Abstract
Monochromatic light is widely applied to promote poultry reproductive performance, yet little is currently known regarding the mechanism by which light wavelengths affect pigeon reproduction. Recently, high-throughput sequencing technologies have been used to provide genomic information for solving this problem. In this study, we employed Illumina Hiseq 2000 to identify differentially expressed genes in ovary tissue from pigeons under blue and white light conditions and de novo transcriptome assembly to construct a comprehensive sequence database containing information on the mechanisms of follicle development. A total of 157,774 unigenes (mean length: 790 bp) were obtained by the Trinity program, and 35.83% of these unigenes were matched to genes in a non-redundant protein database. Gene description, gene ontology, and the clustering of orthologous group terms were performed to annotate the transcriptome assembly. Differentially expressed genes between blue and white light conditions included those related to oocyte maturation, hormone biosynthesis, and circadian rhythm. Furthermore, 17,574 SSRs and 533,887 potential SNPs were identified in this transcriptome assembly. This work is the first transcriptome analysis of the Columba ovary using Illumina technology, and the resulting transcriptome and differentially expressed gene data can facilitate further investigations into the molecular mechanism of the effect of blue light on follicle development and reproduction in pigeons and other bird species.
Introduction
The White King pigeon (Columba) is an important commercial meat pigeon that has become popular in China in recent years [1–2]. Pigeons were probably domesticated in the Mediterranean region at least 3,000–5,000 years ago and possibly even earlier as a food source [3]. Paired pigeons lay only two eggs in a laying period, and newly hatch squabs are fed with crop milk from their parents, which is unique among birds [4]. Furthermore, the duration from egg laying to foraging ability of squabs is at least 38 days. Thus, the low productivity of the pigeon breeding industry hinders its development and needs to be improved.
Light is a major environmental factor affecting the reproductive activity of birds. Different light wavelengths have varying stimulatory effects on the retina and pineal cells of birds, resulting in behavioral changes that affect their physiology and reproduction [5]. Previous studies of the influence of light wavelength on physiological mechanism and its reproductive performance have been conducted in birds. McGinnis (1967) reported the chicken pullets were stimulated early reproductive activity by blue light, while Woodard et al. (1969) confirmed fertility rate of eggs under blue light was significantly lower than under white light [6–7]. Yadav et al. (2015) suggested that long-term exposure to blue light at low intensity may induce gonadal regression, even under long-day conditions [8]. A previous study from our group affirmed that monochromatic light influences the reproductive performance of pigeons [9]. In addition to the effect of light on reproductive activity, Halevy (2006) and Liu et al. (2010) found a stimulatory effect of green light on skeletal muscle development in chicks in ovo [10–11], and Xie et al. (2008) and Sadrzadeh et al. (2011) reported that monochromatic light affected immune function in broiler chickens [12–13]. However, the molecular mechanism by which monochromatic light influences physiological function in pigeons is poorly understood and needs further exploration.
Transcriptome sequencing can be used to discover genes that are functionally active and participate in specific biological processes [14–15]. In particular, Illumina sequencing technology can be used for gene identification with confirmed reliability [16]. Despite improvement in sequencing tools, sequence databases that provide a molecular understanding of bird physiology are still lacking. In the present study, we employed Illumina sequencing technology and de novo assembly to produce a transcriptome of Columba ovary tissue under blue and white light conditions and to annotate the expressed genes without related genome information. Our results not only provide reference information for the molecular mechanism governing reproductive performance under blue light, but also investigate the simple sequence repeats (SSRs) and single nucleotide polymorphisms (SNPs), which are valuable for further research on pigeons and related species.
Experimental Section
Ethics approval
This study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the Department of Animal Science and Technology, Yangzhou University and was performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals (China, 1988). All pigeon procedures were performed according to the Standards for the Administration of Experimental Practices (Jiangsu, China, 2008).
Pigeon rearing and sample preparation
White King pigeons were raised in an isolated loft at the College of Animal Science and Technology, Yangzhou University, Yangzhou. Seventy-two birds were divided into white and blue light groups, with three subgroups, the experiment was 6 months in length. Pigeons were provided with food and water ad libitum. Birds were exposed to white light (400–760 nm) or blue light (480 nm), received 15 h of light exposure (15 h light, 9 h dark). The intensity of light was 15.20 ± 0.65 lux as measured using a TES-1336A light meter (TES Electrical Electronic Crop., Taipei, China).
Six female birds with similar body weights (mean weight: 557.02 g) and similar physiological periods (i.e., the day after the second egg was laid) were anesthetized with sodium pentobarbital at a dosage of 2.5 mg/100 g body weight. All efforts were made to minimize distress. Ovarian samples were rapidly collected, flash frozen in liquid nitrogen, and stored at -80°C.
Total RNA extraction and cDNA library preparation
Ovary tissues of three individuals from two groups were subjected to RNA extraction. Total RNA was extracted from the collected ovaries using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. The samples for transcriptome analysis were prepared using Illumina’s kit following the manufacturer’s instructions. The quantity, purity, and integrity of RNA were measured using a 2100 Bioanalyzer (Agilent Technologies, Wilmington, DE, USA). Oligo (dT) magnetic beads were used to purify 6 μg of total RNA, furthermore, ovary tissues from 6 pigeons were used to establish libraries respectively. Short fragments (approximately 200 bp) were obtained and used as templates for first-stand cDNA synthesis using random hexamer primers. The second strand was synthesized using buffer, dNTPs, RNase H, and DNA polymerase I. The double-stranded cDNA was purified using the Qiaquick PCR extraction kit (Qiagen, Hilden, Germany) and eluted with elution buffer for end-repair and poly(A) addition. Finally, sequencing adaptors were ligated onto the fragments. The cDNA library was sequenced on the Illumina sequencing platform, and raw reads were generated using the Solexa pipeline according to the manufacturer’s recommendations.
De novo transcriptome assembly
Adaptor sequences and low-quality sequences (threshold quality, 20; window size, 5 bp; threshold length, 35 bp) were filtered. The paired-end sequencing strategy was used to better assemble the transcriptome de novo. De novo transcriptome assembly was performed using the Trinity program (version r2013/11/10) [17], and the Trinities were clustered into unigenes using TGICL tools [18].
