Abstract
Clavibacter michiganensis subsp. nebraskensis (Cmn), the causal organism of Goss’s wilt and leaf blight of maize, can be detected in the phyllosphere of its host prior to disease development. We compared the morphology and pathogenicity of 37 putative isolates of Cmn recovered from asymptomatic and symptomatic maize leaves. Thirty-three of the isolates produced mucoid orange colonies, irrespective of the source of isolation and all but four of these isolates were pathogenic on maize. The remaining 4 isolates recovered from asymptomatic leaves had large fluidal yellow colonies, and were non-pathogenic on maize. Isolates varied in their aggressiveness on a susceptible hybrid of maize but no significant differences in aggressiveness were detected between epiphytic isolates and those recovered from diseased maize tissues. The genomics of Cmn is poorly understood; therefore as a first step to determining what genes may play a role in virulence, we compared 33 putative virulence gene sequences from 6 pathogenic and a non-pathogenic isolate recovered from the phyllosphere. Sequence polymorphisms were detected in 5 genes, cellulase A, two endoglucanases, xylanase B and a pectate lyase but there was no relationship with pathogenicity. Further research is needed to determine what genes play a role in virulence of Cmn. Our data show however, that the virulence factors in Cmn likely differ from those reported for the closely related subspecies michiganensis and sepedonicus.
Introduction
Goss's wilt and leaf blight of maize, caused by Clavibacter michiganensis subspecies nebraskensis (Cmn), is economically important in the United States (US) [1]. The disease was first observed affecting maize plants in south central Nebraska in 1969 and soon after was reported from the seven surrounding states [2, 3]. After decades of reduced incidence, the disease recently reemerged throughout the Midwest [1, 4–6] Texas, [7, 8], Louisiana [9, 10], and in Canada [3, 8, 11]. Significant yield losses of up to 50% have been recorded, and usually depend on the crop developmental stage at which the maize hybrid was infected [12].
Cmn can infect maize at any crop developmental stage causing either leaf blight symptoms and/or wilting of the plant. Goss’s leaf blight is more common and is characterized by large elongated leaf lesions with characteristic small (<1mm2) dark green to black water-soaked spots in the periphery of the lesions. Wilt occurs when the bacteria in the vasculature becomes systemic and is characterized by an orange to brownish discoloration of the internal vascular tissue of the stalk and stunting of the maize plant [5]. The wilting is more common on young seedlings and when seed-transmission of the pathogen occurs
Although infected seed can be a source of inoculum [13,14,15], the main source of inoculum for Goss’s wilt and leaf blight is surface Cmn-infested residue within which the bacterium is able to survive for at least 10 months [5, 16]. Smidt and Vidaver [2] detected Cmn on asymptomatic corn leaves in the field and suggested an epiphytic phase for the pathogen that may contribute to disease development. However, limited research has been done to examine this hypothesis. In several other pathosystems, epiphytic bacterial are important in the epidemiology of the disease but the importance of epiphytic Cmn in Goss’s wilt and leaf blight development is unknown.
The genomics of Cmn is poorly understood. Three of the 5 michiganenisis subspecies genomes have been sequenced and recent comparative genome analyses have shown that the genome of Cmn was very similar to that of Cmm and Cms [17, 18]. In the subspecies michiganensis and sepedonicus, major virulence factors responsible for disease induction were plasmid borne while the genes responsible for successful host colonization were chromosomally encoded. The chromosomal virulence factors were identified as serine proteases of the Chp and Ppa family, and a tomatinase A gene, all located on an area of chromosome known as a pathogenicity island [13]. The plasmid-encoded factors were identified as a pat-1 serine protease and a cellulase A gene [18]. Vidaver [2] reported plasmids were not required for pathogenicity in Cmn. Consequently, Eichenlaub and Gartemann [17] suggested that virulence mechanisms in Cmn may be different from those observed in Cmm. Currently, little is known regarding virulence factors of Cmn.
The goals of this study were (i) to compare the morphology and pathogenicity of isolates of Cmn recovered from the phyllosphere of apparently healthy maize leaves with isolates recovered from maize leaves with characteristic Goss’s leaf blight symptoms, and (ii) to assess sequence heterogeneity between a non-pathogenic isolate of Cmn and pathogenic isolates in 33 genes that encode for putative virulence factors.
Materials and Methods
Bacterial strains and morphological characterization
A total of 37 putative Cmn strains were evaluated in this study (Table 1). Ten of the isolates were recovered from corn plants with symptoms of Goss’s leaf blight in 3 fields in Iowa. Briefly, a piece of leaf tissue was cut using a sterile razor blade from the leading edge of a lesion, surface disinfested in 0.525% NaOCl, rinsed in sterile distilled water and placed on CNS medium [15, 19], which is semi-selective for Cmn. The remaining twenty-seven strains were recovered from washes of leaves harvested in 2012 and 2013 from apparently healthy corn plants grown in minimally tilled fields with a history of Goss’s disease in the previous growing season in Iowa (6 fields) and Nebraska (1 field). All culture plates were incubated at 25°C for a minimum of 5 days. Colonies with morphological characteristic of Cmn are typically apricot-orange, small (3–5 mm in diameter) circular, convex, glistening and butyrous with entire margins after 5 to 7 days of growth on Nutrient Broth Yeast Extract (NBY) medium at 22 to 26°C [2]. Putative colonies of Cmn were sub-cultured on NBY medium and incubated under the same conditions. The resultant colonies were streaked on NBY plates a second time to obtain single colonies. A single colony from each plate was used to make inoculum for pathogenicity studies, genomic and plasmid DNA isolation, and stock cultures for long-term storage on silica gel. Two reference strains of Cmm (obtained from E. Braun, Iowa State University, Ames, IA), were included as comparisons (Table 1). The morphological characteristics of each of the 37 strains plus the two Cmm strains in terms of size (mean colony diameter in mm), color and fluidity were recorded after growth on NBY for 5–7 days at 25°C.
