Abstract
Human papillomavirus (HPV) infections are a major human health problem; they are the cause of recurrent benign warts and of several cancers of the anogenital tract and head and neck region. Although there are two prophylactic HPV vaccines that could, if used universally, prevent as many as two-thirds of HPV-induced cancers, as well as several cytotoxic and immunomodulatory agents for localized treatment of infections, there are currently no HPV antiviral drugs in our arsenal of therapeutic agents. This review examines the status of past and ongoing research into the development of HPV antivirals, focused primarily upon approaches targeting the replication of the viral genome. The only HPV enzyme, E1, is a DNA helicase that interfaces with the cellular DNA replication machinery to replicate the HPV genome. To date, searches for small molecule inhibitors of E1 for use as antivirals have met with limited success. The lack of other viral enzymes has meant that the search for antivirals has shifted to a large degree to the modulation of protein–protein interactions. There has been some success in identifying small molecule inhibitors targeting interactions between HPV proteins but with activity against a small subset of viral types only. As noted in this review, it is thought that targeting E1 interactions with cellular replication proteins may provide inhibitors with broader activity against multiple HPV types. Herein, we outline the steps in HPV DNA replication and discuss those that appear to provide the most advantageous targets for the development of anti-HPV therapeutics.
Introduction
Human papillomaviruses (HPVs) are a large family of viruses that predominantly infect epidermal tissues, both on external skin and on mucosal surfaces. Although some HPV types cause lesions (papillomas/warty lesions) that are inconvenient or unpleasant but not life-threatening, others, particularly those that infect the anogenital tract, can cause a variety of deadly human cancers (for reviews see [1,2]). Cervical cancer, virtually all of which are caused by prior HPV infection, is the second most common cancer in women worldwide. In addition, most other anogenital cancers (anal, vulval and penile) and 25–60% of oropharyngeal cancers are caused by HPV [2–5]. Current estimates are that over 5% of all human cancers are a direct result of HPV, making this virus family the second most frequent cause of human cancer after tobacco [4].
There are currently over 180 types of HPV, with more being discovered every year. Most infect the outer skin, although nearly a quarter infect mucosal epidermis, primarily in the anogenital tract. The tropisms of these viruses for specific anatomical regions are likely due to each viral type evolving to utilize cellular transcription factors specific for mucosal versus cutaneous epidermis. Of the mucosal HPVs, approximately 15 have been classified as ‘high risk’ for induction of human cancer; the most common of these being HPV types 16 and 18 in the majority of the world, although epidemiological studies in specific regions of the world, such as the Caribbean and eastern Europe, show high-risk types other than 16 or 18 to be more common. These mucosal types are sexually transmitted and very common, with over 80% of people in their 30s being sero-positive for at least one type [2]. Fortunately, only one in a thousand infected individuals ever progresses to HPV-induced cancers, highlighting the fact that additional environmental and/or genetic factors are required for cancer development. The low-risk sexually transmitted HPVs (primarily HPV 6 and 11), although low risk for the development of cancer, cause recurring condylomas that affect approximately 1% of the population. Although not life-threatening, these HPV infections create a substantial public-health burden. In addition, on rare occasions, these low-risk HPVs can be passed from mother to child during birth and cause a persistent tracheal infection, in which condyloma growth can block the airway. These children often require repeated surgical intervention to clear their airways to allow them to breathe. These treatments are required for years or even decades, and these patients often suffer from tracheal cancers later in life.
Infectious cycle
HPV has an unusual infection cycle in that it relies on the differentiation programme of the infected stratified epithelium. Several studies have led to the following model of HPV replication. The virus infects proliferating basal keratinocytes, accessed through some sort of break or abrasion in the outer layers of the epidermis. Although cutaneous HPVs scattered into the environment have been shown to remain infectious (HPVs are non-enveloped and resistant to drying and many other environmental variations, leading to environmentally stable virus particles), mucosal HPV types are mostly transmitted through direct contact and the resultant microabrasions in the mucosal skin. The cervix, and in particular the transformation zone located at the junction of granular and stratified epithelium, appears to be particularly prone to infection by sexually-transmitted HPV types. HPV virions attach to the cell through their major viral capsid protein, L1. The cellular receptor for HPV has not been unequivocally identified. Both cell surface heparan sulfate proteoglycans and integrin proteins, in particular those up-regulated during wound healing, have been reported to play a role in viral attachment and entry. After being taken up, the viral particles travel to the nuclear membrane, where the major and minor capsid proteins (L1 and L2, respectively) are left behind, and the viral genome – in the form of a chromatinized double-stranded DNA circular episome of approximately 8 kb – is transported into the host cell nucleus.
HPV has evolved such that the early genes are readily transcribed by the host cell transcription machinery without any need for viral protein synthesis. Expression of these early genes establishes a cellular environment conducive to viral replication, in particular by promoting viral DNA replication (E1 and E2), regulation of viral gene transcription (E2), immune evasion (E5, E6 and E7), prevention of cell cycle arrest and apoptosis (E6) and maintenance of cellular proliferation (E7). Although low amounts of these proteins are synthesized, they are sufficient to support viral DNA amplification to levels of approximately 50–200 genome copies per cell and to immortalize infected keratinocytes. In contrast to basal cells from an uninfected epithelium, which stop proliferating and begin to differentiate as they migrate to the supra-basal layers, infected cells are kept in a proliferating mode by the combined action of E6 and E7, whilst retaining the ability to differentiate. This likely accounts for the hyperkeratosis characteristic of HPV infections, and ultimately results in more infected cells producing more virus. As infected cells are displaced away from the basal layer and continue their differentiation programme, the productive phase of the infectious cycle is induced in the most upper layers of the epithelium. This phase is characterized by the expression of the late genes encoding the L1 and L2 major and minor capsid proteins, respectively, and the amplification of the viral episome to thousands of copies per cell. Also characteristic of this part of the infection cycle is the expression in large amounts of the viral E4 protein, which is somehow required for viral DNA amplification, at least for tested high-risk HPV types [6,7]. Following assembly of individual genomes into chromatin (consisting of the core histones H2A, H2B, H3 and H4) and their packaging into L1/L2 empty capsids, the newly biosysthesized virions are then released – in part through the ability of E4 to disrupt the cellular cytokeratin network [8] – and shed with the outer skin squames ready for subsequent infection.
Cancer is not a normal stage in the HPV replicative cycle. During infections, a few HPV genomes become integrated into the host cell’s genome. These are ‘dead end’ events and cannot lead to virus production. The vast majority of the time these integration events have no appreciable biological consequence. However, on the rare occasion, integration can result in overexpression of the viral E6 and E7 oncogenes, such as promoting carcinogenesis. This, for example, can occur if integration takes place into a transcriptionally active region of the host genome in a way that results in disruption and/or decreased expression of the viral E2 protein, a negative regulator (that is, repressor) of E6 and E7 transcription. De-repression of E6 and E7 caused by the loss of E2 promotes cellular proliferation, in part through the ability of E7 to prevent the differentiation-dependent arrest of cells in G0/G1 phase and maintain the cells in a proliferative state and in part through the ability of E6 to prevent cell cycle arrest and apoptosis in response to unscheduled DNA synthesis and/or damage. As a consequence of E6 and E7 action, infected cells are prone to genomic instability, a hallmark of most transformed cells and the source for the many additional genetic changes required for reaching a full cancerous phenotype.
Prophylactic approaches against HPV
There are various prophylactic avenues to prevent anogenital HPV infection. Although the virus particle is small enough (55 nM diameter) to fit through the naturally occurring pores in latex condoms, epidemiological studies have demonstrated that condoms can appreciably reduce HPV transmission. However, barrier protection cannot be complete as HPVs can be transmitted through unprotected areas of the anogential region. Studies have shown that the natural product carrageenan, a sulphated polysaccharide extracted from certain seaweed species, may be an effective microbicide as it inhibits the interaction of HPV with the heparin sulphate proteoglycans, which are HPV’s initial point of cellular attachment [9–11]. Both of these approaches can be of use in decreasing HPV transmission.
