R, Bauer SR, Butler J, Capes-Davis A, Dirks WG, et al. Cell line misidentification: the beginning of the end. Nat Rev Cancer 2010; 10: 441–8. 10.1038/nrc2852 [DOI] [PubMed] [Google Scholar]
- 12. Ben-Bassat H, Polliack A, Shlomai Z, Kohn G, Hadar R, Rabinowitz R, et al. Farage, a novel early B cell lymphoma cell line with trisomy 11. Leuk Lymphoma 1992; 6: 513–21. [Google Scholar]
- 13. Eberle FC, Rodriguez-Canales J, Wei L, Hanson JC, Killian JK, Sun HW, et al. Methylation profiling of mediastinal gray zone lymphoma reveals a distinctive signature with elements shared by classical Hodgkin`s lymphoma and primary mediastinal large B-cell lymphoma. Haematologica 2011; 96: 558–66. 10.3324/haematol.2010.033167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nacheva E, Dyer MJS, Metivier C, Jadayel D, Stranks G, Morilla R, et al. B-cell non-Hodgkin`s lymphoma cell line (Karpas 1106) with complex translocation involving 18q21.3 but lacking BCL2 rearrangement and expression. Blood 1994; 84: 3422–8. [PubMed] [Google Scholar]
- 15. Möller P, Brüderlein S, Sträter J, Leithäuser F, Hasel C, Bataille F, et al. MedB-1, a human tumor cell line derived from a primary mediastinal large B-cell lymphoma. Int J Cancer 2001; 92: 348–53. [DOI] [PubMed] [Google Scholar]
- 16. Sambade C, Berglund M, Lagercrantz S, Sällström J, Reis RM, Eblad G, et al. U-2940, a human B-cell line derived from a diffuse large cell lymphoma sequential to Hodgkin lymphoma. Int J Cancer 2006; 118: 555–63. [DOI] [PubMed] [Google Scholar]
- 17. Lam LT, Davis RE, Ngo VN, Lenz G, Wright G, Xu W, et al. Compensatory IKKalpha activation of classical NK-kappaB signaling during IKKß inhibition by an RNA interference sensitization screen. Proc Natl Acad Sci USA 2008; 105: 20798–803. 10.1073/pnas.0806491106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hailfinger S, Lenz G, Ngo V, Posvitz-Fejfar A, Rebeaud F, Guzzardi M, et al. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc Natl Acad Sci USA 2009; 106: 19946–51. 10.1073/pnas.0907511106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drexler HG. Guide to Leukemia-Lymphoma Cell Lines, 2nd edition. eBook-on-CD, Braunschweig, 2010.
- 20. Dirks WG, Drexler HG. STR DNA typing of human cell lines: detection of intra- and interspecies cross-contamination. Methods Mol Biol 2013; 946: 27–38. 10.1007/978-1-62703-128-8_3 [DOI] [PubMed] [Google Scholar]
- 21. MacLeod RAF, Kaufmann M, Drexler HG. Cytogenetic harvesting of commonly used tumor cell lines. Nature Protocols 2007; 2: 372–82. [DOI] [PubMed] [Google Scholar]
- 22. Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Systematic Biology 2002; 51: 492–508. [DOI] [PubMed] [Google Scholar]
- 23. Wickham H. ggplot2: elegant graphics for data analysis: Springer; New York; 2009. [Google Scholar]
- 24. ISCN 1995: An International System for Human Cytogenetic Nomenclature, Karger, Basel, 1995. [Google Scholar]
- 25. Klijn C, Durinck S, Stawiski EW, Haverty PM, Jiang Z, Liu H, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015; 33: 306–12. 10.1038/nbt.3080 [DOI] [PubMed] [Google Scholar]
- 26. Kimm LR, deLeeuw RJ, Savage KJ, Rosenwald A, Campo E, Delabie J, et al. Frequent occurrence of deletions in primary mediastinal B-cell lymphoma. Genes Chromosomes Cancer 2007; 46: 1090–7. [DOI] [PubMed] [Google Scholar]
- 27. Tiacci E, Döring C, Brune V, van Noesel CJ, Klapper W, Mechtersheimer G, et al. Analyzing primary Hodgkin and Reed-Sternberg cells to capture the molecular and cellular pathogenesis of classical Hodgkin lymphoma. Blood 2012; 120: 4609–20. 10.1182/blood-2012-05-428896 [DOI] [PubMed] [Google Scholar]