Table 1. Morphological and genetic characterization of 37 putative strains of Clavibacter michiganensis subsp. nebraskensis collected from Goss’s leaf blight symptomatic and asymptomatic maize leaves compared with two strains of Clavibacter michiganensis subsp. michiganensis.
| Strain ID | Field, County, State* | Source † | Year | Morphology and color ‡ | Pathogenic | Groups ¶ | PCR-RFLP groups** | Identity †† | Plasmid | |
|---|---|---|---|---|---|---|---|---|---|---|
| recA | rpoD | |||||||||
| C4 | 1, Iowa, IA | Epiphytic | 2012 | Small mucoid, orange | Yes | 1 | I | I | Cmn | - |
| C5 | 1, Iowa, IA | Epiphytic | 2012 | Small mucoid, orange | No | 2 | III | II | Cm | - |
| C12 | 1, Iowa, IA | Epiphytic | 2012 | Small mucoid, orange | Yes | 1 | I | I | Cmn | - |
| FN | 2, Boone, IA | Diseased | 2013 | Small mucoid, orange | Yes | 1 | I | I | Cmn | - |
| HF2 | 3, Story, IA | Epiphytic | 2012 | Large mucoid, orange | Yes | 3 | I | I | Cmn | - |
| HI 11–5 | 4, Story, IA | Epiphytic | 2013 | Small mucoid, orange | Yes | 1 | I | I | Cmn | - |
| NE2 | 5, Grant, NE | Epiphytic | 2012 | Small mucoid, orange | Yes | 1 | I | I | Cmn | + |
| CL1 | 6, Carroll, IA | Diseased | 2013 | Small mucoid, orange | Yes | 1 | I | I | Cmn | + |
| C10 | 1, Iowa, IA | Epiphytic | 2012 | Large mucoid, orange | No | 4 | III | II | Cm | - |
| HF4 | 3, Story, IA | Epiphytic | 2012 | Large mucoid, orange | No | 4 | I | I | Cmn | - |
| NE1 | 5, Grant, NE | Epiphytic | 2012 | Large mucoid, orange | No | 4 | III | II | Cm | + |
| HI 4–5 | 4, Story, IA | Epiphytic | 2013 | Large mucoid, orange | Yes | 3 | I | I | Cmn | - |
| HI 6–5 | 4, Story, IA | Epiphytic | 2013 | Small mucoid, orange | Yes | 1 | I | I | Cmn | - |
| CL4 | 6, Carroll, IA | Diseased | 2013 | Large mucoid, orange | Yes | 3 | I | I | Cmn | - |
| GIL1 | 7, Story, IA | Diseased | 2013 | Large mucoid, orange | Yes | 3 | I | I | Cmn | - |
| GIL3 | 7, Story, IA | Diseased | 2013 | Large mucoid, orange | Yes | 3 | I | I | Cmn | - |
| NE6 | 5, Grant, NE | Epiphytic | 2011 | Large fluidal, yellow | No | 5 | II | na | Cm | - |
| BR2 | 8, Boone, IA | Epiphytic | 2011 | Large fluidal, yellow | No | 5 | II | na | Cm | - |
| BS2 | 9, Boone, IA | Epiphytic | 2011 | Large fluidal, yellow | No | 5 | II | na | Cm | - |
| C8 | 1, Iowa, IA | Epiphytic | 2012 | Small mucoid, orange | No | 2 | IV | na | Cm | - |
| G1 | 10, Story, IA | Epiphytic | 2012 | Large fluidal, yellow | No | 5 | II | na | Cm | - |
| C2 | 1, Iowa, IA | Epiphytic | 2012 | Small mucoid, orange | Yes | 1 | I | I | Cmn | - |
| C3 | 1, Iowa, IA | Epiphytic | 2012 | Small mucoid, orange | Yes | 1 | I | I | Cmn | - |
| C7 | 1, Iowa, IA | Epiphytic | 2012 | Large mucoid, orange | Yes | 3 | I | I | Cmn | - |
| HF1 | 3, Story, IA | Epiphytic | 2012 | Large mucoid, orange | Yes | 3 | I | I | Cmn | - |
| HI 2-AN | 4, Story, IA | Epiphytic | 2013 | Large mucoid, orange | Yes | 3 | I | I | Cmn | - |
| HI 2-BS | 4, Story, IA | Epiphytic | 2013 | Small mucoid, orange | Yes | 1 | I | I | Cmn | - |
| HI 2–4 | 4, Story, IA | Epiphytic | 2013 | Large mucoid, orange | Yes | 3 | I | I | Cmn | - |
| HI 11–7 | 4, Story, IA | Epiphytic | 2013 | Large mucoid, orange | Yes | 3 | I | I | Cmn | - |
| HI 11–8 | 4, Story, IA | Epiphytic | 2013 | Large mucoid, orange | Yes | 3 | I | I | Cmn | - |
| HI 6–4 | 4, Story, IA | Epiphytic | 2013 | Large mucoid, orange | Yes | 3 | I | I | Cmn | - |
| HI 7–7 | 4, Story, IA | Epiphytic | 2013 | Large mucoid, orange | Yes | 3 | I | I | Cmn | - |
| NE3 | 5, Grant, NE | Epiphytic | 2012 | Small mucoid, orange | Yes | 1 | I | I | Cmn | - |
| NE4 | 5, Grant, NE | Epiphytic | 2012 | Small mucoid, orange | Yes | 1 | I | I | Cmn | - |
| CL2 | 6, Carroll, IA | Diseased | 2013 | Small mucoid, orange | Yes | 1 | I | I | Cmn | - |
| CL3 | 6, Carroll, IA | Diseased | 2013 | Large mucoid, orange | Yes | 3 | I | I | Cmn | - |
| GIL2 | 7, Story, IA | Diseased | 2013 | Large mucoid, orange | Yes | 3 | I | I | Cmn | - |
| BR-4 (Cmm ‡‡ ) | - | Large mucoid, yellow | No | 6 | nr | III | Cmm | + | ||
| DR-60 (Cmm) | - | Large mucoid, yellow | No | 6 | V | III | Cmm | + | ||
Pathogenicity tests were done on corn plants that were inoculated with each isolate at the V3–V4 crop developmental stage. Foliar blight severity was rated as the proportion of leaf area affected six days after inoculation.