Immunological prophylaxis is also of great value. Two anti-HPV vaccines are currently available: the quadrivalent Merck vaccine, against HPV low-risk types 6 and 11 and high-risk types 16 and 18; and the GlaxoSmithKline bivalent vaccine against types 16 and 18. These vaccines are based on empty ‘ghost’ virus-like particles made up of the L1 capsid proteins from each viral type. Optimal protection requires three injections, ideally given prior to the individual becoming sexually active. These vaccines have been shown to be highly protective against the early stage transformed-cell lesions that represent the precursors of cervical cancer. As HPV 16 and 18 currently represent the two most common high-risk HPVs in most areas, universal vaccine adoption would protect against over two-thirds of HPV-induced cancers. However, both societal and economic issues currently prevent broad HPV vaccine adoption in most populations. Research continues to attempt to develop additional prophylactic vaccines, using either L1 capsomeres or by addition of virus-like particles from other prevalent HPV types, such as the V503 nonavalent vaccine currently used in clinical studies, which would cover nine HPV types [12]. One vaccine being studied will attempt to use a more conserved and cross-reacting neutralizing epitope in the minor capsid protein L2 [13–17]. For further information on prophylactic vaccines, we direct readers to several excellent recently published reviews on this topic [18–22]. The current anti-HPV vaccines are useful tools in the battle against HPV infections and cancers, but do not provide a complete or long-term answer to eradication of HPV diseases. By their very nature of being capable of inducing a strong and protective humoral – but not cellular – host immune response, they are also of little therapeutic value to already infected patients.
Several vaccines are also currently under development to treat HPV-induced cancers, rather than to prevent initial viral infections. These vaccines would not be used in a prophylactic manner, but as a therapeutic vaccine for patients battling early- or late-stage HPV cancers. In general, these development-stage therapeutic vaccines are based on using the HPV oncoproteins E6 and/or E7 as cancer antigens and would be used to specifically target the host immune response to the HPV-transformed cells (for reviews see [19,23,24]).
Therapeutic approaches against HPV infection
Because HPV infections are not systemic and often localized to easily accessible regions of the skin and mucosa, various cytopathic options are available for topical treatment of HPV-induced lesions (reviewed in [1]). Some simple options include surgical removal of the infected area and cryogenic destruction of the local epidermis. Cytotoxic agents are also used to attempt to kill the focus (foci) of infection, and these range from simple chemicals, like phenol and salicylic acid (in over-the-counter products) or trichloroacetic acid and silver nitrate, to more complex natural products, such as podophyllotoxin (a mitotic cell cycle inhibitor). However, unless the cells within the infected area are removed/killed in a fairly extensive manner, the rate of recurrence with all these treatments can be substantial, often leading to the necessity of repeated and costly treatments.
Localized immunomodulation has proven helpful to aid in the normal immunological clearance/suppression of HPV infections. Topical application of imiquimod, the active agent in the Aldara cream, is a Toll-like receptor 7 agonist, which activates dendritic cells, macrophages and keratinocytes to release type I interferons and proinflammatory cytokines. Imiquimod is licensed to treat external genital warts [25]. As anticipated from its proinflammatory action, there are negative side effects to the use of imiquimod, including severe irritation at the application site. Polyphenon E is a standardized extract of green tea leaves that has also been shown to stimulate clearance of HPV apparently through immunomodulation [26].
Currently no HPV-specific antiviral agents are available to treat HPV infections. This is in direct contrast to many other viruses for which antiviral agents are available (for example, HIV, herpes virus, hepatitis B and C, cytomegalovirus, influenza virus and others). Why is this so? For many viruses there are specific viral functions, often enzymes, that can be attacked. For example, HIV encodes three enzymes, a reverse transcriptase (RT), an integrase (IN) and a protease (PR) that catalyse the conversion of the RNA genome into DNA, the integration of the viral DNA into the host genome and the cleavage of the encoded HIV polyprotein into the individual viral enzymes/proteins required for various viral functions, respectively. Anti-HIV drugs have been developed that act against each of these viral enzymes [27]. Similarly, drugs have been developed against key events of the influenza life cycle that target the acidification/viral uncoating step or the neuraminidase/viral release process [28]. The most common antiviral agents are nucleoside analogues that preferentially inhibit the viral polymerase responsible for viral genome synthesis, such as HIV RT and the herpes virus DNA polymerases. Nucleoside analogues are screened for those that inhibit the viral enzyme at much lower concentrations than those required to inhibit host polymerase activity (exhibit a therapeutic window) and/or require activation/phosphorylation by a viral enzyme in order to become an effective analogue. Nucleoside analogue antivirals are available against HIV, herpes virus types 1 and 2, cytomegalovirus and hepatitis B and C amongst others. Because HPV encodes only a single enzyme, E1 (a NTPase/helicase), and utilizes the cellular DNA polymerases for synthesis of HPV genomes, this severely constrains the number of more conventional antiviral targets. To date, there has been little progress in developing therapeutic agents that target the HPV E1 protein.
As a result of the lack of enzymatic targets, several current anti-HPV strategies aim to modulate various protein–protein interactions required for the HPV infection cycle. These include inhibitors of the E1–E2 interaction to inhibit the initiation of viral DNA replication, inhibitors of interactions between E2 and cellular transcription factors, such as Brd4 involved in E2’s transcriptional activity and genome maintenance function, and interactions between E6 and E7 and their cellular partners, such as E6AP, p53, PDZ domain proteins and retinoblastoma protein family members. Targeting the latter interactions would be particularly useful as they might also be effective against HPV-induced cancers, which remain reliant on expression of E6 and E7 to maintain their viability and transformed phenotype [29,30]. This has made these two oncoproteins the prime antigens for therapeutic vaccine development, which remains an active and promising area of research [19,23,24].
For therapies targeting an active HPV infection, we believe it is advisable to develop a therapeutic that inhibits HPV DNA replication, as this would be expected to suppress HPV integration events, which would presumably lead to decreased HPV-induced cancers. A concern of targeting other aspects of the HPV viral life cycle, such as E6 or E7, is that this could allow HPV to continue to replicate its DNA in the infected basal cells. As basal cells are stem cells and are inherently immortal, long-term replication of HPV in basal cells is theoretically possible, even if E6/E7 functions are blocked. As such, these basal cells would not be targeted by therapies against E6/E7. These cells may even escape immune-based therapies as basal cells express exceedingly low levels of HPV viral antigens, which would be required for immune detection. Such a situation of long-term HPV DNA replication in basal cells alone would increase the likelihood of viral genome integration into host cell chromosomes, possibly leading to cellular transformation and HPV-dependent cancer years later. It has been clearly shown that expression of HPV E1 and E2 alone are sufficient to induce integration of HPV DNA [31,32], and this is a particular danger in the presence of DNA damage [33]. Hence, anti-HPV agents that do not block viral DNA synthesis could, over time, favour cancer development by increasing HPV genome integration.
Although anti-HPV replication drugs would be more attractive for ongoing infections, these drugs are not likely to be useful against high-grade cervical lesions because the viral genome would have already been integrated. For high-grade lesions with integrated HPV genomic therapeutics, targeting the viral oncoproteins would be more appropriate. A target that would reintroduce p53 function would be more apt for treatment of high-grade lesions and HPV-induced cancers. There are several possible approaches under investigation that could stabilize p53 and induced apoptosis in such cell types [30,34–36]. Likewise, there may even be concerns about using anti-HPV replication drugs alone to treat lesions containing a mixture of episomal and integrated viral genomes as loss of the episomes and E2 expression might result in de-repression of E6 and E7 from the integrated genomes, thereby stimulating carcinogenesis. Although theoretically possible, it is unclear if episomal and integrated copies of the viral genome can coexist for long in the same cell, as opposed to different cells in the same lesion, given that integrated genomes would be subject to ‘onion-skin’ replication driven by episome-encoded E1 and E2, which would induce cellular DNA damage responses. And, at this point, it is difficult to evaluate whether any potential negative effects of decreased E2 levels would be outweighed by the prevention of further viral integration.
HPV DNA replication
To highlight potential therapeutic targets within HPV DNA replication let us first outline the HPV DNA replication process (reviewed in [31,37,38]).
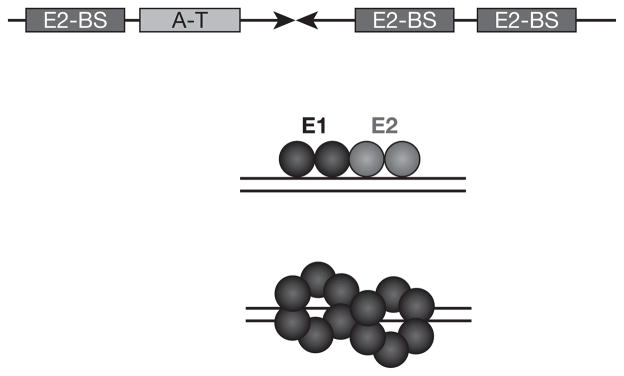
HPV DNA replication is initiated upon the binding of the viral E2 protein to the E2 binding sites (E2–BS) within the origin and the concomitant recruitment of the HPV E1 protein through a protein–protein interaction between E1 and E2 to the adjacent E1 binding site (Figure 1). This E1–E2 interaction has been shown to be vital for efficient HPV DNA synthesis in many systems and is, therefore, a potential target for antiviral therapy. Origin binding and ATP binding induce E1 to assemble into higher order structures, ultimately resulting in a double hexameric E1 DNA helicase [38]. This is the form of E1 that drives the HPV bi-directional DNA replication fork and recruits, and productively interacts with, the various cellular DNA replication factors that are pirated by HPV for its own genome synthesis (Figure 2).