- 28. Schmitz R, Hansmann ML, Bohle V, Martin-Subero JI, Hartmann S, Mechtersheimer G, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med 2009; 206: 981–9. 10.1084/jem.20090528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Melzner I, Bucur AJ, Brüderlein S, Dorsch K, Hasel C, Barth TFE, et al. Biallelic mutation of SOCS-1 impairs JAK2 degradation and sustains phospho-JAK2 action in the MedB-1 mediastinal lymphoma line. Blood 2005; 105: 2535–42. [DOI] [PubMed] [Google Scholar]
- 30. Melzner I, Weniger MA, Bucur AJ, Brüderlein S, Dorsch K, Hasel C, et al. Biallelic deletion within 16p13.13 including SOCS-1 in Karpas1106P mediastinal B-cell lymphoma line is associated with delayed degration of JAK2 protein. Int J Cancer 2006; 118: 1941–4. [DOI] [PubMed] [Google Scholar]
- 31. Mestre-Escorihuela C, Rubio-Moscardo F, Richter JA, Siebert R, Climent J, Fresquet V, et al. Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood 2007; 109: 271–80. [DOI] [PubMed] [Google Scholar]
- 32. Bertolo C, Roa S, Sagardoy A, Mena-Varas M, Robles EF, Martinez-Ferrandis JI, et al. LITAF, a BCL6 target gene, regulates autophagy in mature B-cell lymphomas. Br J Haematol 2013;162: 621–30. 10.1111/bjh.12440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou J, Yang Z, Tsuji T, Gong J, Xie J, Chen C, et al. LITAF and TNFSF15 two downstream targets of AMPK exert inhibitory effects on tumor growth. Oncogene 2011; 30: 1892–900. 10.1038/onc.2010.575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu D, Guo R, Sobeck A, Bachrati CZ, Yang J, Enomoto T, et al. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev 2008; 22: 2843–55. 10.1101/gad.1708608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Challa-Malladi M, Lieu YK, Califano O, Holmes AB, Bhagat G, Murty VV, et al. Combined genetic inactivation of β2-Microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell 2011; 20: 728–40. 10.1016/j.ccr.2011.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Savage KJ, Monti S, Kutok JL, Cattoretti G, Neuberg D, De Leval L, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood 2003; 102: 3871–9. [DOI] [PubMed] [Google Scholar]
- 37. Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature 2012; 481: 90–93. 10.1038/nature10688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de Miranda NF, Peng R, Georgiou K, Wu C, Falk Sörqvist E, Berglund M, et al. DNA repair genes are selectively mutated in diffuse large B cell lymphomas. J Exp Med 2013; 210: 1729–42. 10.1084/jem.20122842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Enciso-Mora V, Broderick P, Ma Y, Jarrett RF, Hjalgrim H, Hemminki K, et al. A genome-wide association study of Hodgkin's lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3). Nat Genet 2010; 42: 1126–30. 10.1038/ng.696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Taskinen M, Louhimo R, Koivula S, Chen P, Rantanen V, Holte H, et al. Deregulation of COMMD1 is associated with poor prognosis in diffuse large B-cell lymphoma. PLoS One 2014;9: e91031 10.1371/journal.pone.0091031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bartuzi P, Hofker MH, van de Sluis B. Tuning NF-κB activity: a touch of COMMD proteins. Biochim Biophys Acta 2013; 1832: 2315–21. 10.1016/j.bbadis.2013.09.014 [DOI] [PubMed] [Google Scholar]
- 42. Weniger MA, Gesk S, Ehrlich S, Martin-Subero JI, Dyer MJS, Siebert R, et al. Gains of REL in primary mediastinal B-cell lymphoma coincide with nuclear accumulation of REL protein. Genes Chromosomes Cancer 2007; 46: 406–15. [DOI] [PubMed] [Google Scholar]