* GPS co-ordinates of nearest town to which fields were located are provided in S1 Table.
† Epiphytic, strain recovered from asymptomatic maize leaves; diseased, strain recovered from maize leaves with symptoms of Goss’s leaf blight
‡ Colony size was measured after 5 days growth on CNS medium [12] using a ruler and divided into three groups as A, small mucoid orange, (2–3 mm); B, large mucoid orange (3–4 mm) and C, large fluidal yellow (3–4 mm or larger in diameter) (see Fig 1).
¶ Groups based on colony morphology after 5 days’ growth on NBY medium and pathogenicity on maize
** PCR-RFLP of housekeeping genes recA and rpoD [24]. Numbers in columns represent fragment patterns; na, not amplified; nr, not restricted.
††Strains identified as Clavibacter michiganensis subsp. nebraskensis based on PCR-RFLP pattern of recA and rpoD genes [24], all belonged to Groups 1 and 3; Cm stands for Clavibacter michiganensis.
‡‡ Clavibacter michiganensis subsp. Michiganensis
Pathogenicity and comparative aggressiveness of isolates
All 37 putative strains of Cmn and the two Cmm strains were tested for pathogenicity and aggressiveness on the susceptible maize hybrid DKC55-09 in the greenhouse. The Cmn strain 91-R (obtained from C. Block, USDA Plant Introduction Station, Ames, IA), which is a rifampicin-tolerant derivative, was included as a reference for pathogenicity. Inoculum for pathogenicity tests was prepared by flooding 3 day-old cultures of the bacterium on NBY plates with 10 ml of phosphate-buffered saline (PBS) and gently scraping the bacterial cells off the surface of the media. Each bacterial suspension was adjusted to an OD600nm of 0.04 (106 CFUs ml-1). Two seeds were sown per pot (25.4 cm in diameter) in a peat moss: metro mix: coarse perlite (4:3:4) and plants were grown to the V3 crop developmental stage before inoculation [20]. The third leaf of each plant was wounded by making a cut across 3 veins on one side of the midrib, half way between the leaf sheath and the tip of the leaf. Ten microliters of the inoculum was dropped on the wound using a 25 μl 702N Hamilton syringe (Hamilton CO. Reno, Nevada). Only the third leaf was inoculated per plant and six plants were inoculated per strain. The experimental setup was a completely randomized design and the plants were maintained at a temperature of 25–30°C with a 14 h photoperiod on a greenhouse bench. Disease severity was defined by the proportion of the leaf area affected, and disease ratings started at 6 days after inoculation (DAI) and thereafter were done on every fourth day for 28 DAI. Throughout the experiments, pots were watered daily and plants were fertilized once a week with a liquid fertilizer (NPK: 15–5–15; Miracle-Gro, The Scotts Co., Marysville, OH) that was supplemented with Ca(NO3)2 and MgSO4 micronutrients at the rate of 43 g/L and 22 g/L, respectively. This experiment was conducted twice and the bacterial strains were re-isolated from the diseased leaves at the end of each experiment to fulfill Koch’s postulates [21].
The leaf blight disease severity ratings were converted to area under the disease progress curve (AUDPC) using the trapezoidal method [22] and analysis of variance was performed using PROC GLIMMIX of SAS software (version 9.3, SAS Institute, Cary, NC), in which plant and leaf were considered random. Mean comparisons were done using Tukey’s test.
DNA extraction and plasmid isolation
Genomic DNA of each isolate was extracted from a loopful of 6 day old cultures grown on NBY suspended in 1 ml of PBS using a PowerLyser™ UltraClean® Microbial DNA Isolation Kit (MOBIO, Cleveland, CA) following the manufacturer’s instructions. Cultures for the isolation of plasmids were grown overnight in 150 ml of TBY plus glucose media [23] on a 250rpm shaker at 25°C. The cells were then pelleted by centrifugation and the plasmids were isolated using the Qiagen plasmid mini kit (Qiagen Sciences, Germantown, MD) following the manufacturers instructions with a few modifications. The bacterial pellet was resuspended in 10ml of buffer PI containing 1mg/ml of lysozyme and incubated at 37°C for 37 mins. Similarly in subsequent steps, 10 ml of buffers P2 and P3 were used. Plasmid DNA was eluted with 30 μl of TE buffer warmed to 65°C.
Confirmation of identification with PCR-RFLP using housekeeping genes recA and rpoD
The housekeeping genes recA and rpoD were amplified using the following primers: for recA, the forward primer recAF (5’-TCGGCAAGGGCTCGGTCATGC- 3’) and reverse primer recAR (5’-GGTCGCCRTCGTASGTGTACCA- 3’). The forward and reverse primers used for rpoD were rpoDF (5’-ATGGTGCTGTCGAACAAGGA- 3’) and rpoDR (5’-CGATCTGGTCGAGSGTCTT- 3’), respectively [24]. PCR amplification was performed using a TopTaq master mix kit in 50 μl reactions. The DNA concentration was adjusted to 20 ng/ul and amplifications were performed as described by Waleron and colleagues [24] in a BIO RAD T100™ thermocycler with the following conditions: an initial denaturation at 95°C for 3 min, 32 cycles of denaturation at 94°C for 1 min, annealing at 62°C, for 1 min for recA, and 58°C or 60°C for 1 min for rpoD, extension at 72°C for1min, and a final extension at 72°C for 5min. For most of the PCR reactions an annealing temperature of 62°C and 58°C was used for recA and rpoD respectively. Following PCR amplification, the products were restricted with the endonuclease BstUI, which is an isoschizomer of FnuDII [24]. Digestion was carried out in a total volume of 50 μl containing 1 μl of BstUI, 5 μl of 10X NEBuffer, 24 μl of molecular grade water and 20 μl of DNA template. The reaction mixture was incubated for 30 min at 60°C. After digestion, the reaction mixture was concentrated for 20 min at medium heat (43°C) in a DNA concentrator to half the volume. The resultant fragments of DNA were separated in a 12% polyacrylamide gel at 200 V for 5–6 h in 0.5X TBE buffer and visualized with UV light after staining in ethidium bromide (0.5 μg ml-1) [24]. The PCR-RFLP using polyacrylamide gel was carried out twice under same conditions.