Figure 1.
Depiction of HPV origin recognition by E1 and E2
Papillomavirus origin recognition. BS, binding sites; HPV, human papillomavirus.
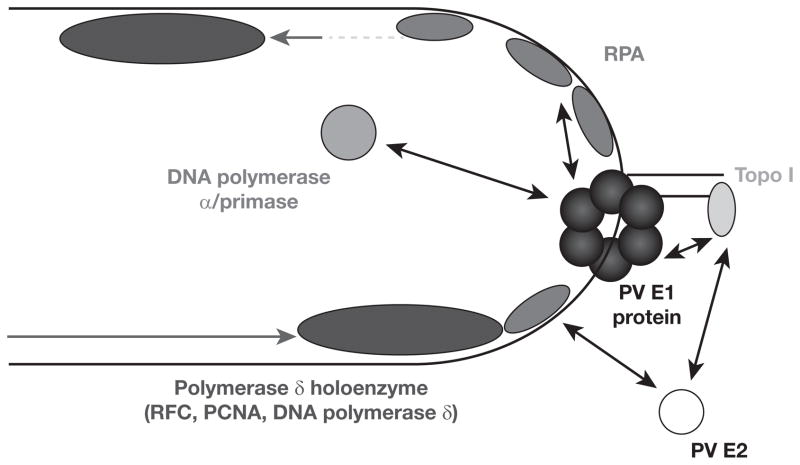
Figure 2.
Known interactions between E1 or E2 and cellular DNA replication factors
Papillomavirus (PV) DNA replication fork. PCNA, proliferating cell nuclear antigen; RFC, replication factor C; RPA, replication protein A; Topo I, topoisomerase I.
The HPV E1 protein also interacts with human topoisomerase I (topo I) [39]. The significance of a physical interaction with a topoisomerase, an enzyme that can act at a distance to release torsional stress on double-stranded DNA caused by progression of a DNA replication fork, was originally unclear. However, the interaction of topo I with the related polyomavirus replicative DNA helicase, SV40 large T-antigen (Tag), has been shown to play roles very early in SV40 DNA replication, including assisting with origin recognition by Tag [40,41]. There are many similarities in how polyomaviruses and papillomaviruses (PVSs) utilize the cellular DNA replication machinery to replicate their viral genomes; hence, studies on PV DNA replication often look to what is known about SV40 DNA replication for additional insights. Based on the SV40 findings, we investigated whether topo I affects E1’s interaction with the origin. We discovered that topo I does indeed assist the interaction and assembly of the BPV1 E1 helicase at the PV origin [42]. Furthermore, we discovered that the interaction of E1 with topo I has a substantial effect on topo I’s enzymatic activity, stimulating its DNA super-coil relaxation activity several-fold [39]. After years of study, Simmon’s group [43] has shown that individual point mutations in Tag that weaken, but do not totally abrogate, Tag’s interaction with topo I have a significant effect on SV40 DNA replication levels in vitro and decrease virus production by 200–2,000-fold. This indicates that the interaction between Tag and topo I is critical for efficient SV40 DNA synthesis and, therefore, for SV40 propagation. Although not yet reported, it is possible that the HPV E1–topo I interaction may be similarly vital for HPV DNA synthesis.
The PV E1 protein has also been shown to interact with the human polymerase α–primase complex [44–48]. The interaction of SV40 Tag with the human polymerase α–primase complex has been found to play an important role in synthesizing primers during SV40 DNA replication [49]. Very recent work has shown that, like the Tagtopo I interaction, individual point mutants in Tag that compromise its interaction with polymerase α–primase inhibit SV40 DNA replication [50]. Although the functional consequences of the E1-polymerase α-primase interaction are less well-studied than that of the Tag–polymerase α–primase interaction, it seems likely that this interaction would be vital for primer synthesis during HPV DNA replication.
HPV E1 also interacts with the cellular single-stranded (ss)DNA binding complex replication protein A (RPA) required for DNA replication [51]. We have demonstrated that this interaction modulates the ability of RPA to bind to ssDNA. Based on a wealth of biochemical data, we proposed a model whereby E1 binds to free RPA in the nucleoplasm and actively loads it onto the ssDNA being extruded from the E1 DNA helicase [52]. Subsequent studies on SV40 Tag provided further support of this model for RPA loading by DNA helicases [53]. In addition, the interaction of SV40 Tag with RPA has been shown to be important for primer synthesis by the polymerase α–primase complex through de-repressing RPA’s capacity to out-compete polymerase α–primase for the lagging strand ssDNA template [54,55]. Although this has not been shown for the E1–RPA interaction yet, it would not be surprising to find the E1–RPA interaction to be important for de-repression of primer synthesis during HPV DNA replication.
Elongation at the SV40 DNA replication fork occurs via the action of replication factor C (RFC), which loads the processivity factor proliferating cell nuclear antigen PCNA and DNA polymerase δ onto RNA–DNA primers synthesized by DNA polymerase–α primase for both leading- [56] and lagging-strand synthesis [57]. Although it has been proposed that DNA polymerase epsilon plays the primary role in leading-strand replication for eukaryotic chromosomal DNA replication with DNA polymerase δ assigned to the lagging strand [58], for both SV40 and PV DNA replication there has been no apparent role demonstrated for DNA polymerase epsilon. For reasons that remain unclear, it appears that both virus families have evolved to primarily utilize DNA polymerase δ for elongation on both strands [59]. Although the action of RFC, PCNA and polymerase δ exhibit substantial intra-molecular interplay, to date, there is no published evidence of either of the PV DNA replication proteins, E1 or E2, or SV40 Tag interacting with any of these DNA replication elongation factors. It is possible that no such interactions are necessary because RPA and DNA polymerase α–primase play roles in directing RFC to newly synthesized RNA–DNA primers. However, further research on this area is merited.
To complete synthesis of newly replicated daughter molecules requires the removal of the RNA primers and the RNA–DNA junction, the synthesis of the resultant gap and the ligation of the completed nascent strands. This has been shown to be accomplished by RNase H, the nuclease FEN1, DNA polymerase δ and DNA ligase I for SV40 DNA replication [57], although others have shown that there are alternate processing pathways for the RNA primer by DNA helicase 2, depending on the sequence and possibly secondary structure of the primer flap [60,61]. Presumably these primer processing factors all play similar roles in HPV DNA replication.
Interactions have also been reported to occur between E2 and the cellular DNA replication proteins RPA and topo I [62,63]. And although there is a few-fold stimulation of topo I activity by E2, no biochemical effect has been published resulting from the E2–RPA interaction, and neither of these interactions were shown to be vital for PV DNA synthesis. Also, the lack of a strict requirement for E2 in HPV DNA replication systems suggests that targeting interactions between E2 and cellular replication proteins may not be as productive or potentially broad-range as targeting interactions between E1 and cellular factors [64,65].
HPV DNA replication and the DNA damage response
Many small DNA tumour viruses manipulate the cellular DNA damage response (DDR), either taking advantage of it or inhibiting it to promote their replication. HPV is no exception. Although an ATM-dependent DDR had no effect on HPV genome replication in undifferentiated cells, it was found to be important for amplification of the viral genome in differentiated keratinocytes [66]. In these cells, the viral oncoprotein E6 was found to be sufficient to induce a DDR [66]. Interestingly, parallel studies in cultured cells showed that nuclear accumulation of E1 is sufficient to induce an ATM-dependent DDR, in part through the ability of this helicase to induce DNA double strand breaks in the host genome [67–69]. This E1-induced DDR is accompanied by a block in cellular proliferation caused by cell-cycle arrest in S phase [67,69]. Viral DNA replication still proceeds unaffected despite the induction of DNA damage and a concomitant check point response, although cellular DNA synthesis is stopped as expected [69]. This failure of HPV DNA replication to be regulated by a check point response was first noticed by King et al. [33], who also observed that this behaviour contrasts with other viruses, such as SV40, whose genome replication does arrest upon DNA damage signalling, likely by inhibitory phosphorylation of the large Tag helicases by DDR kinases [33]. Although induction of a DDR is important for HPV amplification, it is not required for maintenance of the viral episome in undifferentiated immortalized keratinocytes, therefore, DDR would not be an optimal target for the development of an HPV antiviral for reasons discussed herein.
Targeting HPV DNA replication for development of antivirals
HPV DNA replication is an attractive target for HPV antivirals because inhibition of HPV DNA replication would result in fewer genomes available for viral protein synthesis, as well as fewer genomes for integration – a critical step in the development of HPV-dependent cancers. This section will explore various possibilities for targeting HPV DNA replication, both those attempted and theoretical, and discuss their pros and cons.