- 43. Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 2009; 459: 717–21. 10.1038/nature07968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carbone A, Gloghini A, Kwong YL, Younes A. Diffuse large B cell lymphoma: using pathologic and molecular biomarkers to define subgroups for novel therapy. Ann Hematol 2014; 93: 1263–77. 10.1007/s00277-014-2116-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lenz G, Wright GW, Emre TNC, Kohlhammer H, Dave SS, Davis RE, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci USA 2008; 105: 13520–5. 10.1073/pnas.0804295105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guiter C, Dusanter-Fort I, Copie-Bergman C, Boullard ML, Le Gouvello S, Gaulard P, et al. Constitutive STAT6 activation in primary mediastinal large B-cell lymphoma. Blood 2004; 104: 543–9. [DOI] [PubMed] [Google Scholar]
- 47. Weniger MA, Melzner I, Menz CK, Wegener S, Bucur AJ, Dorsch K, et al. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene 2006; 25: 2679–84. [DOI] [PubMed] [Google Scholar]
- 48. Hao Y, Chapuy B, Monti S, Sun HH, Rodig SJ, Shipp MA. Selective JAK2 inhibition specifically decreases Hodgkin lymphoma and mediastinal large B-cell lymphoma growth in vitro and in vivo. Clin Cancer Res 2014; 20: 2674–83. 10.1158/1078-0432.CCR-13-3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Skinnider BF, Elia AJ, Gascoyne RD, Patterson B, Trumper L, Kapp U, et al. Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood 2002; 99: 618–26. [DOI] [PubMed] [Google Scholar]
- 50. Ritz O, Rommel K, Dorsch K, Kelsch E, Melzner J, Buck M, et al. STAT6-mediated BCL6 repression in primary mediastinal B-cell lymphoma (PMBL). Oncotarget 2013; 4: 1093–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med 2000; 192: 1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Homet Moreno B, Ribas A. Anti-programmed cell death protein-1/ligand-1 therapy in different cancers. Br J Cancer 2015; 112: 1421–7. 10.1038/bjc.2015.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: Transcriptional profiling of PMBL and other hematopoietic cell lines was used to construct a cluster diagram–(updated from ref. 10)—of mRNA expression. The AU value (printed red) gives the "approximately unbiased" p-value, which is calculated by multiscale bootstrap resampling. The bootstrap probability value is less stringent than AU value when testing significance. Clusters (edges) with high AU values are strongly correlated. Results are shown as a dendrogram, showing different expression profiles as early dividing branches. B: PCA plot of microRNA expression. Note discrete clustering of both mRNA and miR expression showing that PMBL cell lines occupy a unique niche apart from other hematopoietic entities. Abbreviations: AML, acute myeloid leukemia; DLBCL, diffuse large B-cell lymphoma; erythro-megakaryocytic leukemia; ITL, immortalized T-cell; PMBL, primary mediastinal B-cell lymphoma; TCL, T-cell lymphoma.
(TIF)
Shows absence of product yielded for ACTR2-RAF1 fusion suggested by genomic breakpoints and reported recently [25]. Amplification of ETV6 served as positive control for confirmation of cDNA quality. NTC: no template control. Control cell line HL-60 is derived from a patient with acute myeloid leukemia.
(TIF)
Shows global microarray expression for select genes in PMBL patients compared to cHL. Data extracted from ref [27].
(PDF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.