The protocol of Waleron et al. [24] was later modified in that resolution of the restricted fragments was done in a 4% agarose gel prepared in sodium borate solution [25]. The DNA fragments were electrophoresed at 100 V for 2 h and visualized with UV light after staining in ethidium bromide (0.5 μg ml-1). This experiment was done twice.
Comparative sequence analyses of putative virulence genes
Published virulence factors in the subspecies sepedonicus and michiganensis [17] were used to search for corresponding homologs in the genome of Cmn in order to differenciate between the non-pathogenic and pathogenic strains of Cmn. In addition, several serine proteases that encode for toxin-antitoxin systems and the chloride anion channel gene [26, 27] were selected for sequencing. The sequences of these putative virulence factors were identified in the genome of the Cmn isolate NCPP 2581 (NCBI Reference Sequence: NC_020891.1, released but not yet published) found on the NCBI nucleotide database (http://www.ncbi.nlm.nih.gov/nuccore/473832060?report=graph). Primer pairs, designed with sequences of genomic loci that flanked each gene using the DNADynamo sequence analysis program (Blue tractor software Ltd, UK), were used to amplify the complete sequences of the target genes. The PCR products were sequenced at the DNA facility of Iowa State University. Sequences were aligned and manually edited using the biological editor, BioEdit [28] before they were translated and aligned using the DNADynamo sequence analysis program.
Results
Morphological characteristics of isolates, pathogenicity, and aggressiveness on maize
The 37 putative strains of Cmn were classified into six groups based on colony morphology after 5 days’ growth on NBY medium (Fig 1) and pathogenicity on maize (Table 1). Thirteen strains that produced small, mucoid orange colonies and were pathogenic on maize, were classified into Group 1. Ten of these strains were recovered from asymptomatic maize leaves and the remaining three from Goss’s leaf blight lesions. Group 2 contained two epiphytic strains that were morphologically similar to strains from Group 1 but were not pathogenic on maize. Group 3 contained strains recovered from asymptomatic (10 strains) and symptomatic maize leaves (5 strains) that produced large, mucoid, orange colonies and were all pathogenic on maize. Three epiphytic strains also produced large, mucoid orange colonies but did not cause disease symptoms on maize and were classified as Group 4. Four strains, that produced large fluidal yellow colonies and were not pathogenic on maize, were classified as Group 5. All four strains were recovered from asymptomatic leaves. The two Cmm reference strains included in this study produced large, mucoid yellow colonies on NBY and were non-pathogenic on maize (Group 6). Thus, based on colony morphology (orange mucoid) and pathogenicity on maize, a total of 28 strains (20 epiphytic and 8 from diseased maize leaves) were identified as Cmn.
Fig 1. Examples of colony morphology after 5 days growth on nutrient broth yeast extract (NBY) medium of 37 putative isolates of Clavibacter michiganensis subsp. nebraskensis.
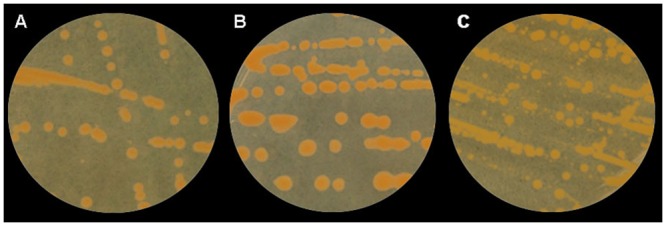
A small mucoid orange colony; B large mucoid orange colony; and C large fluidal yellow colony.
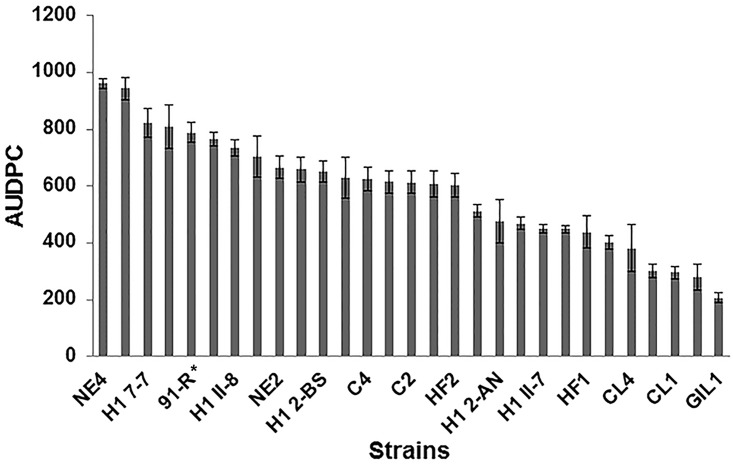
Analysis of variance showed significant differences in aggressiveness among the 28 Cmn strains (P < 0.0001). Disease severity (proportion of the leaf area affected) ranged from 16.0 to 78%. Strain NE4 was the most aggressive while GIL 1 was the least aggressive on the maize hybrid DKC55-09 (Fig 2).