Because HPV does not encode its own polymerase, this eliminates the possibility of using the most common approach to developing antivirals, that of screening nucleoside analogues exhibiting a useful therapeutic window. Several large pharmaceutical companies have invested substantial resources in identifying active site inhibitors, either nucleoside analogues or small molecules that target the single enzyme encoded by HPV E1 ATPase/helicase. Although small molecules that inhibit E1 ATPase and DNA helicase activities have been identified [70,71], unfortunately none of these projects have resulted in a viable therapeutic. This may be in part because the E1 nucleoside triphosphate (NTP) binding site is not particularly stringent (unlike most ATPases, E1 can utilize a wide variety of NTPs [72–74]). E1’s flexible NTP binding domain may make it more difficult to identify inhibitors that can discriminate between E1 and the wide variety of NTPases present in human cells. Conversely, the difficulty in translating E1 inhibitors into therapeutics may have been a result of simple issues, such as the lack of activity against many HPV types, cellular uptake or of other drug-like properties. With the lack of success in identifying inhibitors that specifically abrogate broad-range HPV E1 activity, the absence of other HPV viral enzyme targets has shifted attention to the approach of targeting molecular interactions as the major avenue for development of anti-HPV therapeutics.
One obvious molecular interaction target for anti-HPV development would be the recognition of the HPV ori by E1 and E2. Recently, some success has been reported using pyrrole-imidazole polyamides, a class of sequence-specific DNA binding compounds designed to target AT-rich regions of DNA. It was shown that specific polyamides designed to target the HPV ori could interfere with the maintenance of HPV 16, 18 and 31 viral episomes in immortalized keratinocytes, without promoting viral DNA integration and, importantly, with no apparent sign of cellular toxicity [75]. Presumably, the compounds act by binding to the ori and prevent the binding of E1 and/or E2, although this has not yet been formally examined. Polyamides have also been shown to reduce HPV DNA copy number in organotypic rafts treated topically. These encouraging results have motivated the further investigation of these ori-binding compounds as topical agents for the treatment of HPV infections.
In recent years, antiviral research has begun turning to the modulation of vital protein–protein interactions as a potential therapeutic avenue. This is a relatively new paradigm but has led to promising lead compounds for the development of anti-herpes simplex virus (HSV)2 drugs that modulate the interaction of the HSV DNA helicase and primase enzymes [76,77], as well as possible drugs to treat HPV condylomal disease [78,79]. It is of course important that the interaction being targeted for disruption is both vital for HPV DNA synthesis and susceptible to molecular interference (for example, a very stable protein interaction that is not dynamic during the course of HPV genome synthesis would not become accessible to pharmacological interference; conversely, a more transient, although still vital, interaction would provide an opportunity for a drug to bind and interfere. For a general discussion of protein–protein interactions as drug targets, see Fry [80].
The first protein–protein interaction shown to be essential for HPV DNA synthesis was the interaction between the HPV E1 and E2 proteins. With the role that E2 plays in helping E1 to bind to the HPV origin and assemble into a homomultimeric DNA helicase (reviewed in [38]), it was reasonable to hypothesize that the E1–E2 interaction might be vital for HPV DNA replication. This hypothesis was validated by mutations in E2 that abrogate E1 binding [81–87]. Small molecule inhibitors of the E1–E2 interaction were identified that could indeed inhibit HPV DNA replication in cultured cells, but, unfortunately, these compounds were highly specific, only affecting viral DNA replication for the low-risk HPV types 6 and 11 [78,79]. Determination of the co-crystal structure of one of these inhibitors bound to E2 led to an understanding of the HPV-type specificity [88]. Although it is unfortunate that these compounds were not effective against all HPV types, it should be noted that HPV 6 and 11 cause the great majority (approximately 90%) of condylomas, which makes targeting these HPV types of value to human health. In retrospect, it is not altogether surprising that an inhibitor of the E1–E2 interaction might be fairly specific because the protein sequences of E1 and E2 can vary significantly between different HPV types. The process of HPV evolution, maintaining the structure, function and interactions of the viral proteins while allowing the precise amino acid residues to vary, means that the E1–E2 interaction will vary slightly from type to type. This limitation in targeting interactions between viral proteins is also seen with the drugs developed against the HSV2 helicase–primase interaction; it has been shown that HSV2 develops resistance to these drugs at a rate similar to that for valacyclovir [89,90]. Double mutations in the helicase and the primase alter the interaction sufficiently to confer resistant to the drug but are compensatory with regard to the nature of the critical protein–protein interaction. In these two cases, the limited HPV type spectrum of the E1–E2 interaction inhibitors and the development of drug resistance to the anti-HSV drugs demonstrate how the variability of viral proteins and viral protein–protein interactions can be a significant limitation in developing broad-spectrum antivirals. One way of possibly minimizing these issues would be to target a vital protein interaction between a viral protein and a cellular protein.
An advantage in targeting a viral–host protein interaction for development of antivirals relies upon the relative genetic stability of the cellular genome. Because the amino acid sequence of the human protein is virtually inviolate and not subject to the natural variation or rapid mutation exhibited by many viral proteins, this means the cellular protein provides a ‘molecular anchor’ to the interaction, inherently limiting the degree to which the viral protein (such as HPV E1, for example) can vary and still interact productively with the cellular protein. A potential disadvantage of targeting such an interaction is that there is a chance that a small molecule that interferes with a viral protein–cellular protein interaction might also interfere with the interaction between that cellular protein and one of its normal cellular interaction partners. This might or might not be the case because small molecule inhibitors can be very specific (case in point being the E1–E2 inhibitors that showed high specificity between HPV isotypes). But with this concern in mind, design of second-generation inhibitory small molecules, utilizing crystal structure information on the protein–protein and drug–protein interactions, could be used to preferentially target drugs to the conserved interacting pockets/surfaces on the viral protein, thereby decreasing the likelihood of secondary effects on cellular interaction partners.
To identify viral–cellular protein interactions suitable for the discovery of drugs that inhibit HPV genome replication, ideally one should focus on those involving host DNA replication factors. Below we explore the various interactions between E1 and cellular DNA replication proteins that are attractive candidates for pharmacological intervention.
Polymerase α-primase interaction
Although we have no direct evidence of E1 stimulating the activity of DNA polymerase α–primase, there is large amount of data on the importance of the interaction between SV40 Tag and DNA polymerase α–primase for primer synthesis during SV40 DNA replication. Recently, several amino acid residues on SV40 Tag have been identified that compromise its interaction with the p70 subunit of DNA polymerase α–primase, and these Tag mutations show severely compromised SV40 DNA replication in vitro and in cultured cells [50]. A parallel situation for HPV DNA replication would imply that the E1 interaction with DNA polymerase α–primase would be vital for HPV DNA synthesis, and we, therefore, anticipate that this would be a valid potential target for developing an anti-HPV DNA replication therapeutic. In support of this suggestion, E1 was found to bind to the p70 subunit of DNA polymerase α–primase. Furthermore, it was found that a fragment of p70 that can compete the interaction of E1 with the holoenzyme could effectively inhibit E1-catalysed cell-free DNA replication [44]. We anticipate that the E1 interaction with DNA polymerase α–primase will be an excellent target for development of antivirals.
Replication protein A interaction
There is evidence of E1 affecting the function of RPA and of E1 potentially acting to load RPA onto ssDNA being extruded from the E1 helicase [52]. Although there is no direct evidence of this interaction being essential for HPV DNA replication, it is known that Escherichia coli single-stranded DNA binding protein cannot replace RPA for ssDNA binding function in the DNA replication of many small DNA viruses, including PVs [54,91–93], implying that a specific interaction is required between E1 and RPA. Whether this specificity is at the level of RPA loading onto ssDNA or at the level of primer synthesis with the E1–RPA interaction playing a role in primer synthesis, as has been shown for the SV40 Tag–RPA interaction [54], is currently unknown. Regardless, the absolute requirement for RPA in HPV DNA replication suggests that the E1–RPA interaction is likely vital for this process and, therefore, could potentially be an effective target for development of anti-HPV antivirals.
Topo I interaction
The interaction of HPV E1 with human topo I would at first glance not be expected to be a good target for development of an HPV antiviral. Topoisomerases, by acting on the topology of the DNA helix, can act at a distance from where the replication fork actually is and, furthermore, for a long time it was believed that different types of topoisomerases were able to partially compensate for loss of another type [94]. With these assumptions, a strict requirement for a specific topoisomerase for a particular function was considered unlikely. However, there has been mounting evidence for many years for a specific and vital interaction between topo I and the polyomavirus DNA helicase SV40 Tag [40,95]. Furthermore, this interaction has been shown to have effects on Tag at the earliest stages of SV40 DNA replication [41]. Likewise, we have shown that E1 interacts with topo I [39], and that this interaction stimulates the interaction of E1 with the PV origin of replication [42]. The discovery that point mutations in SV40 Tag that weaken the interaction with topo I have a hundred- to thousand-fold effects on SV40 DNA replication [43] indicates that this interaction is critical and suggests that, in general, viral helicase–topo I interactions may be excellent targets for the development of antiviral therapeutics. Indeed, we have recently shown that single amino acid substitutions in HPV E1 that weaken the interaction with topo I are also severely compromised for HPV DNA replication (unpublished observations). We are currently investigating the feasibility of targeting this E1–topo I interaction for therapeutic intervention.