Functional annotation of unigenes
The assembled unigenes were searched against the BLAST NR protein sequence database (http://blast.ncbi.nlm.nih.gov), the Swiss-Prot database (http://www.expasy.ch/sprot), and the KEGG database (http://www.genome.jp/kegg) using the BLASTx algorithm. A typical cut-off value of E<1e-5 was used. The unigenes were sorted to recover proteins with the most similarity to the unigenes with putative functional annotations. GO annotation of the unigenes was obtained at the second level according to biological process, component function, and cellular component. Furthermore, eukaryotic orthologous groups were used to predict and classify unigene functions.
Molecular markers detection
MIcroSAtellite (http://pgrc.ipk-gatersleben.de/misa) was used to identify putative SSRs in the unigenes from the assembled transcripts. The minimum number of repeat units for mono-, di-, tri-, tetra-, penta- and hexa-nucleotide motifs were set as 10, 6, 5, 5, 5 and 5, respectively. Samtools and bcftools were applied to identify putative SNPs in this transcriptome.
Validation of RNA-seq results by qRT-PCR
Total RNA was isolated by TRIzol reagent (Invitrogen), and the concentration of RNA was diluted to 1 μg/μl to be used for first-strand cDNA synthesis using the Fast Quant RT Kit (Tiangen). qRT-PCR was performed using SuperReal PreMix (SYBR Green, Tiangen) according to the manufacturer’s protocol. To validate the assembled unigenes, 8 unigenes were selected for qRT-PCR (S1 Table). All measurements were conducted in triplicates. The pigeon glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as an internal control. The 2-ΔΔCT method was used to analyze relative RNA expression [19].
Results and Discussion
De novo transcriptome sequencing and assembly
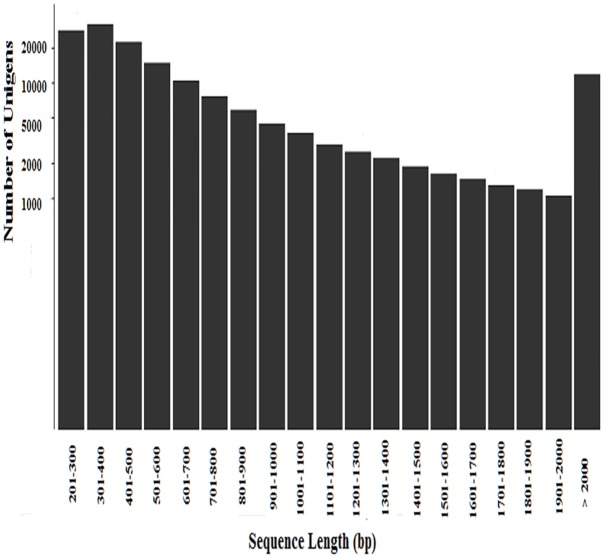
From our reproductive data, the total egg production of pigeons (32.67±1.67 monthly) under blue light which was significantly lower than white light (68.16±3.45 monthly). This results induced us to explore differences in pigeon ovary transcriptome under blue and white light conditions, three pigeons under blue light (B1, B2, and B3) and three pigeons under white light (W1, W2, and W3) were selected for analysis. The cDNA of pigeons was sequenced by Illumina Hiseq 2000, and transcriptome sequencing generated 14.07 million and 11.90 million reads from ovaries under blue and white light conditions, respectively. Detailed results of sequencing and assembly are shown in Table 1. Trinity software was applied to assemble the reads into a transcriptome, as no reference genome is yet available for Columba [20–21]. The numbers of clean reads in the ovary transcriptome libraries for blue and white light conditions were 17.39 Gb and 14.69 Gb, respectively. We obtained 157,774 unigenes (mean length: 790 bp) with an N50 of 1,108 bp. The length of the unigenes ranged from 201 to 17,581 bp, and the lengths of 11,781 unigenes greater than 2,000 bp (Fig 1). Notably, the unigenes obtained in the present study were longer than those obtained in previous studies.
Table 1. Summary of Illumina sequencing and transcriptome assembly.
| Condition | Sample name | Raw reads | Clean reads | Clean bases (bp) | Valid rate (%) | Q30 (%) | GC content (%) |
|---|---|---|---|---|---|---|---|
| Blue light | B1 | 41550778 | 41229474 | 5140724575 | 98.97 | 94.23 | 53.00 |
| Blue light | B2 | 55086542 | 54601944 | 6810865040 | 98.88 | 93.86 | 53.00 |
| Blue light | B3 | 43980608 | 43620184 | 5437497814 | 98.90 | 94.00 | 53.00 |
| White light | W1 | 42935054 | 42523434 | 5298145890 | 98.71 | 93.79 | 55.00 |
| White light | W2 | 33365916 | 33090892 | 4132171509 | 99.07 | 93.2 | 54.00 |
| White light | W3 | 42670976 | 42283004 | 5258718979 | 98.59 | 92.07 | 55.00 |
Fig 1. Length distribution and frequency of unigenes in Columba.
Our analysis indicated that, in all samples, 84.41% of the left and 91.02% of the right reads could be mapped back to the assembled transcriptome, with 65.53% of proper pairs mapped for a representative sample. Unmapped sequences were due to read orphans, poor-quality reads, or incomplete transcripts. Until now, no standard criteria have been applied to evaluate the quality of transcriptome assemblies [22]. Our results indicate that the Illumina-generated dataset has high reliability and covers most transcriptome sequences, which can be valuable for further research. The sequences of unigenes were deposited in the NCBI Transcriptome Shortgun Assembly Sequence Database (B1: SRR2094734, B2: SRR2094746, B3: SRR2094764; W1: SRR2094777, W2: SRR2094789, W3: SRR2094799.).