Fig 2. Mean aggressiveness (n = 12 plants) of 28 Clavibacter michiganensis subsp. nebraskensis strains estimated from foliar blight severity on maize hybrid DKC 55–09.
Plants were inoculated at the V3–V4 crop developmental stage and foliar blight severity was rated as the proportion of leaf area affected six days after inoculation. Bars represent standard errors of the mean. *Strain 91-R was included as a reference for pathogenicity in this experiment.
Among the 37 putative Cmn strains, only NE1, NE2 and CL1 contained single large plasmids that were estimated to be 70 kb (Table 1).
Identification of Cmn using PCR-RFLP analysis
The rpoD and recA genes were successfully amplified from all 37 putative Cmn strains and the size of each amplicon was consistent with that reported by Waleron et al. [24]. Digestion of the rpoD and recA amplicons using BstU1 resulted in two and four restriction patterns, respectively (Table 1, Fig 3). When the amplified recA fragment was digested with BstU1, RFLP pattern I was observed for all 27 strains that were pathogenic as well as strain HF4, which was not pathogenic on maize. Six bands of size ranging from 50 to 140 bp were observed on the polyacrylamide gel while only five bands of the same size range were observed with 4% agarose gel. RFLP pattern I was similar to PCR-RFLP pattern 3 reported by Waleron et al. [24] for Cmn. Three additional RFLP patterns, II, III and IV, were observed for strains representative of Groups 2, 4 and 5 respectively, that were not pathogenic on maize. The RFLP pattern observed with BstU1digestion of recA amplicon of Cmm strain DR-60 was consistent with that reported by Waleron et al. [24]. The recA amplicon of Cmm strain BR-4 could not be digested with the enzyme (data not shown).
Fig 3. Representative RFLP patterns obtained after restriction of rpoD and recA with BstU1 for 37 putative isolates of Clavibacter michiganensis subsp. nebraskensis and two strains of Clavibacter michiganensis subsp. michiganensis.
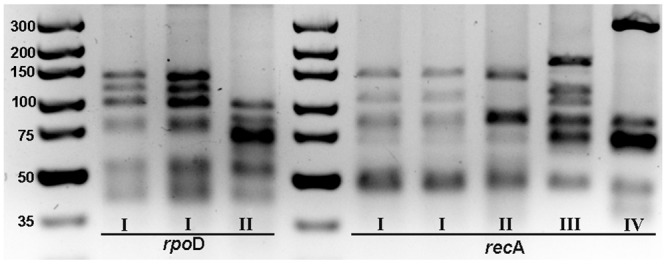
Roman numerals correspond to pattern types observed. Molecular size marker: GeneRuler 100 bp DNA Ladder.
For BstU1digestion of rpoD fragment, two RFLP patterns, I and II, were observed for all the 37 putative Cmn strains. RFLP pattern I that had six bands (40 to 140 bp) on both polyacrylamide and agarose gels was observed for all strains that were pathogenic on maize, as well as HF4 (Table 1; Fig 3). The first three bands of the pattern on both gels were larger (approximately 40, 30 and 20 bp for the first, second and third band, respectively) than those reported by Waleron et al. [24], while the smaller bands were smaller than reported. Pattern II was obtained for three non-pathogenic strains (Table 1). The RFLP patterns observed for BstU1digestion of rpoD for the two strains of Cmm that were included in the study were consistent with those reported by Waleron et al. [24].
Comparative sequence analyses of putative virulence genes
Thirty-three genes that encode for putative virulence factors identified in the Cmn genome database were successfully amplified and sequenced for Cmn strain GIL1 (pathogenic) and Cmn strain HF4 (non-pathogenic on maize). The pathogenic strain GIL1 had identical gene sequences with the reference Cmn strain NCPPB 2581. When the GIL1 gene sequences were compared to those of HF4, polymorphisms were detected in the Cellulase A, two endoglucanase genes, the xylanase B gene, and the pectate lyase gene (Table 2; S1 File). The cellulase A gene in strain HF4 was 1100 base pairs long with a 18 nucleotide insertion that was absent in strain GIL1. An in silico translation of the nucleotide sequence revealed the 18 nucleotide insertion was a repeat that corresponded to the amino acid sequence “PTPPSQ” (S2 File). This amino acid sequence was repeated three times in the reference strain NCPPB 2581 and GIL1, while HF4 contained an additional repeat of the sequence bringing the total to four. One of the endoglucanase genes (Cmn 2651) was 1386 nucleotides in length with a 66 nucleotide insertion located between nucleotides 419–486 in strain HF4 that was absent in strain GIL1. An in silico translation of the gene sequence showed the insertion corresponded to 22 additional amino acids in which no repeats could be identified (S2 File). The second endoglucanase (Cmn 2650), a xylanase B gene and a pectate lyase gene (Cmn 2654) differentiated the two strains by 3, 6 and 1 single nucleotide polymorphisms (SNPs) respectively. Only one of the SNPs in Cmn 2650 resulted in a change in amino acid sequence from A to G. Four of the SNPs in the Xylanase B gene resulted in the amino acids A, D, P and L instead of S, Y, L, and V respectively in the reference strain and the rest were silent mutations (S2 File).