E1 has been shown to interact with all of the cellular complexes required for the earliest stages of PV and polyomvirus DNA replication (topo I, RPA and DNA polymerase α–primase) and, as a result of their early and vital role in the DNA replication process, these three interactions would likely be excellent opportunities for drug targeting. However, it is reasonable to speculate that other cellular proteins that interact with E1 might also provide opportunities for pharmacological intervention.
Targeting of other cellular partners
In addition to the cellular factors directly involved in DNA replication, E1 has also been found to interact with several cellular proteins of diverse functions, including the heat shock proteins Hsp40 and Hsp70 [96,97], histone H1 [98], SNF5 [99], E1-BP [100], Ubc9 [100–104], p80/Uaf1 [105,106], cyclin A/E-Cdk2 [107–110] and the interferon-stimulated protein p56/IFIT1 [111,112]. However, for many of these interactions, their relevance for viral DNA replication remains to be clearly established. Amongst the E1-interacting proteins not directly involved in DNA synthesis, the interactions of E1 with p80/Uaf1 and with cyclin A/E-Cdk2 appear to be the most promising targets from an antiviral drug discovery perspective, for reasons discussed below.
Binding to p80/Uaf1 has been detected for the E1 proteins from anogenital HPV types but not those of cutaneous viruses, suggesting that this interaction plays a specific role in the replication of mucosal HPVs [105]. P80/Uaf1 associates with the N-terminal 40 amino acids of E1, and mutations within this region that prevent p80 binding greatly reduce transient HPV DNA replication in transfected cells and abolish maintenance of the viral episome in immortalized keratinocytes [105]. Furthermore, it was shown that the E1–p80 interaction can be antagonized in vivo by expression in trans of a peptide corresponding to the N-terminal 40 amino acids of E1 [106]. This peptide prevents E1-dependent recruitment of p80 to the viral origin, resulting in a significant reduction in viral DNA replication [106]. These findings provided evidence that antagonizing the E1–p80 interaction could be a tenable antiviral strategy. Although the exact function of p80/Uaf1 in viral DNA replication remains unclear, this WD40 repeats-containing protein is known to interact with the de-ubiquitinating enzymes Usp1, Usp12 and Usp46, whose substrates include PCNA, histones H2A and H2B and Fanconi anaemia complementation group D type 2, a component of the Fanconi anaemia pathway [113,114].
The interaction of E1 with cyclin A-E/cdk2 also appears attractive as an antiviral target because it tightly regulates the shuttling of E1 between the nucleus and cytoplasm [108,109]. Specifically, it has been shown that E1 interacts with cyclin A-E/cdk2 through a highly conserved cyclin-binding motif (RxL) and is a substrate of these cell-cycle regulatory kinases [107–110]. Phosphorylation of either HPV11 E1 or HPV31 E1 on amino acid residues near or within their nuclear-export sequences abolishes Crm1-dependent nuclear export, thus promoting E1 accumulation in the nucleus during S phase when viral DNA replication takes place [108,109]. In principle, therefore, interfering with the interaction of E1 with cyclin A-E/cdk2, or with the activity of cdk2 directly, would be expected to trigger export of this helicase to the cytoplasm and prevent viral DNA replication occuring.
Based on the work mentioned above showing that HPV E1 sequestered in the nucleus is able to induce an S phase check point and block cellular proliferation, it is tempting to speculate that promoting the sustained nuclear accumulation of E1 by pharmacological means could be a useful strategy to hinder the proliferation of HPV-infected cells. A finding consistent with this concept is the observation that cells maintaining HPV episomes can be induced to apoptose by treatment with the Crm1-exporting inhibitor leptomycin B [115], which should trigger nuclear accumulation of E1 in these cells (although that was not directly tested in this study). Obviously, many other Crm1 cargo proteins should also be mislocalized upon leptomycin B treatment, making it hazardous to infer that E1 nuclear accumulation is the only mechanism involved in this apoptotic response. However, pharmacological induction of E1 nuclear accumulation may cause more harm than good if it results in increased viral DNA replication and induction of a DDR. It can easily be imagined that the ability of E1 to induce DNA double-strand breaks [32,69], coupled with the intrinsic resistance of HPV DNA replication to check point signalling [33], would result in the accumulation of viral DNA replication intermediates that are recombinagenic and prone to integration into the host genome. Because integration of the viral DNA can result in overexpression of the viral oncogenes and promotion of cancer development, it may, therefore, be unwise to contemplate nuclear accumulation of E1 as a therapeutic strategy.
Current perspectives
Although it is possible that useful active site inhibitors of E1 can still be developed, after several large-scale attempts have failed, this approach seems to have largely been abandoned. Conversely, the era of developing small molecule inhibitors of protein–protein interactions as therapeutics is in its infancy. Although there have been issues that have mitigated success, such as issues of toxicity for the HSV2 helicase–primase interaction drugs or issues of narrow-type specificity for the small molecules developed to inhibit the low-risk HPV E1–E2 interaction, these initial studies have provided the important proof-of-principle that such small molecule inhibitors can be developed to successfully modulate protein–protein interactions. For HPV in particular, with its single virally encoded enzyme, modulation of protein–protein interactions is likely to be the major avenue for the development of antiviral agents. We believe this to be true for drugs targeted not only at HPV DNA replication but also towards other HPV proteins. As noted above, there are likely several HPV proteins whose protein–protein interactions could be effectively targeted for therapeutic intervention; however, because of the ability of E1 and E2 alone to induce HPV DNA integration, targeting HPV DNA replication through E1’s interactions with cellular replication proteins is likely to be one of the best approaches for developing an HPV antiviral that also prevents integration of HPV into host cell chromosomes.
Acknowledgments
We apologize to those whose work was not included because of space considerations or whose papers were inadvertently omitted. We thank members of the Archambault and Melendy laboratories for their helpful discussions. Work in the authors’ laboratories is supported by grants from the Canadian Institutes of Health Research (CIHR), the Canadian Cancer Society Research Institute (CCSRI), the Cancer Research Society (CRS) and the National Institutes of Health (NIH – AI095632).
Footnotes
Disclosure statement
JA participated in the studies that led to the identification of the E1 ATPase inhibitors and HPV E1–E2 interaction inhibitors mentioned in this review, is an inventor on one patent related to this subject and has provided advice to companies interested in anti-HPV drug discovery. TM led the studies that identified the interactions of E1 with RPA and topo I and E2 with topo I, and has provided advice to companies interested in anti-HPV drug discovery.
References
- 1.Yanofsky VR, Patel RV, Goldenberg G. Genital warts: a comprehensive review. J Clin Aesthet Dermatol. 2012;5:25–36. [PMC free article] [PubMed] [Google Scholar]
- 2.Stanley MA. Genital human papillomavirus infections: current and prospective therapies. J Gen Virol. 2012;93:681–691. doi: 10.1099/vir.0.039677-0. [DOI] [PubMed] [Google Scholar]
- 3.Wang XI, Thomas J, Zhang S. Changing trends in human papillomavirus-associated head and neck squamous cell carcinoma. Ann Diagn Pathol. 2012;16:7–12. doi: 10.1016/j.anndiagpath.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Crow JM. HPV: The global burden. Nature. 2012;488:S2–S3. doi: 10.1038/488S2a. [DOI] [PubMed] [Google Scholar]
- 5.Rampias T, Sasaki C, Weinberger P, Psyrri A. E6 and E7 gene silencing and transformed phenotype of human papillomavirus 16-positive oropharyngeal cancer cells. J Natl Cancer Inst. 2009;101:412–423. doi: 10.1093/jnci/djp017. [DOI] [PubMed] [Google Scholar]
- 6.Wilson R, Fehrmann F, Laimins LA. Role of the E1--E4 protein in the differentiation-dependent life cycle of human papillomavirus type 31. J Virol. 2005;79:6732–6740. doi: 10.1128/JVI.79.11.6732-6740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson R, Ryan GB, Knight GL, Laimins LA, Roberts S. The full-length E1E4 protein of human papillomavirus type 18 modulates differentiation-dependent viral DNA amplification and late gene expression. Virology. 2007;362:453–460. doi: 10.1016/j.virol.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Roberts S, Ashmole I, Gibson LJ, Rookes SM, Barton GJ, Gallimore PH. Mutational analysis of human papillomavirus E4 proteins: identification of structural features important in the formation of cytoplasmic E4/cytokeratin networks in epithelial cells. J Virol. 1994;68:6432–6445. doi: 10.1128/jvi.68.10.6432-6445.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buck CB, Thompson CD, Roberts JN, Muller M, Lowy DR, Schiller JT. Carrageenan is a potent inhibitor of papillomavirus infection. PLoS Pathog. 2006;2:e69. doi: 10.1371/journal.ppat.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marais D, Gawarecki D, Allan B, et al. The effectiveness of Carraguard, a vaginal microbicide, in protecting women against high-risk human papillomavirus infection. Antivir Ther. 2011;16:1219–1226. doi: 10.3851/IMP1890. [DOI] [PubMed] [Google Scholar]
- 11.Roberts JN, Buck CB, Thompson CD, et al. Genital transmission of HPV in a mouse model is potentiated by nonoxynol-9 and inhibited by carrageenan. Nat Med. 2007;13:857–861. doi: 10.1038/nm1598. [DOI] [PubMed] [Google Scholar]
- 12.Clinical Trials Database. [Accessed 4 October 2012];A study of V503 in preadolescents and adolescents. Updated 2 May 2012. Available from http://clinicaltrials.gov/ct2/show/NCT00943722?term=NCT00943722&rank=1.