Functional annotation and classification
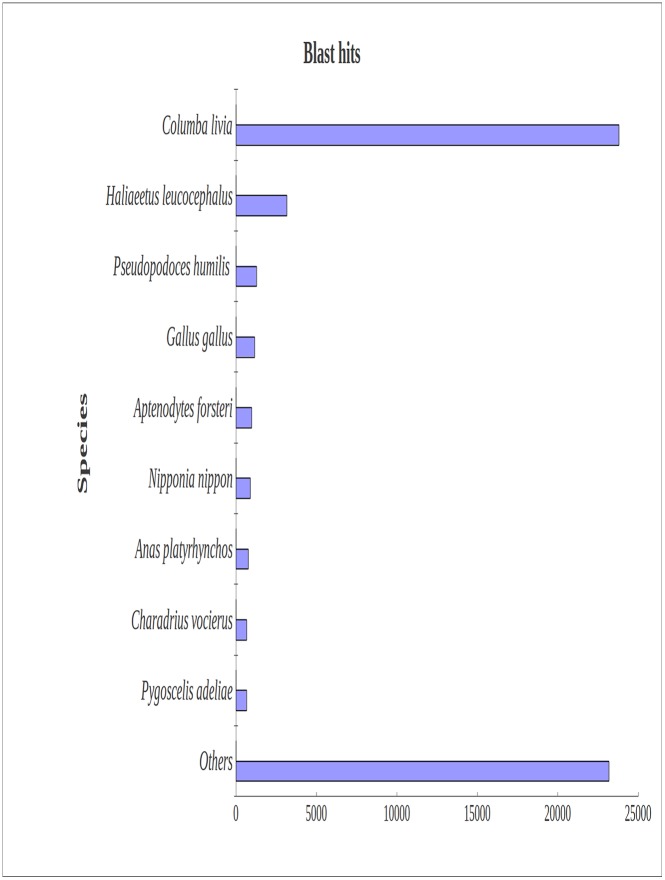
Blast2GO was used to annotate the unigenes [23]. We carried out a BLASTx search against NR (non-redundant) and Swiss-prot protein databases with a cut-off E-value of 10−5 or lower. Short assembled Trinity transcripts were difficult to match with known genes; 56,530 unigenes (35.83% of all distinct sequences) and 45,797 unigenes (29.03% of all distinct sequences) were determined as significant hits in the NR and Swiss-prot databases, respectively (Table 2). Many sequences did not have BLASTx hits. Most (42.10%) BLASTx-hit transcripts matched Columba livia, followed by Haliaeetus leucocephalus, Pseudopodoces humilis, Gallus gallus, and Aptenodytes forsteri (Fig 2).
Table 2. List of annotations.
| Annotation database | Annotation number | Percent of annotation (%) |
|---|---|---|
| Total unigenes | 157774 | 100 |
| NR | 56530 | 35.8297 |
| Swiss-prot | 45797 | 29.0270 |
| COG | 34655 | 21.9650 |
| KEGG | 18203 | 11.5374 |
| GO | 18833 | 11.9367 |
NR, non-redundant; COG, cluster of orthologous group; KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, gene ontology.
Fig 2. Species distribution of BLASTx matches for ovary transcriptome unigenes.
More than 40% of identified transcripts had the highest homology with Columba livia, and about 6% of top hits matched Haliaeetus leucocephalus.
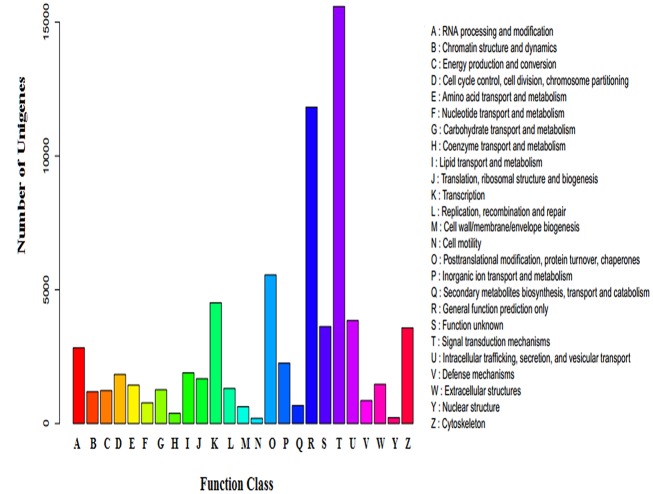
The cluster of orthologous group (COG) classifications of annotated sequences were used to determine the effectiveness of the annotation process and the completion of the transcriptome library. A total of 34,655 sequences were aligned to the COG database (Fig 3). The cluster for ‘signal transduction mechanisms’ represented the largest group (15,587, 44.98%), followed by ‘general function prediction’ (11,827, 34.13%) and ‘posttranslational modification, protein turnover, chaperones’ (5555, 16.03%). ‘Nuclear structure’ (220, 0.0063%) and ‘cell motility’ (198, 0.0057%) were the smallest groups.
Fig 3. COG classifications.
A total of 34,655 sequences were grouped into 25 COG categories.
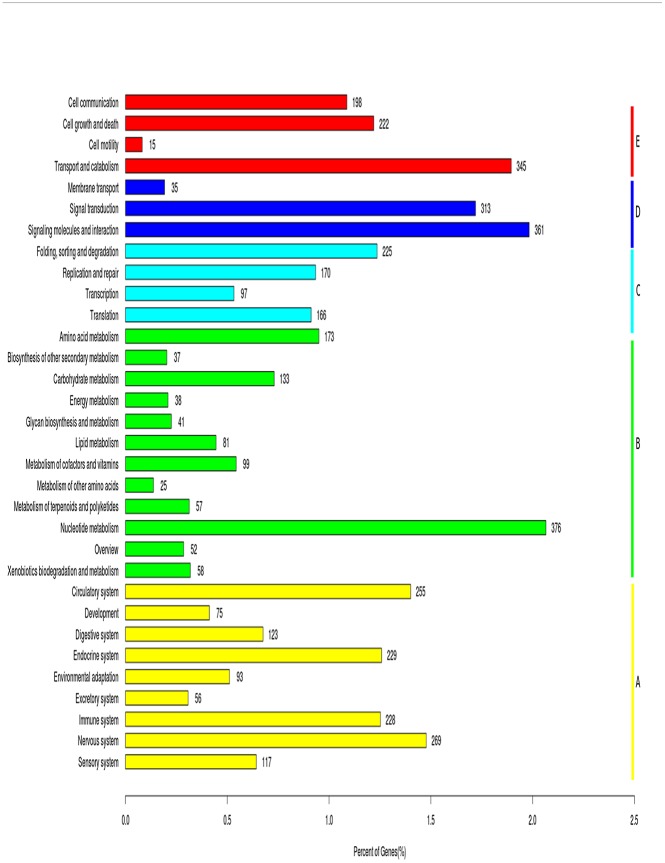
All unigenes were mapped to reference canonical pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Fig 4) [24]. A total of 18,203 unigenes were matched to 356 KEGG pathways (S2 Table). The most highly enriched pathways were those related to PI3K-Akt signaling (n = 998), MAPK signaling (n = 801), and focal adhesion (n = 772). Enriched pathways also included those involved in reproduction and circadian rhythm, such as ovarian steroidogenesis, Wnt signaling, estrogen signaling, and circadian rhythm and entrainment [25–26].