Table 2. Nucleotide polymorphisms in the sequences of putative virulence factors found in the genome of Clavibacter michiganensis subsp. nebraskensis strains GIL1 and HF4. PCR primers flanking each locus were used in sequencing reactions to obtain the complete sequence of each gene.
| Locus in Cmn genome (Isolate NCPPB 2581) | Putative function | Nucleotide differences |
|---|---|---|
| Cmn 00734 | Chloride anion channel | No difference |
| Cmn 00144 | Secreted cellulase A | Indel (18 nucleotides) |
| Cmn 02650 | Endoglucanase | 3 SNPs |
| Cmn 02651 | Endoglucanase | Indel (66 nucleotides) |
| Cmn 00792 | Translocase glycosyl hydrolase | No difference |
| Cmn 01115 | Polysaccharide deacetylase | No difference |
| Xys A | Endo-1, 4-beta xylanase A | No difference |
| Xys B | Endo-1, 4-beta xylanase B | 6 SNPs |
| Pga A | Polygalacturonase A | No difference |
| Cmn 01173 | Secreted serine peptidase | No difference |
| Cmn ppaF | Secreted serine peptidase | No difference |
| Cmn 457 | Secreted serine peptidase | No difference |
| Cmn sbtC | Serine peptidase | No difference |
| Cmn sbtB | Serine peptidase | No difference |
| Cmn 2417 | Secreted serine peptidase | No difference |
| Cmn 2381 | Secreted serine peptidase | No difference |
| Cmn 2235 | Secreted serine peptidase | No difference |
| Cmn 1337 | Secreted serine peptidase | No difference |
| Cmn 1248 | Secreted serine peptidase | No difference |
| Cmn 00106 | Toxin-antitoxin system | No difference |
| Cmn 00626 | Toxin-antitoxin system | No difference |
| Cmn 00771 | Toxin-antitoxin system | No difference |
| Cmn 01077 | Toxin-antitoxin system | No difference |
| Cmn 01078 | Toxin-antitoxin system | No difference |
| Cmn 02136 | RTX toxin | No difference |
| Cmn 02626 | Toxin component | No difference |
| Cmn 02669 | Toxin component | No difference |
| Cmn 02707 | Toxin gene | No difference |
| Cmn 00414 | Protein kinase | No difference |
| Cmn 0118 | Exported toxin | No difference |
| Cmn 02101 | TetR lipase/esterase | No difference |
| Cmn 00283 | Glycosyl transferase | No difference |
| Cmn 02654 | Pectate lyase | I SNP |
Sequences were aligned and examined for nucleotide sequence differences. Primers were designed from reference strain NCPPB 2581 genomic sequence.
To further characterize the nucleotide differences we observed in the cellulose A and endogluconase (Cmn 2651) genes, we sequenced each loci in five additional Cmn strains (FN, C4, NE3, GIL3, and 91-R) and a non-pathogenic Clavibacter michiganensis strain (NE1). The same indel in the cellulase A gene sequence was found. The avirulent strain NE1 and the virulent strains NE3, FN, GIL3, and 91-R were similar and lacked the 18 nucleotide insertion, while the virulent strains C4 and the avirulent strain HF4 contained the 18 nucleotide insertion. Similarly, the sequence of the endoglucanase gene (Cmn 2651) in the virulent strains C4, GIL3, and NE3 and the avirulent strain NE1 contained the 66 nucleotide insertion, while the sequence in the two virulent strains FN and 91-R was the same as in the avirulent strain HF4.
Discussion
In this study we compared the morphology and aggressiveness of 37 strains of putative Cmn recovered from maize leaves with Goss’s leaf blight symptoms or asymptomatic leaves from maize plants grown in fields with a history of Goss’s wilt and leaf blight in Iowa and Nebraska. The bacterium was always recovered from symptomatic tissues but the recovery rate of Cmn from asymptomatic tissues was lower (approximately 0 to 50 percent). This may have been a function of our sampling technique. There are few data regarding epiphytic colonization of the plant canopy, or individual leaves, by Cmn. Such data would enable more targeted sample collection rather than our sampling method that consisted of collecting arbitrary leaves from various positions within the canopy of numerous plants within a field with a history of the disease. We identified 28 of the strains as Cmn based on colony morphology, pathogenicity on maize, and PCR-RFLP of housekeeping genes. Of these 28 strains, 20 had been recovered from the phyllosphere of maize leaves.
We found no relationship between colony morphology of putative Cmn strains and pathogenicity on maize confirming that it is difficult to identify Cmn purely on colony morphology on CNS [15, 29]. In environments where it is important to verify the organism, for example seed laboratories where testing maize seed for which quarantine restrictions have been placed to prevent the introduction of Cmn, this is obviously a concern. Consequently, additional tests that usually include pathogenicity assays on maize in the greenhouse are always required to confirm if a putative Cmn colony is the pathogen of interest.
We found the PCR-RFLP method described by Waleron et al. [24] was able to correctly identify putative Cmn colonies within a couple of days compared to greenhouse pathogenicity tests that take several weeks. This suggests that this method could be useful for diagnosis of Cmn. However, there are limitations with the method that should be addressed. Strain HF4 was identified as Cmn using this assay, although no symptoms developed when it was inoculated onto maize seedlings indicating it was non-pathogenic. The occurrence of non-pathogenic variants within subspecies of C. michiganensis is of great concern especially for hosts that are of phytosanitary importance. Detection of non-pathogenic Cmn in corn seed that may pose minimal risk to the importing country could result in exclusion of the seed. The existence of non-pathogenic variants in the subspecies michiganensis has also been very problematic in the design of assays for the detection and quantification of the pathogen in tomato seed [30]. Louws et al., [31] were unable to distinguish between virulent and avirulent C. michiganensis subsp. michiganensis strains using rep-PCR. When box-PCR and AFLP primers were used to genotype Cmn strains, pathogenic and non-pathogenic variants of Cmn were indistinguishable [32]. These results are an indication of how closely related the pathogenic and non-pathogenic strains of Cmn are and the differences may lie in virulence genes or promoters.
Modification of the PCR-RFLP method in which a 4% agarose gel prepared in sodium borate solution was used rather than a polyacrylamide gel to visualize the digested fragments, makes this C. michiganensis identification method user-friendly since agarose is easier to use, cheaper and less toxic than acrylamide. However, resolution of bands that differ in size by a few base pairs can be difficult on an agarose gel [33]. This was evident in our study where the restriction pattern we observed for recA had five bands rather than six as was observed on the polyacrylamide gel. Thus on the agarose gel we were unable to resolve the third and fourth bands that Waleron et al. [24] reported. Nevertheless this method could be useful to identify subspecies of C. michiganensis from environmental samples.