- 13.Alphs HH, Gambhira R, Karanam B, et al. Protection against heterologous human papillomavirus challenge by a synthetic lipopeptide vaccine containing a broadly cross-neutralizing epitope of L2. Proc Natl Acad Sci U S A. 2008;105:5850–5855. doi: 10.1073/pnas.0800868105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gambhira R, Karanam B, Jagu S, et al. A protective and broadly cross-neutralizing epitope of human papillomavirus L2. J Virol. 2007;81:13927–13931. doi: 10.1128/JVI.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jagu S, Karanam B, Gambhira R, et al. Concatenated multitype L2 fusion proteins as candidate prophylactic pan-human papillomavirus vaccines. J Natl Cancer Inst. 2009;101:782–792. doi: 10.1093/jnci/djp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pastrana DV, Gambhira R, Buck CB, et al. Cross-neutralization of cutaneous and mucosal Papillomavirus types with anti-sera to the amino terminus of L2. Virology. 2005;337:365–372. doi: 10.1016/j.virol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 17.Roden RB, Yutzy WHt, Fallon R, Inglis S, Lowy DR, Schiller JT. Minor capsid protein of human genital papillomaviruses contains subdominant, cross-neutralizing epitopes. Virology. 2000;270:254–257. doi: 10.1006/viro.2000.0272. [DOI] [PubMed] [Google Scholar]
- 18.Campo MS, Roden RB. Papillomavirus prophylactic vaccines: established successes, new approaches. J Virol. 2010;84:1214–1220. doi: 10.1128/JVI.01927-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frazer IH, Leggatt GR, Mattarollo SR. Prevention and treatment of papillomavirus-related cancers through immunization. Annu Rev Immunol. 2011;29:111–138. doi: 10.1146/annurev-immunol-031210-101308. [DOI] [PubMed] [Google Scholar]
- 20.Gersch ED, Gissmann L, Garcea RL. New approaches to prophylactic human papillomavirus vaccines for cervical cancer prevention. Antivir Ther. 2012;17:425–434. doi: 10.3851/IMP1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiller JT, Lowy DR. Understanding and learning from the success of prophylactic human papillomavirus vaccines. Nat Rev Microbiol. 2012;10:681–692. doi: 10.1038/nrmicro2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanley M. Prospects for new human papillomavirus vaccines. Curr Opin Infect Dis. 2010;23:70–75. doi: 10.1097/QCO.0b013e328334c0e1. [DOI] [PubMed] [Google Scholar]
- 23.Nieto K, Gissmann L, Schadlich L. Human papillomavirus-specific immune therapy: failure and hope. Antivir Ther. 2010;15:951–957. doi: 10.3851/IMP1665. [DOI] [PubMed] [Google Scholar]
- 24.van der Burg SH, Melief CJ. Therapeutic vaccination against human papilloma virus induced malignancies. Curr Opin Immunol. 2011;23:252–257. doi: 10.1016/j.coi.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 25.Novak N, Yu CF, Bieber T, Allam JP. Toll-like receptor 7 agonists and skin. Drug News Perspect. 2008;21:158–165. [PubMed] [Google Scholar]
- 26.Hoy SM. Polyphenon E 10% ointment: in immunocompetent adults with external genital and perianal warts. Am J Clin Dermatol. 2012;13:275–281. doi: 10.2165/11209370-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 27.Hashimoto C, Tanaka T, Narumi T, Nomura W, Tamamura H. The successes and failures of HIV drug discovery. Expert Opin Drug Discov. 2011;6:1067–1090. doi: 10.1517/17460441.2011.611129. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh HP, Hsu JT. Strategies of development of antiviral agents directed against influenza virus replication. Curr Pharm Des. 2007;13:3531–3542. doi: 10.2174/138161207782794248. [DOI] [PubMed] [Google Scholar]
- 29.Butz K, Ristriani T, Hengstermann A, Denk C, Scheffner M, Hoppe-Seyler F. siRNA targeting of the viral E6 oncogene efficiently kills human papillomavirus-positive cancer cells. Oncogene. 2003;22:5938–5945. doi: 10.1038/sj.onc.1206894. [DOI] [PubMed] [Google Scholar]
- 30.Hall AH, Alexander KA. RNA interference of human papillomavirus type 18 E6 and E7 induces senescence in HeLa cells. J Virol. 2003;77:6066–6069. doi: 10.1128/JVI.77.10.6066-6069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kadaja M, Silla T, Ustav E, Ustav M. Papillomavirus DNA replication - from initiation to genomic instability. Virology. 2009;384:360–368. doi: 10.1016/j.virol.2008.11.032. [DOI] [PubMed] [Google Scholar]
- 32.Kadaja M, Sumerina A, Verst T, Ojarand M, Ustav E, Ustav M. Genomic instability of the host cell induced by the human papillomavirus replication machinery. EMBO J. 2007;26:2180–2191. doi: 10.1038/sj.emboj.7601665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King LE, Fisk JC, Dornan ES, Donaldson MM, Melendy T, Morgan IM. Human papillomavirus E1 and E2 mediated DNA replication is not arrested by DNA damage signalling. Virology. 2010;406:95–102. doi: 10.1016/j.virol.2010.06.033. [DOI] [PubMed] [Google Scholar]
- 34.Baleja JD, Cherry JJ, Liu Z, et al. Identification of inhibitors to papillomavirus type 16 E6 protein based on three-dimensional structures of interacting proteins. Antiviral Res. 2006;72:49–59. doi: 10.1016/j.antiviral.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green KL, Brown C, Roeder GE, Southgate TD, Gaston K. A cancer cell-specific inducer of apoptosis. Hum Gene Ther. 2007;18:547–561. doi: 10.1089/hum.2006.042. [DOI] [PubMed] [Google Scholar]
- 36.Hwang ES, Naeger LK, DiMaio D. Activation of the endogenous p53 growth inhibitory pathway in HeLa cervical carcinoma cells by expression of the bovine papillomavirus E2 gene. Oncogene. 1996;12:795–803. [PubMed] [Google Scholar]
- 37.D’Abramo CM, Archambault J. Small molecule inhibitors of human papillomavirus protein - protein interactions. Open Virol J. 2011;5:80–95. doi: 10.2174/1874357901105010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stenlund A. Initiation of DNA replication: lessons from viral initiator proteins. Nat Rev Mol Cell Biol. 2003;4:777–785. doi: 10.1038/nrm1226. [DOI] [PubMed] [Google Scholar]
- 39.Clower RV, Fisk JC, Melendy T. Papillomavirus E1 protein binds to and stimulates human topoisomerase I. J Virol. 2006;80:1584–1587. doi: 10.1128/JVI.80.3.1584-1587.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmons DT, Trowbridge PW, Roy R. Topoisomerase I stimulates SV40 T antigen-mediated DNA replication and inhibits T antigen’s ability to unwind DNA at nonorigin sites. Virology. 1998;242:435–443. doi: 10.1006/viro.1997.9024. [DOI] [PubMed] [Google Scholar]
- 41.Trowbridge PW, Roy R, Simmons DT. Human topoisomerase I promotes initiation of simian virus 40 DNA replication in vitro. Mol Cell Biol. 1999;19:1686–1694. doi: 10.1128/mcb.19.3.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y, Clower RV, Melendy T. Cellular topoisomerase I modulates origin binding by bovine papillomavirus type 1 E1. J Virol. 2006;80:4363–4371. doi: 10.1128/JVI.80.9.4363-4371.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khopde S, Simmons DT. Simian virus 40 DNA replication is dependent on an interaction between topoisomerase I and the C-terminal end of T antigen. J Virol. 2008;82:1136–1145. doi: 10.1128/JVI.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amin AA, Titolo S, Pelletier A, Fink D, Cordingley MG, Archambault J. Identification of domains of the HPV11 E1 protein required for DNA replication in vitro. Virology. 2000;272:137–150. doi: 10.1006/viro.2000.0328. [DOI] [PubMed] [Google Scholar]
- 45.Bonne-Andrea C, Santucci S, Clertant P, Tillier F. Bovine papillomavirus E1 protein binds specifically DNA polymerase alpha but not replication protein A. J Virol. 1995;69:2341–2350. doi: 10.1128/jvi.69.4.2341-2350.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conger KL, Liu JS, Kuo SR, Chow LT, Wang TS. Human papillomavirus DNA replication. Interactions between the viral E1 protein and two subunits of human DNA polymerase alpha/primase. J Biol Chem. 1999;274:2696–2705. doi: 10.1074/jbc.274.5.2696. [DOI] [PubMed] [Google Scholar]