Fig 4. Pathway assignment based on the KEGG database.
Classification based on (A) organismal system categories, (B) metabolism categories, (C) genetic information processing categories, (D) environmental information processing categories, and (E) cellular process categories.
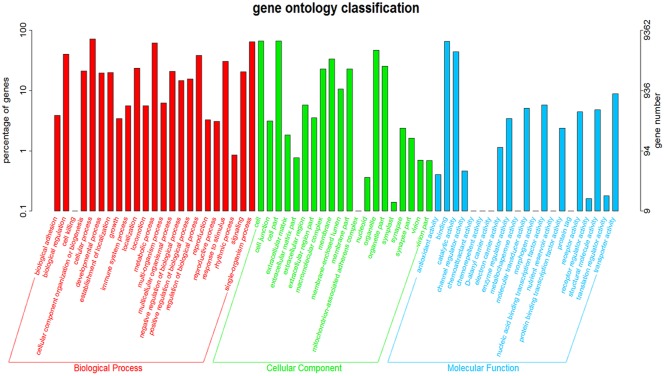
When gene ontology (GO) assignment programs were applied for functional categorization, a total of 18,833 unigenes were classified into 63 functional groups (Fig 5) [27]. Biological process, cellular component, and molecular function were the three main ontologies. As expected, the reproductive process, reproduction, and the rhythmic process were found among the biological process categories (S3 Table). This GO assignment results are similar to the previously sequenced Anser cygnoides ovary transcriptome [28]. The KEGG and GO annotations are valuable for identifying potential genes from the vast transcriptome database, providing a helpful information on the mechanisms by which light wavelength may influence the bird ovary.
Fig 5. GO classification map.
The x-axis indicates the next level GO term of the three GO categories: biological process, cellular component, and molecular function.
Analysis of differentially expressed genes (DEGs)
The light spectrum affects the reproduction of pigeons due to changes in reproductive processes and circadian rhythm [10, 29]. Although the effect of monochromatic light on the reproductive performance and growth of birds have been widely studied, the molecular mechanisms regulating biosynthesis and reproductive function remain unknown. We used the reads per kilobase per million method to calculate gene expression filtered by a false discovery rate of <1 and log2 (fold change) of >1 [30]. A total of 6,831 DEGs were identified between blue and white light conditions, with 3,305 up-regulated and 3,526 down-regulated genes. These genes include those involved in hormone synthesis, oocyte meiosis, and circadian rhythm (S4 Table). As the number of unigenes with no homologs in the NR database was 45%, this suggests that some of the DEGs may be expressed specifically in the pigeon ovary and involved in ovulation. In our study, a number of genes involving in the circadian rhythm, and genes regulating the synthesis and metabolism of hormones and vital components for oocyte maturation. The further studies are necessary to identify these unknown genes which will facilitate molecular mechanism of blue light transduction in pigeons.
The circadian clock, regulates the circadian rhythm [31], and circadian rhythm is a 24 hr rhythms of behavior and other physiological function which are based upon an endogenous self-sustained oscillation [32]. Wunderer et al. (2013) suggested that clock genes and their protein products may be directly involved in the photoperiod-dependent regulation and adaptation of hormone synthesis and release [33]. The pathway of circadian entertainment controls the pathway of circadian rhythm, light induces the presynaptic retinal ganglion cells (RGC) neurons and postsynaptic suprachiasmatic nucleus (SCN) neurons, which trigger the clock genes immediate early genes and the initial of circadian rhythm. The PER and CRY synthesize heterodimer which works as negative component while the BMAL1 and CLOCK heterodimer is positive component [34–35], PER/CRY heterodimers inhibit CLOCK and BMAL1 expression, forming a loop or cycle of CLOCK/BMAL1-PER/CRY [36]. The Dec, Ror and CK1 genes regulate the circadian rhythm which were found in this transcriptome. AMPK modulated the degradation of CRY, was also identified in this transcriptome [37]. Furthermore, our previous studies showed the BMAL1 which was a core component of the circadian rhythm, correlated with birth rate of pigeons under the monochromatic lights supplement [9].
Steroid hormones, such as estrogen, androgen and progestin, have been studied in pigeons, regulated vitellogenesis, incubation, and oviduct development [38–41]. The enzymes involved in steroid biosynthesis pathway are being recognized as important target for endocrine-disrupting, which would impair reproduction [42], the 3β-hydroxysteroid dehydrogenase (3-β HSD) and aromatase cytochrome P450 enzymes which are essential for the biosynthesis of all classes of steroid hormones, were detected in this transcriptome. All active steroid hormones need to be converted from 3-β HSD precursors to hormonally active 3-ketosteroid, which is Δ5–3β-hydroxysteroids converted into Δ4-3-ketosteroids [43], a reaction step catalyzed by 3-β HSD [44]. P450arom catalyzes the conversion of androgens to estrogen, which is a key step in estrogen biosynthesis [45–46]. Furthermore, placental P450arom converts androstenedione (Δ4A) and testosterone (T) derived from fetal and maternal adrenal dehydroepiandrosterone sulfate (DHEA-S) to estrone (E1) and estradiol (E2) [47]. Apart from the genes involved in steroid hormone biosynthesis, genes encoding their receptor were also detected, such as estrogen receptor β (ERβ), mice lacking ERβ have fewer and smaller litters than wild-type mice [48]. The expression of ERβ and 3-β HSD in ovary was significantly lower in blue light, which in accord with the egg production of this experiment.