It is unclear why HF4 was non-pathogenic on maize despite it being so similar morphologically and genetically to other Cmn strains in our study. Although the presence of plasmids has been associated with virulence in Cmm and Cms, this is not the case for Cmn [19, 34]. Earlier studies have shown that there were no differences in pathogenicity between Cmn strains that contained plasmids and those without plasmids [2,13]. In the present study, strains NE2 and CL1 were both pathogenic while NE1 was non-pathogenic, but all three contained plasmids, while the rest of the pathogenic strains were devoid of plasmids. Although these plasmids are yet to be characterized, our data further confirm that Cmn virulence factors are not plasmid-borne.
Toxicity genes have been hypothesized to play a role in virulence of Cmn, but only one of twelve putative toxin genes in the genome of Cmn has been partially characterized. This membrane-active component called the chloride anion channel (CAC) protein was shown to form anion channels in planar lipid bilayers in vitro. The activity of this protein was shown to be similar to that of both colicins and the Hm-T toxins. However, its direct role in pathogenicity on maize was not demonstrated [26, 27, 35,36]. In an effort to understand why HF4 was non-pathogenic on maize, we sequenced 33 genes that encode putative virulence factors, including the CAC gene, and compared the sequences to those of other pathogenic strains, and the reference strain NCPP2581 whose genome has been sequenced. Differences in sequence data were detected only in 5 of the 33 genes, but the sequence polymorphisms observed were unrelated to pathogenicity. Moreover, we found no sequence polymorphism within the CAC gene in both pathogenic and non-pathogenic strains. Similarly, there was no relationship between sequence polymorphisms observed in four putative virulence genes and the ability of the strains to be pathogenic on maize. However, these data do not eliminate the role of CAC or the other putative virulence factors in the disease causing process. Gene regulation was not tested in this study. Thus it is possible that differences at the level of regulation of gene expression may be responsible for virulence. These data further confirm reports that pathogenicity determinants in Cmn may be different from those in its close relatives, Cmm and Cms [17]. Currently in a collaborative project, the genomic sequence of strain HF4 has been completed and is being annotated. Comparative genomics and subsequent functional analysis should enable the virulence factors in Cmn to be identified.
Supporting Information
(PDF)
(PDF)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no support or funding to report.
References
- 1. Jackson TA, Harveson RM, Vidaver AK. Reemergence of Goss’s wilt and blight of corn to the central High Plains. Online. Plant Health Progress 10.1094/PHP-2007-0919-01-BR 2007. [DOI] [Google Scholar]
- 2. Vidaver AK, Mandel M. Corynebacterium nebraskense, a new, orange-pigmented phytopathogenic species. Int J Syst Bacteriol. 1974; 24: 482–485. [Google Scholar]
- 3. Wysong DS, Vidaver AK, Stevens H, Stenberg D. Occurrence and spread of an undescribed species of Corynebacterium pathogenic on corn in the western corn belt. Plant Dis Report. 1973; 57: 291–294. [Google Scholar]
- 4.Bradley C. Uncommon diseases of corn observed. The Bulletin. 22 July 2010. Available at http://bulletin.ipm.illinois.edu/print.php?id=1385. Accessed 22 July 2015.
- 5. Jackson TA, Harveson RM, Vidaver AK. Goss’s bacterial wilt and leaf light of corn NebGuide G1675. Lincoln; Nebraska Institute of Agriculture and Natural Resources: 2007. [Google Scholar]
- 6.Robertson A. Unusual foliar diseases showing up in Iowa corn. Integrated Crop Management News. 2008. Available at http://www.extension.iastate.edu/CropNews/2008/0718robertson.htm. Accessed 22 July 2015.
- 7. Malvick D, Syversen R, Mollov D, Ishimaru CA. Goss’s bacterial blight and wilt of corn caused by Clavibacter michiganensis subsp. nebraskensis occurs in Minnesota. Plant Dis. 2010; 94: 1064.1. [DOI] [PubMed] [Google Scholar]
- 8. Ruhl G, Wise K, Cresswll T, Leonberger A, Speers C. First report of Goss’s bacterial wilt and leaf blight on corn caused by Clavibacter michiganensis subsp. nebraskensis in Indiana. Plant Dis. 2009; 93: 841. [DOI] [PubMed] [Google Scholar]
- 9. Hollier CA, Singh RA, Frazier R. Goss’s wilt in Louisiana: Incidence, severity and loss. Phytopathology 2014; 104 Supplement 2: S2:5. [Google Scholar]
- 10. Korus KA, Timmerman AD, French-Monar RD, Jackson TA. First report of Goss’s bacterial wilt and leaf blight (Clavibacter michiganensis subsp. nebraskensis) of corn in Texas. Plant Dis. 2011; 95: 73.2. [DOI] [PubMed] [Google Scholar]
- 11. Howard RJ, Harding MW, Lynn J, Kawchuck LM, Rasmussen NM. First report of Clavibacter michiganensis subsp. nebraskensis in Alberta, Canada. Plant Disease 10.1094/PDIS-11-14-1117-PDN [DOI] [Google Scholar]
- 12. Claflin LE. Goss’s bacterial wilt and blight Pages 4–5 In: White DG, editor. Compendium of corn diseases 3rd Ed. St. Paul: MN; American Phytopathological Society; 1999. [Google Scholar]
- 13. Bentley SD, Corton C, Brown SE, Barron A, Clark L, Doggett J, et al. Genome of the actinomycete plant pathogen Clavibacter michiganensis subsp. sepedonicus suggests recent niche adaptation. J. Bacteriol. 2008; 190(6): 2150–2160. 10.1128/JB.01598-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Biddle JA, McGee DC, Braun EJ. Seed transmission of Clavibacter michiganensis in corn. Plant Dis. 1990; 74, 908–911. [Google Scholar]
- 15.Shepherd L. Detection and transmission of Clavibacter michiganensis subsp. nebraskensis of corn. MS Thesis. Iowa State University; 1999.