- 47.Masterson PJ, Stanley MA, Lewis AP, Romanos MA. A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase alpha-primase p68 subunit. J Virol. 1998;72:7407–7419. doi: 10.1128/jvi.72.9.7407-7419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park P, Copeland W, Yang L, Wang T, Botchan MR, Mohr IJ. The cellular DNA polymerase alpha-primase is required for papillomavirus DNA replication and associates with the viral E1 helicase. Proc Natl Acad Sci U S A. 1994;91:8700–8704. doi: 10.1073/pnas.91.18.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins KL, Kelly TJ. Effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase alpha-primase. Mol Cell Biol. 1991;11:2108–2115. doi: 10.1128/mcb.11.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou B, Arnett DR, Yu X, et al. Structural basis for the interaction of a hexameric replicative helicase with the regulatory subunit of human DNA polymerase alpha-primase. J Biol Chem. 2012;287:26854–26866. doi: 10.1074/jbc.M112.363655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han Y, Loo YM, Militello KT, Melendy T. Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J Virol. 1999;73:4899–4907. doi: 10.1128/jvi.73.6.4899-4907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loo YM, Melendy T. Recruitment of replication protein A by the papillomavirus E1 protein and modulation by single-stranded DNA. J Virol. 2004;78:1605–1615. doi: 10.1128/JVI.78.4.1605-1615.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang X, Klimovich V, Arunkumar AI, et al. Structural mechanism of RPA loading on DNA during activation of a simple pre-replication complex. EMBO J. 2006;25:5516–5526. doi: 10.1038/sj.emboj.7601432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melendy T, Stillman B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J Biol Chem. 1993;268:3389–3395. [PubMed] [Google Scholar]
- 55.Schneider C, Weisshart K, Guarino LA, Dornreiter I, Fanning E. Species-specific functional interactions of DNA polymerase alpha-primase with simian virus 40 (SV40) T antigen require SV40 origin DNA. Mol Cell Biol. 1994;14:3176–3185. doi: 10.1128/mcb.14.5.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsurimoto T, Melendy T, Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990;346:534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- 57.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 58.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zlotkin T, Kaufmann G, Jiang Y, et al. DNA polymerase epsilon may be dispensable for SV40- but not cellular-DNA replication. EMBO J. 1996;15:2298–2305. [PMC free article] [PubMed] [Google Scholar]
- 60.Balakrishnan L, Bambara RA. Eukaryotic lagging strand DNA replication employs a multi-pathway mechanism that protects genome integrity. J Biol Chem. 2011;286:6865–6870. doi: 10.1074/jbc.R110.209502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Balakrishnan L, Gloor JW, Bambara RA. Reconstitution of eukaryotic lagging strand DNA replication. Methods. 2010;51:347–357. doi: 10.1016/j.ymeth.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clower RV, Hu Y, Melendy T. Papillomavirus E2 protein interacts with and stimulates human topoisomerase I. Virology. 2006;348:13–18. doi: 10.1016/j.virol.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 63.Li R, Botchan MR. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 64.Narahari J, Fisk JC, Melendy T, Roman A. Interactions of the cellular CCAAT displacement protein and human papillomavirus E2 protein with the viral origin of replication can regulate DNA replication. Virology. 2006;350:302–311. doi: 10.1016/j.virol.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 65.Gopalakrishnan V, Khan SA. E1 protein of human papillomavirus type 1a is sufficient for initiation of viral DNA replication. Proc Natl Acad Sci U S A. 1994;91:9597–9601. doi: 10.1073/pnas.91.20.9597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moody CA, Laimins LA. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 2009;5:e1000605. doi: 10.1371/journal.ppat.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sakakibara N, Mitra R, McBride AA. The papillomavirus E1 helicase activates a cellular DNA damage response in viral replication foci. J Virol. 2011;85:8981–8995. doi: 10.1128/JVI.00541-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kadaja M, Isok-Paas H, Laos T, Ustav E, Ustav M. Mechanism of genomic instability in cells infected with the high-risk human papillomaviruses. PLoS Pathog. 2009;5:e1000397. doi: 10.1371/journal.ppat.1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fradet-Turcotte A, Bergeron-Labrecque F, Moody CA, Lehoux M, Laimins LA, Archambault J. Nuclear accumulation of the papillomavirus E1 helicase blocks S-phase progression and triggers an ATM-dependent DNA damage response. J Virol. 2011;85:8996–9012. doi: 10.1128/JVI.00542-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faucher AM, White PW, Brochu C, Grand-Maitre C, Rancourt J, Fazal G. Discovery of small-molecule inhibitors of the ATPase activity of human papillomavirus E1 helicase. J Med Chem. 2004;47:18–21. doi: 10.1021/jm034206x. [DOI] [PubMed] [Google Scholar]
- 71.White PW, Faucher AM, Massariol MJ, et al. Biphenylsulfonacetic acid inhibitors of the human papillomavirus type 6 E1 helicase inhibit ATP hydrolysis by an allosteric mechanism involving tyrosine 486. Antimicrob Agents Chemother. 2005;49:4834–4842. doi: 10.1128/AAC.49.12.4834-4842.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rocque WJ, Porter DJ, Barnes JA, et al. Replication-associated activities of purified human papillomavirus type 11 E1 helicase. Protein Expr Purif. 2000;18:148–159. doi: 10.1006/prep.1999.1182. [DOI] [PubMed] [Google Scholar]
- 73.Titolo S, Pelletier A, Pulichino AM, et al. Identification of domains of the human papillomavirus type 11 E1 helicase involved in oligomerization and binding to the viral origin. J Virol. 2000;74:7349–7361. doi: 10.1128/jvi.74.16.7349-7361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.White PW, Pelletier A, Brault K, et al. Characterization of recombinant HPV6 and 11 E1 helicases: effect of ATP on the interaction of E1 with E2 and mapping of a minimal helicase domain. J Biol Chem. 2001;276:22426–22438. doi: 10.1074/jbc.M101932200. [DOI] [PubMed] [Google Scholar]
- 75.Edwards TG, Koeller KJ, Slomczynska U, et al. HPV episome levels are potently decreased by pyrrole-imidazole polyamides. Antiviral Res. 2011;91:177–186. doi: 10.1016/j.antiviral.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crute JJ, Grygon CA, Hargrave KD, et al. Herpes simplex virus helicase-primase inhibitors are active in animal models of human disease. Nat Med. 2002;8:386–391. doi: 10.1038/nm0402-386. [DOI] [PubMed] [Google Scholar]
- 77.Kleymann G, Fischer R, Betz UA, et al. New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nat Med. 2002;8:392–398. doi: 10.1038/nm0402-392. [DOI] [PubMed] [Google Scholar]
- 78.White PW, Titolo S, Brault K, et al. Inhibition of human papillomavirus DNA replication by small molecule antagonists of the E1-E2 protein interaction. J Biol Chem. 2003;278:26765–26772. doi: 10.1074/jbc.M303608200. [DOI] [PubMed] [Google Scholar]
- 79.Yoakim C, Ogilvie WW, Goudreau N, et al. Discovery of the first series of inhibitors of human papillomavirus type 11: inhibition of the assembly of the E1-E2-Origin DNA complex. Bioorg Med Chem Lett. 2003;13:2539–2541. doi: 10.1016/s0960-894x(03)00510-9. [DOI] [PubMed] [Google Scholar]
- 80.Fry DC. Small-molecule inhibitors of protein-protein interactions: how to mimic a protein partner. Curr Pharm Des. 2012;18:4679–84. doi: 10.2174/138161212802651634. [DOI] [PubMed] [Google Scholar]
- 81.Abbate EA, Berger JM, Botchan MR. The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev. 2004;18:1981–1996. doi: 10.1101/gad.1220104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abroi A, Kurg R, Ustav M. Transcriptional and replicational activation functions in the bovine papillomavirus type 1 E2 protein are encoded by different structural determinants. J Virol. 1996;70:6169–6179. doi: 10.1128/jvi.70.9.6169-6179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brokaw JL, Blanco M, McBride AA. Amino acids critical for the functions of the bovine papillomavirus type 1 E2 transactivator. J Virol. 1996;70:23–29. doi: 10.1128/jvi.70.1.23-29.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cooper CS, Upmeyer SN, Winokur PL. Identification of single amino acids in the human papillomavirus 11 E2 protein critical for the transactivation or replication functions. Virology. 1998;241:312–322. doi: 10.1006/viro.1997.8941. [DOI] [PubMed] [Google Scholar]