Oocyte maturation, immature oocytes become fertilizable eggs through meiotic maturation [49], in Xenopus oocytes, maturation is thought to be initiated by steroid hormone progesterone [50], the progesterone-mediated oocyte maturation pathway was found here. There is some evidence that progesterone receptor in oocytes is a membrane-bound receptor, possibly coupled to heterotrimeric G-proteins, which inhibit adenylate cyclase [51]. Gβγ-subunit activates meiotic maturation, which may be mediated by PI3K activation. Previous study have showed phosphatidylinositol 3-kinase (PI3K) is known to play critical roles in signal transduction processes related to a variety of cellular activities [52], which participate in mouse meiotic maturation and also implicate in progesterone-induced maturation [53]. Furthermore, class 1A PI3K catalytic subunits (locus name PIK3c) are associated with a regulatory subunit (PIK3r) which participate in phosphorylation reaction [54], both genes were identified in this transcriptome. The gene protein kinase Mos and anaphase-promoting complex regulated the oocyte maturation were identified through the Illumina analysis [55–56], which will enable us to inspect the molecular mechanism of oocyte maturation which affect the reproductive process of pigeon.
In this de novo transcriptome assembly, we obtained a number of genes involved in the regulation of light spectra (blue and white light) in pigeons, most of these genes were discovered in pigeons for the first time, such as RORβ, 3β-HSD and CDC27. Further studies are required to elaborate their roles in reproductive process under the blue light in pigeons.
DEGs in the ovary under blue and white light conditions
As it is important to understand the mechanisms of the effect of light wavelengths on ovulation, we characterized annotated DEGs related to ovulation, which included genes involved in ovarian steroidogenesis (bone morphogenetic protein 15 (BMP15), 3-β-hydroxysteroid dehydrogenase (3β-HSD)), oocyte meiosis (cell division cycle 27 (CDC27) and mitotic spindle assembly checkpoint), and cell cycle (transforming growth factor β2 (TGF-β2), E2F transcription factor 1 (E2F1)). In addition to genes related to ovulation, we also characterized several genes previously found to play a role in the effects of blue light, including estrogen receptor β (ERβ), mitogen-activated protein kinase kinase kinase 1 (MAP3K1), nuclear receptor RORβ, and melatonin receptor.
MAP3K1, an important component of the MAPK pathway, cooperates with other factors in steroid-dependent transcription [57]. In the present study, MAP3K1 expression in the ovary was up-regulated under blue light, indicating its potential involvement in the effects of monochromatic light. Previous studies show that BMP15 enhances oocyte development and regulates oocyte developmental programming, and BMP15 mutation causes monoovulatory cycles in humans and reduces ovulation rate in mice [58–59]. In the present study, BMP15, ERβ, and 3β-HSD gene expression in the ovary were down-regulated under blue light, which is also consistent with the reproduction data in our experiment, which is in accordance with the results of Foss and White (1983), showing that a long wavelength of light might increase egg production in brown egg laying hens [60].
CDC27 is a part of the anaphase-promoting complex, which plays a vital role in mitosis [61]. E2F1 is thought to act as a transcriptional activator of progression through the G1/S transition, and loss of this factor abolishes the ability of mouse embryonic fibroblasts to enter S phase and progress through the cell cycle [62–63]. TGF-β2 also affects cell cycle. In the present study, we observed changes in the expression of TGF-β2 and E2F1 under blue and white light conditions, which are in accordance with the change of these genes in the cell cycle pathways. Also, as RORβ contributes to the peripheral circadian clock [64–65], the higher expression of RORβ observed in the present study suggests that circadian clock genes involved in the blue light mechanism on pigeons.
To validate the expression profiles obtained from Illumina sequencing analysis, eight DEGs chosen for their closely relation to ovulation and circadian rhythm were analyzed by quantitative real-time polymerase chain reaction (qRT-PCR) analysis. We observed that the trend of DEGs expressions were similar to the sequencing data (Table 3), providing further evidence of the credibility of the sequencing database.
Table 3. Real-time PCR confirmation of DEGs in ovaries between blue and white light conditions (log2fold-change).
| Sequence ID | Gene | Illumina sequencing | Real-time PCR |
|---|---|---|---|
| CL15329Contig1 | estrogen receptor β | -1.50 | -1.18 |
| CL10084Contig1 | cell division cycle 27 | 4.48 | 0.81 |
| CL3334Contig2 | mitogen-activated protein kinase kinase kinase 1 | 2.58 | 1.47 |
| CL9880Contig1 | bone morphogenetic protein 15 | -1.82 | -0.92 |
| CL12107Contig1 | transforming growth factor β2 | 2.15 | 1.77 |
| CL2Contig675 | nuclear receptor RORβ | -1.72 | -0.67 |
| CL8711Contig1 | 3-β-hydroxysteroid dehydrogenase | -2.33 | -1.60 |
| CL8514Contig1 | E2F transcription factor 1 | -1.76 | -0.97 |
Putative molecular markers
A total of 533,887 potential SNPs were identified among all of the unigens using the Poly Bayers (Table 4), including 400,607 (75.04%) transitions. The greatest amount of transitional base changes (53,809 A-G, 56,616 G-A, 72,756 C-T and 32,829 T-C) were from blue light, while the white light samples had (41,726 A-G, 51,989 G-A, 62,991 C-T and 27,891 T-C). 133,280 (24.96%) tranversions, including 3.43% A-C, 2.43% A-T, 2.94% C-A, 4.68% C-G, 3.41% G-C, 4.16% G-T, 1.68% T-A and 2.22% T-G. To determine the quality of the unigene database, 157,774 unigenes were assembled in this study to detect potential SSRs. A total of 17,574 potential SSRs were identified, which included two types of mononucleotide, four types of dinucleotide, ten types of trinucleotide and tetranucleotide, pentanucleotide, hexanucleotide SSRs. Mononucleotides SSRs were the most abundant microsatellite repeats unites (11,221, 63.85%), dinucleotide SSRs were second (2673 15.21%), followed by trinucleotide (3314, 18.86%), tetranucleotide (308, 1.75%), pentanucleotide (45, 0.26%), hexanucleotide (13, 0.07%) (Table 5). It was obvious that A/T accounted for 74.21% of the mononucleotide SSRs, AC/GT accounted for 44.56% of the dinucleotide SSRs, AGG/CCT accounted for 38.71% of the trinucleotide SSRs. The validation of the putative SNPs not only showed the utility of Illumina sequence for SNPs, but also identified a large number of SNPs and SSRs. This huge data will provide a valuable resources for the further genetic study of pigeons.