- 16. Schuster ML. Leaf freckles and wilt of corn incited by Corynebacterium nebraskense Schuster, Mandel, Lazar In: Schuster ML, editor. UNL Res Bull. University of Nebraska-Lincoln: Institute of Agriculture and Natural Resources; 1975. [Google Scholar]
- 17. Eichenlaub R, Gartemann K-H. The Clavibacter michiganensis subspecies: Molecular investigation of Gram-positive bacterial plant pathogens. Ann. Rev. Phytopathol. 2011; 40: 445–464. [DOI] [PubMed] [Google Scholar]
- 18. Zaloga J, Stragier P, Baeyen S, Haegeman A, Van Vaerenbergh J, Maes M. et al. Comparative genome analysis of pathogenic and non-pathogenic Clavibacter strains reveals adaptation to their lifestyle. BMC Genomics 2014; 15: 392 http://www.biomedcentral.com/1471-2164/15/392. 10.1186/1471-2164-15-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gross DC, Vidaver AK, Keralis MB. Indigenous plasmids from phytopathogenic Corynebacterium species. J Gen Microbiol. 1979; 115: 479–489. [Google Scholar]
- 20. Abendroth LJ, Elmore RW, Boyer MJ, Marlay SK. Corn growth and development. Ames, IA: Iowa State Extension; 2011. [Google Scholar]
- 21. Louws FJ, Bell J, Medina-Mora CM, Smart CD, Opgenorth D, Ishimaru CA, et al. rep-PCR-mediated genomic fingerprinting: A rapid and effective method to identify Clavibacter michiganensis . Phytopathology. 1998; 88(8): 862–868. 10.1094/PHYTO.1998.88.8.862 [DOI] [PubMed] [Google Scholar]
- 22. Madden LV, Hughes G, van den Bosch F, editors. The Study of Plant Disease Epidemics. St. Paul: MN: APS Press; 2007. [Google Scholar]
- 23. Meletzus D, Eichenlaub R. Transformation of the phytopathogenic bacterium Clavibacter michiganensis subsp. michiganensis by electroporation and development of a cloning vector. J. Bacteriol. 1991; 197(1): 187–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Waleron M, Waleron K, Kamasa J, Przewodowski W, Lojkowska E. Polymorphism analysis of housekeeping genes for identification and differentiation of Clavibacter michiganensis subspecies. Eur J Plant Pathol. 2011. 131: 341–354. [Google Scholar]
- 25. Brody JR, Kern SE. Sodium boric acid: a tris-free, cooler conductive medium for DNA electrophoresis. Bioinformatics. 2011; 36: 214–216. [DOI] [PubMed] [Google Scholar]
- 26. Schürholz T, Wilimzig M, Katsiou E, Eichenlaub E. Anion channel forming activity from the plant pathogenic bacterium Clavibacter michiganensis ssp. nebraskense . J Membr Biol. 1991; 123, 1–8. [DOI] [PubMed] [Google Scholar]
- 27. Schürholz T, Dloczik L, Neumann E. Single-channel analysis of the anion channel-forming protein from the plant pathogenic bacterium Clavibacter michiganensis ssp. nebraskense . Biophysics Journal. 1993; 64: 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT.; Nucl. Acids Symp.1999; Ser. 41:95–98. [Google Scholar]
- 29.Korus KA. Evaluating commercially available diagnostic tests for the detection of Clavibacter michiganensis subsp. nebraskensis, cause of Goss’s wilt and leaf blight in corn. MS Thesis. University of Nebraska; 2011. Available at http://digitalcommons.unl.edu/agronhortdiss/22
- 30. Cho MS, Lee JH, Her NH, Kim CK, Seol Y-J, Hahn JH., et al. A quantitative and direct PCR assay for the subspecies-specific detection of Clavibacter michiganensis subsp. michiganensis based on a ferredoxin reductase gene. The J. Microbiol. 2012; 50(3): 496–501. 10.1007/s12275-012-1611-x [DOI] [PubMed] [Google Scholar]
- 31. Louws FJ, Bell J, Medina-Mora CM, Smart CD, Opgenorth D, Ishimaru CA, et al. rep-PCR-mediated genomic fingerprinting: A rapid and effective method to identify Clavibacter michiganensis . Phytopathology. 1998; 88(8): 862–868. 10.1094/PHYTO.1998.88.8.862 [DOI] [PubMed] [Google Scholar]
- 32. Agarkova IV, Lambrecht PA, Vidaver AK. Genetic diversity and population structure of Clavibacter michiganensis subsp. nebraskensis . Can. J. Microbiol. 2011; 57: 366–374. 10.1139/W11-016 [DOI] [PubMed] [Google Scholar]
- 33. Stewart S, Wickramasinghe D, Dorrance AE, Robertson AE. Comparison of three microsatellite analysis methods for detecting genetic diversity in Phytophthora sojae (Stramenopila: Oomycete). Biotechnology Letters 2011; 33: 2217–2223 10.1007/s10529-011-0682-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eichenlaub R, Gartemann K-H, Burger A. Clavibacter michiganensis, a group of gram-positive phytopathogenic bacteria In: Gnanamanickam SS. editor. Plant-Associated Bacteria Springer; 2007. p. 385–421. [Google Scholar]
- 35. Michalke A, Galla H-J, Steinem C. Channel activity of a phytotoxin of Clavibacter michiganensis ssp. nebraskense in tethered membranes. Eur. Biophys. Journal 2001; 30: 421–429. [DOI] [PubMed] [Google Scholar]
- 36. Michalke A, Schürholz T, Galla H-J, Steinem C. Membrane activity of an anion channel from Clavibacter michiganensis ssp. nebraskense . Langmuir 2001; 17: 2251–2257. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.