- 85.Ferguson MK, Botchan MR. Genetic analysis of the activation domain of bovine papillomavirus protein E2: its role in transcription and replication. J Virol. 1996;70:4193–4199. doi: 10.1128/jvi.70.7.4193-4199.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grossel MJ, Sverdrup F, Breiding DE, Androphy EJ. Transcriptional activation function is not required for stimulation of DNA replication by bovine papillomavirus type 1 E2. J Virol. 1996;70:7264–7269. doi: 10.1128/jvi.70.10.7264-7269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sakai H, Yasugi T, Benson JD, Dowhanick JJ, Howley PM. Targeted mutagenesis of the human papillomavirus type 16 E2 transactivation domain reveals separable transcriptional activation and DNA replication functions. J Virol. 1996;70:1602–1611. doi: 10.1128/jvi.70.3.1602-1611.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Y, Coulombe R, Cameron DR, et al. Crystal structure of the E2 transactivation domain of human papillomavirus type 11 bound to a protein interaction inhibitor. J Biol Chem. 2004;279:6976–6985. doi: 10.1074/jbc.M311376200. [DOI] [PubMed] [Google Scholar]
- 89.Biswas S, Field HJ. Herpes simplex virus helicase-primase inhibitors: recent findings from the study of drug resistance mutations. Antivir Chem Chemother. 2008;19:1–6. doi: 10.1177/095632020801900101. [DOI] [PubMed] [Google Scholar]
- 90.Biswas S, Swift M, Field HJ. High frequency of spontaneous helicase-primase inhibitor (BAY 57-1293) drug-resistant variants in certain laboratory isolates of HSV-1. Antivir Chem Chemother. 2007;18:13–23. doi: 10.1177/095632020701800102. [DOI] [PubMed] [Google Scholar]
- 91.Melendy T, Sedman J, Stenlund A. Cellular factors required for papillomavirus DNA replication. J Virol. 1995;69:7857–7867. doi: 10.1128/jvi.69.12.7857-7867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Müller F, Seo YS, Hurwitz J. Replication of bovine papillomavirus type 1 origin-containing DNA in crude extracts and with purified proteins. J Biol Chem. 1994;269:17086–17094. [PubMed] [Google Scholar]
- 93.Stracker TH, Cassell GD, Ward P, et al. The Rep protein of adeno-associated virus type 2 interacts with single-stranded DNA-binding proteins that enhance viral replication. J Virol. 2004;78:441–453. doi: 10.1128/JVI.78.1.441-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326:414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- 95.Wun-Kim K, Upson R, Young W, Melendy T, Stillman B, Simmons DT. The DNA-binding domain of simian virus 40 tumor antigen has multiple functions. J Virol. 1993;67:7608–7611. doi: 10.1128/jvi.67.12.7608-7611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lin BY, Makhov AM, Griffith JD, Broker TR, Chow LT. Chaperone proteins abrogate inhibition of the human papillomavirus (HPV) E1 replicative helicase by the HPV E2 protein. Mol Cell Biol. 2002;22:6592–6604. doi: 10.1128/MCB.22.18.6592-6604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu JS, Kuo SR, Makhov AM, et al. Human Hsp70 and Hsp40 chaperone proteins facilitate human papillomavirus-11 E1 protein binding to the origin and stimulate cell-free DNA replication. J Biol Chem. 1998;273:30704–30712. doi: 10.1074/jbc.273.46.30704. [DOI] [PubMed] [Google Scholar]
- 98.Swindle CS, Engler JA. Association of the human papillomavirus type 11 E1 protein with histone H1. J Virol. 1998;72:1994–2001. doi: 10.1128/jvi.72.3.1994-2001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee D, Sohn H, Kalpana GV, Choe J. Interaction of E1 and hSNF5 proteins stimulates replication of human papillomavirus DNA. Nature. 1999;399:487–491. doi: 10.1038/20966. [DOI] [PubMed] [Google Scholar]
- 100.Yasugi T, Vidal M, Sakai H, Howley PM, Benson JD. Two classes of human papillomavirus type 16 E1 mutants suggest pleiotropic conformational constraints affecting E1 multimerization, E2 interaction, and interaction with cellular proteins. J Virol. 1997;71:5942–5951. doi: 10.1128/jvi.71.8.5942-5951.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fradet-Turcotte A, Brault K, Titolo S, Howley PM, Archambault J. Characterization of papillomavirus E1 helicase mutants defective for interaction with the SUMO-conjugating enzyme Ubc9. Virology. 2009;395:190–201. doi: 10.1016/j.virol.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rangasamy D, Wilson VG. Bovine papillomavirus E1 protein is sumoylated by the host cell Ubc9 protein. J Biol Chem. 2000;275:30487–30495. doi: 10.1074/jbc.M003898200. [DOI] [PubMed] [Google Scholar]
- 103.Rangasamy D, Woytek K, Khan SA, Wilson VG. SUMO-1 modification of bovine papillomavirus E1 protein is required for intranuclear accumulation. J Biol Chem. 2000;275:37999–38004. doi: 10.1074/jbc.M007777200. [DOI] [PubMed] [Google Scholar]
- 104.Yasugi T, Howley PM. Identification of the structural and functional human homolog of the yeast ubiquitin conjugating enzyme UBC9. Nucleic Acids Res. 1996;24:2005–2010. doi: 10.1093/nar/24.11.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Côté-Martin A, Moody C, Fradet-Turcotte A, et al. Human papillomavirus E1 helicase interacts with the WD repeat protein p80 to promote maintenance of the viral genome in keratinocytes. J Virol. 2008;82:1271–1283. doi: 10.1128/JVI.01405-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lehoux M, Fradet-Turcotte A, Lussier-Price M, Omichinski JG, Archambault J. Inhibition of human papillomavirus DNA replication by an E1-derived p80/UAF1-binding peptide. J Virol. 2012;86:3486–3500. doi: 10.1128/JVI.07003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cueille N, Nougarede R, Mechali F, Philippe M, Bonne-Andrea C. Functional interaction between the bovine papillomavirus virus type 1 replicative helicase E1 and cyclin E-Cdk2. J Virol. 1998;72:7255–7262. doi: 10.1128/jvi.72.9.7255-7262.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Deng W, Lin BY, Jin G, et al. Cyclin/CDK regulates the nucleocytoplasmic localization of the human papillomavirus E1 DNA helicase. J Virol. 2004;78:13954–13965. doi: 10.1128/JVI.78.24.13954-13965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fradet-Turcotte A, Moody C, Laimins LA, Archambault J. Nuclear export of human papillomavirus type 31 E1 is regulated by Cdk2 phosphorylation and required for viral genome maintenance. J Virol. 2010;84:11747–11760. doi: 10.1128/JVI.01445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ma T, Zou N, Lin BY, Chow LT, Harper JW. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc Natl Acad Sci U S A. 1999;96:382–387. doi: 10.1073/pnas.96.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Saikia P, Fensterl V, Sen GC. The inhibitory action of P56 on select functions of E1 mediates interferon’s effect on human papillomavirus DNA replication. J Virol. 2010;84:13036–13039. doi: 10.1128/JVI.01194-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Terenzi F, Saikia P, Sen GC. Interferon-inducible protein, P56, inhibits HPV DNA replication by binding to the viral protein E1. EMBO J. 2008;27:3311–3321. doi: 10.1038/emboj.2008.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cohn MA, Kee Y, Haas W, Gygi SP, D’Andrea AD. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J Biol Chem. 2009;284:5343–5351. doi: 10.1074/jbc.M808430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cohn MA, Kowal P, Yang K, et al. UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell. 2007;28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 115.Jolly CE, Gray LJ, Parish JL, Lain S, Herrington CS. Leptomycin B induces apoptosis in cells containing the whole HPV 16 genome. Int J Oncol. 2009;35:649–656. doi: 10.3892/ijo_00000377. [DOI] [PubMed] [Google Scholar]