Table 4. Summary of single nucleotide polymorphisms (SNPs) of transcriptomic data from Columba.
| SNP Type | Blue | White | Total |
|---|---|---|---|
| Transition | 216010 | 184597 | 400607 |
| A-G | 53809 | 41726 | 95535 |
| G-A | 56616 | 51989 | 108605 |
| C-T | 72756 | 62991 | 135747 |
| T-C | 32829 | 27891 | 60720 |
| Transversion | 74836 | 58444 | 133280 |
| A-C | 10545 | 7778 | 18323 |
| A-T | 7472 | 5504 | 12976 |
| C-A | 8515 | 7176 | 15691 |
| C-G | 14036 | 10961 | 24997 |
| G-C | 10053 | 8162 | 18215 |
| G-T | 12634 | 9592 | 22226 |
| T-A | 5054 | 3932 | 8986 |
| T-G | 6527 | 5339 | 11866 |
| Total | 290846 | 243041 | 533887 |
Table 5. Summary of SSRs identified from the ovary transcriptomme of Columba.
| SSR type | Repeats | Total number | Proportion of total SSRs(%) |
|---|---|---|---|
| Mononucleotide | Total | 11221 | 63.85 |
| A(T) | 8327 | 47.38 | |
| C(G) | 2894 | 16.47 | |
| Dinucleotide | Total | 2673 | 15.21 |
| AC(GT) | 1191 | 6.78 | |
| AG(CT) | 535 | 3.04 | |
| AT(AT) | 932 | 5.30 | |
| CG(CG) | 15 | 0.09 | |
| Trinucleotide | Total | 3314 | 18.86 |
| AAC(GTT) | 155 | 0.88 | |
| AAG(CTT) | 210 | 1.19 | |
| AAT(ATT) | 251 | 1.43 | |
| ACC(GGT) | 213 | 1.21 | |
| ACG(CGT) | 10 | 0.06 | |
| ACT(AGT) | 13 | 0.07 | |
| AGC(CTG) | 753 | 4.28 | |
| AGG(CCT) | 1283 | 7.30 | |
| ATC(ATG) | 177 | 1.01 | |
| CCG(CGG) | 249 | 1.42 | |
| Tetranucleotide | Total | 308 | 1.75 |
| Pentanucleotide | Total | 45 | 0.26 |
| Hexanucleotide | Total | 13 | 0.07 |
Conclusions
In this study, we generated the first reference sequence for Columba by RNA sequencing of ovarian tissue under blue or white light conditions and cloud-based de novo transcriptome assembly. We obtained 157,774 unigenes with 56,530 sequences with a cut-off E-value greater than 10−5. This transcriptome database provides genomic information on the mechanisms of ovulation under different light wavelengths in pigeons. Considering the reliability of Illumina sequencing analysis for the detection of DEGs, potential SSRs and SNPs, our findings should be useful for future functional studies of genes involved in bird reproduction.
Supporting Information
(XLS)
(XLS)
(XLS)
(XLS)
Acknowledgments
This research was funded by grants from A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions and The Cultivate Funding of Six Kinds of High-level Personnel Selection in Jiangsu Province (2014—NY—036). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
The sequences of unigenes were deposited in the NCBI Transcriptome Shotgun Assembly Sequence Database (B1: SRR2094734, B2: SRR2094746, B3: SRR2094764; W1: SRR2094777, W2: SRR2094789, W3: SRR2094799).
Funding Statement
This research was funded by grants from A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions and The Cultivate Funding of Six Kinds of High-level Personnel Selection in Jiangsu Province (2014 - NY - 036). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gu HC. Encyclopedia of pigeon breeding. 1st ed. Shanghai: Shanghai dictionary press; 2004. p. 2–8. [Google Scholar]
- 2. Zhao WL. Production of special economic poultry. 2nd ed. Beijing: Agriculture publishing company; 1993. p. 12–23. [Google Scholar]
- 3. Driscoll CA, Macdonal DW, O’Brien SJ. From wild animals to domestic pets, an evolutionary view of domestication. Proc Natl Acad Sci U S A. 2009; 106: 9971–8. 10.1073/pnas.0901586106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Horseman ND, Buntin JD. Regulation of pigeon cropmilk secretion and parental behaviors by prolactin. Ann Rev Nutr. 1995; 15(1): 213–8. [DOI] [PubMed] [Google Scholar]
- 5. Campbell CL, Colton S, Haas R, Rice M, Porter A, Schenk A, et al. Effects of different wavelengths of light on the biology, behavior, and production of grow-out Pekin ducks. Poult Sci. 2015; pev166. [DOI] [PubMed] [Google Scholar]
- 6. McGinnis J. Effect of colored lights on growing pullets and laying hens Pacific Poultryman, 1967: 24–44. [Google Scholar]
- 7. Woodard AE, Moore JA, Wilson WO. Effect of wave length of light on growth and reproduction in Japanese quail (Coturnix coturnix japonica). Poul. Sci. 1969; 48: 118–23. [DOI] [PubMed] [Google Scholar]
- 8. Yadav S, Chaturvedi CM. Light colour and intensity alters reproductive/ seasonal responses in Japanese quail. Physiol Behav. 2015; 147: 163–8. 10.1016/j.physbeh.2015.04.036 [DOI] [PubMed] [Google Scholar]
- 9. Wang Y, Ding JT, Yang HM, Cao W, Li YB. The effect of new monochromatic light regimes on egg production and expression of the circadian gene BMAL1 in pigeons. Poult Sci. 2015; pev057. [DOI] [PubMed] [Google Scholar]
- 10. Halevy O, Piestun Y, Rozenboim I, Yablonka-Reuveni Z. In ovo exposure to monochromatic green light promotes skeletal muscle cell proliferation and affects myofiber growth in posthatch chicks. Am J Physiol Regul Integr Comp Physiol. 2006; 290(4): 1062–70. [DOI] [PubMed] [Google Scholar]
- 11. Liu W, Wang Z, Chen Y. Effects of monochromatic light on developmental changes in satellite cell population of pectoral muscle in broilers during early posthatch period. Anat Rec. 2010; 293(8): 1315–24. [DOI] [PubMed] [Google Scholar]
- 12. Xie D, Wang ZX, Dong YL, Cao J, Wang JF, Chen JL, et al. Effects of monochromatic light on immune response of broilers. Poult Sci. 2008; 87(8): 1535–9. 10.3382/ps.2007-00317 [DOI] [PubMed] [Google Scholar]
- 13. Sadrzadeh A, Brujeni GN, Liv M, Nazari MJ, Sharif MT, Hassanpour H, et al. Cellular immune response of infectious bursal disease and Newcastle disease vaccinationsin broilers exposed to monochromatic lights. Afr J Biotechnol. 2011; 10(24): 9528–32. [Google Scholar]
- 14. Teaniniuraitemoana V, Huvet A, Levy P, Klopp C, Lhuillier E, Gaertner-Mazouni N, et al. Gonad transcriptome analysis of pearl oyster Pinctada margaritifera: identification of potential sex differentiationand sex determining genes. BMC genomics. 2014; 15(1): 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Künstner A, Wolf JB, Backström N, Whitney O, Balakrishnan CN, Day L, et al. Comparative genomics based on massive parallel transcriptome sequencing reveals patterns of substitution and selection across 10 bird species. Mol Ecol. 2010; 19(s1): 266–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rosenkranz R, Borodina T, Lehrach H, Himmelbauer H. Characterizing the mouse ES cell transcriptome with Illumina sequencing. Genomics. 2008; 92(4): 187–94. 10.1016/j.ygeno.2008.05.011 [DOI] [PubMed] [Google Scholar]
- 17. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 2011; 29(7): 644–52. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pertea G, Huang X, Liang F, Valentin A, Sultana R, Karamycheva S, et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19(5): 651–2. [DOI] [PubMed] [Google Scholar]
- 19. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001; 25(4): 402–8. [DOI] [PubMed] [Google Scholar]
- 20. Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J,C et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013; 8(8): 1494–512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao QY, Wang Y, Kong YM, Da L, Li X, Hao P. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 2011; 12: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martin J, Bruno VM, Fang Z, Meng X, Blow M, Zhang T, et al. Rnnotator: an automated de novo transcriptome assembly pipeline from stranded RNA-Seq reads. BMC Genomics. 2010; 11(1): 663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005; 21(8): 3674–6. [DOI] [PubMed] [Google Scholar]
- 24. Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008; 36(s1): 480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boyer A, Goff AK, Boerboom D. WNT signaling in ovarian follicle biology and tumorigenesis.Trends Endocrin Met. 2010; 21(1): 25–32. [DOI] [PubMed] [Google Scholar]
- 26. Nakao N, Yasuo S, Nishimura A, Yamamura T, Watanabe T, Anraku T, et al. Circadian clock gene regulation of steroidogenic acute regulatory protein gene expression in preovulatory ovarian follicles. Endocrinology. 2007; 148(7): 3031–8. [DOI] [PubMed] [Google Scholar]
- 27. Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006; 34(s2): 293–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu Q, Zhao WM, Chen Y, Tong YY, Rong GH, Huang ZY, et al. Transcriptome profiling of the goose (Anser cygnoides) ovaries identify laying and broodiness phenotypes. PloS one. 2013; 8: e55496 10.1371/journal.pone.0055496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lewis PD, Morris TR. Poultry and coloured light. Worlds Poult Sci J. 2000; 56(3): 189–207. [Google Scholar]
- 30. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008; 5(7): 625–8. [DOI] [PubMed] [Google Scholar]
- 31. Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, et al. Mop3 is an essential components of the master circadian pacemaker in mammals. Cell. 2000; 103: 1009–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aschoff J. Circadian activity pattern with two peaks. Ecology. 1966; 47(4): 657–62. [Google Scholar]
- 33. Wunderer F, Kühne S, Jilg A, Ackermann K, Sebesteny T, Maronde E, et al. Clock gene expression in the human pituitary gland. Endocrinology. 2013; 154(6): 2046–57. 10.1210/en.2012-2274 [DOI] [PubMed] [Google Scholar]
- 34. Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for thedrosophilacircadian clock. Nature. 1996; 380: 129–35. [DOI] [PubMed] [Google Scholar]
- 35. Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting the drosophilaclock by photic regulation of PER and a PER-TIM complex. Science. 1996; 271: 1740–44. [DOI] [PubMed] [Google Scholar]
- 36. Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, et al. Disruption of the CLOCK components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010; 466(7306): 627–31. 10.1038/nature09253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009; 326(5951): 437–40. 10.1126/science.1172156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goerlich VC, Dijkstra C, Groothuis TGG. Effects of in vivo testosterone manipulation on ovarian morphology, follicular development, and follicle yolk testosterone in the homing pigeon. J Exp Zool A Ecol Genet Physiol. 2010; 313(6): 328–38. 10.1002/jez.600 [DOI] [PubMed] [Google Scholar]
- 39. Silver R. Prolactin and parenting in the pigeon family. J Exp Zool. 1984; 232(3): 617–25. [DOI] [PubMed] [Google Scholar]
- 40. Riddle O. Prolactin or progesterone as key to parental behaviour: A review. Anim Behav. 1963; 11(4): 419–32. [DOI] [PubMed] [Google Scholar]
- 41. Zhang H, Chen F, Li GL, Ding YY, Tao ZR, Li JJ, et al. Molecular cloning, expression, and regulation of estrogen receptors in pigeon oviduct epithelial cells. GMR. 2014; 13(1): 1926–37. 10.4238/2014.March.17.20 [DOI] [PubMed] [Google Scholar]
- 42. Sanderson JT. The steroid hormone biosynthesis pathway as a target for endocrine-disrupting chemicals. Toxicol Sci. 2006; 94(1): 3–21. [DOI] [PubMed] [Google Scholar]
- 43. Simard J, Ricketts ML, Gingras S, Soucy P, Feltus FA, Melner MH. Molecular biology of the 3β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase gene family. Endocr Rev. 2005; 26: 525–82 [DOI] [PubMed] [Google Scholar]
- 44. Yamamura K, Doi M, Hayashi H, Ota T, Murai I, Hotta Y, et al. Immunolocalization of murine type VI 3β-hydroxysteroid dehydrogenase in the adrenal gland, testis, skin, and placenta. Mol Cell Endocrinol. 2014; 382(1): 131–8. 10.1016/j.mce.2013.09.014 [DOI] [PubMed] [Google Scholar]
- 45. Simpson ER, Mahendroo MS, Means GD, Kilgore MW, Hinshelwood MM, Graham-Lorence S, et al. Aromatase Cytochrome P450, The Enzyme Responsible for Estrogen Biosynthesis. Endocr Rev. 1994; 15(3): 342–55. [DOI] [PubMed] [Google Scholar]
- 46. Nebert DW, Adesnik M, Coon MJ, Estabrook RW, Gonzalez FJ, Guengerich FP, et al. The P450 gene superfamily: recommended nomenclature. DNA. 1987; 6(1): 1–13. [DOI] [PubMed] [Google Scholar]
- 47. Mullis PE, Yoshimura N, Kuhlmann B, Lippuner K, Jaeger P, Harada H. Aromatase deficiency in a female who is compound heterozygote for two new point mutations in the P450arom gene: impact of estrogens on hypergonadotropic hypogonadism, multicystic ovaries, and bone densitometry in childhood. J Clin Endocrinol Metab. 1997; 82(6): 1739–45. [DOI] [PubMed] [Google Scholar]
- 48. Krege JH, Hodgin JB, Couse JF, Enmark E, Warner M, Mahler JF, et al. Generation and reproductive phenotypes of mice lacking estrogen receptorβ. Proc Natl Acad Sci U S A. 1998; 95(26): 15677–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schmitt A, Nebreda AR. Signalling pathways in oocyte meiotic maturation. J Cell Sci. 2002; 115(12): 2457–9. [DOI] [PubMed] [Google Scholar]
- 50. Wasserman WJ, Richter JD, Smith LD. Protein synthesis during maturation promoting factor-and progesterone-induced maturation in Xenopus oocytes. Dev Biol. 1982; 89(1): 152–8. [DOI] [PubMed] [Google Scholar]
- 51. Thomas P, Dressing G, Pang Y, Berg H, Tubbs C, Benninghoff A, et al. Progestin, estrogen and androgen G-protein coupled receptors in fish gonads. Steroids 2006; 71(4): 310–6. [DOI] [PubMed] [Google Scholar]
- 52. Liu K, Rajareddy S, Liu L, Jagarlamudi K, Boman K, Selstam G, et al. Control of mammalian oocyte growth and early follicular development by the oocyte PI3 kinase pathway: new roles for an old timer. Dev Biol. 2006; 299(1): 1–11. [DOI] [PubMed] [Google Scholar]
- 53. Hoshino Y, Yokoo M, Yoshida N, Sasada H, Matsumoto H, Sato E. Phosphatidylinositol 3-kinase and Akt participate in the FSH-induced meiotic maturation of mouse oocytes. Mol Reprod Dev. 2004; 69(1): 77–86. [DOI] [PubMed] [Google Scholar]
- 54. Wymann MP, Marone R. Phosphoinositide 3-kinase in disease: timing, location, and scaffolding. Curr Opin Cell Biol. 2005; 17(2): 141–9. [DOI] [PubMed] [Google Scholar]
- 55. Choi T, Fukasawa K, Zhou R, Tessarollo L, Borror K, Resau J, et al. The Mos/mitogen-activated protein kinase (MAPK) pathway regulates the size and degradation of the first polar body in maturing mouse oocytes. Proc Natl Acad Sci. 1996; 93(14): 7032–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, et al. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003; 22(24): 6598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995; 270(5241): 1491–4. [DOI] [PubMed] [Google Scholar]
- 58. Yoshino O, McMahon HE, Sharma S, Shimasaki S. A unique preovulatory expression pattern plays a key role in the physiological functions of BMP-15 in the mouse. Proc Natl Acad Sci U S A. 2006; 103(28): 10678–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hussein TS, Thompson JG, Gilchrist RB. Oocyte-secreted factors enhance oocyte developmental competence. Dev Biol. 2006; 296(2): 514–21. [DOI] [PubMed] [Google Scholar]
- 60. Foss DC, White JL. Early sexual maturity of brown-egg pullets cage-grown in narrow-band light with high nutrient density diets. Poult. Sci. 1983; 62(7): 1424. [Google Scholar]
- 61. Gieffers C, Dube P, Harris JR, Stark H, Peter JM. Three-dimensional structure of the anaphase-promoting complex. Mol Cell. 2001; 7(4): 907–13. [DOI] [PubMed] [Google Scholar]
- 62. Polager S, Ginsberg D. E2F-at the crossroads of life and death. Trends Cell Biol. 2008; 18(11): 528–35. 10.1016/j.tcb.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 63. Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001; 414(6862): 457–62. [DOI] [PubMed] [Google Scholar]
- 64. Maurer G, Portugal SJ, Cassey R. Review: an embryo's eye view of avian eggshell pigmentation. J Avian Biol. 2011; 42(6): 494–504. [Google Scholar]
- 65. Yang X, Downes M, Ruth TY, Bookout AL, He WM, Straume M, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006; 126(4): 801–10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLS)
(XLS)
Data Availability Statement
The sequences of unigenes were deposited in the NCBI Transcriptome Shotgun Assembly Sequence Database (B1: SRR2094734, B2: SRR2094746, B3: SRR2094764; W1: SRR2094777, W2: SRR2094789, W3: SRR2094799).