Abstract
Peroxisome proliferator-activated receptor delta (PPARD) is a key regulator of lipid metabolism, insulin sensitivity, cell proliferation and differentiation. In this study, we identified two Single Nucleotide Polymorphisms (SNPs, g.1015 A>G and g.1018 T>C) constituting four haplotypes (GT, GC, AC and AT) in the 5’ regulatory region of porcine PPARD gene. Functional analysis of the four haplotypes showed that the transcriptional activity of the PPARD promoter fragment carrying haplotype AC was significantly lower than that of the other haplotypes in 3T3-L1, C2C12 and PK-15 cells, and haplotype AC had the lowest binding capacities to the nuclear extracts. Transcription factor 7-like 2 (TCF7L2) enhanced the transcription activities of promoter fragments of PPARD gene carrying haplotypes GT, GC and AT in C2C12 and 3T3-L1 cells, and increased the protein expression of PPARD gene in C2C12 myoblasts. TCF7L2 differentially bound to the four haplotypes, and the binding capacity of TCF7L2 to haplotype AC was the lowest. There were significant associations between -655A/G and fat deposition traits in three pig populations including the Large White × Meishan F2 pigs, France and American Large White pigs. Pigs with genotype GG had significantly higher expression of PPARD at both mRNA and protein level than those with genotype AG. These results strongly suggested that the SNPs in 5’ regulatory region of PPARD genes had significant impact on pig fat deposition traits.
Introduction
Peroxisome proliferator-activated receptors (PPARs) are lipid-activated nuclear receptors belonging to the nuclear hormone receptor superfamily [1]. Ligand-activated PPARs form heterodimers with retinoic X receptors (RXRs) which bind to PPAR response elements (PPREs) and positively regulate transcription of target genes [2]. PPARs are involved in a number of biological processes, including lipid metabolism [3], insulin sensitivity, inflammation [4], cell proliferation and/or differentiation [5]. PPARD gene is widely expressed in the tissues including liver, hearts, adrenal, intestine and adipose in mice and rats [6, 7]. It enhances gene transcriptions that are involved in the fatty acid transport, oxidation, energy uncoupling, mitochondrial respiration and thermogenesis [1, 8]. PPARD inhibits the onset of oxidative stress-induced apoptosis in H9c2 cells [2], cell proliferation in keratinocytes [5], vascular smooth muscle cells [9], lung fibroblasts [10], and cardiac fibroblasts [11, 12]. In preadipocytes, PPARD gene begins to be expressed during the early periods of induced differentiation in vitro [13], playing important roles in the regulation of adipogenesis by fatty acid [14]. PPARD-deficient mice showed multiple developmental and homeostatic abnormalities, including placental defects causing frequent embryonic lethality, decreased adipose mass, altered skin inflammatory responses, and impaired wound healing [15–17]. In human, DNA variations within the PPARD genomic sequence are significantly associated with body mass index, high-density lipoprotein cholesterol, leptin, skeletal muscle glucose uptake, homeostasis model assessment of insulin resistance (HOMA-IR), adiposity measures or fasting serum lipids and height [18–21].
We are interested in studying pig fat deposition because it plays important roles in animal agriculture and can be used as biomedical model for human obesity. Pig carcass composition such as backfat thickness and lean meat percentage are controlled by polygenes with pleiotropic effects. Identification of these polygenes or linked markers is necessary for understanding the genetic basis of carcass traits and the application of marker assisted selection (MAS) in breeding programs [22]. A significant quantitative trait loci (QTL) for backfat thickness (BFT) has been consistently mapped to sus scrofa chromosomes 7 (SSC7) p1.1-q1.4 in several pig populations [23–31]. Using the F2 resource population derived from the intercross of Large White boars and Meishan dams, we have identified significant QTLs for carcass and meat quality traits on SSC7 [23, 32]. Haplotypes of the porcine PPARD gene are associated with backfat thickness [33], and PPARD gene also functions in fatty acid metabolism and fat metabolism [34]. Therefore, PPARD gene was considered to be a promising positional candidate gene for the fat deposition traits. Previous studies also showed that one missense mutation within the coding sequence of PPARD gene was significantly associated with ear size [35, 36]. However, whether and how the genetic variants within upstream regulatory region of PPARD gene affected fat deposition traits were still unknown in pigs. In this study, we identified two functional SNPs (GU565976.1: g.1015 A>G and g.1018 T>C) in the 5’ regulatory region of PPARD which altered the binding capacity transcription factor TCF7L2 to the promoter region, and found that the SNP g.1015A>G was significantly associated with fat deposition traits in three pig populations.
Materials Methods
Animals and trait measurement
All animal procedures were performed according to protocols approved by the Animal Care and Use Committee of Huazhong Agricultural University, Hubei province, P. R. China. Pigs from thirteen pig populations (12 Chinese Bamei pigs, 19 Jianli pigs, 16 Exihei pigs, 44 Meishan pigs, 34 Erhualian pigs, 19 Wannan pigs, 27 Huainan pigs, 31 English Large White pigs, 34 Landrace pigs, 19 Yangxin pigs, 409 American Large White pigs, 710 France Large White pigs, and 274 Large White × Meishan F2 pigs) were used to investigate the distribution of allele and genotype frequency of SNP g.1015A>G. The association analyses were conducted in three pig populations including 274 Large White × Meishan F2 pigs, 409 American Large White pigs and 710 France Large White pigs. All traits were measured and recorded according to the Principles and methods of swine testing [37]. The fat deposition and carcass traits were as following: backfat thickness at shoulder (BFT1, cm), backfat thickness at thorax-waist (BFT2, cm), backfat thickness at buttock (BFT3, cm), average backfat thickness at shoulder, thorax-waist and buttock (ABT, cm), leaf fat wight (LFW, kg), backfat thickness between 6th and 7th ribs (67RIBBF, cm), rib number (RN), carcass length from the first cervical vertebra to anterior border of pubic symphysis (CL1, cm), carcass length from the first thoracic vertebra to anterior border of pubic symphysis (CL2, cm), Internal fat rate (IFR). The live backfat thickness were measured by B-ultrasound machine (ESAOTE, Mylob Touch VET) between 85 kg and 115 kg body weight and then corrected to the backfat thickness at 100 kg body weight according to the national standard of swine performance testing of P. R. China (NY/T822-2004). Sequencing and haplotypes inference were conducted in 90 pigs from seven breeds, including 9 Chinese Bamei pigs, 6 Erhualian pigs, 4 Huainan pigs, 5 Duroc pigs, 20 American Large White pigs, 25 France Large White pigs, and 21 Large White × Meishan F2 pigs.
SNP detection and haplotype inference
In order to detect SNPs in the PPARD 5’ regulatory region, we sequenced the promoter region (GenBank accession no. GU565976.1) from five Meishan pigs and five Large White pigs, respectively. PCR products of 544 bp, 568 bp and 757 bp spanning nucleotides − 1415 bp to 464 bp (1880 bp length) were sequenced to detect the SNPs in the 5’ regulatory region. The mix of PCR contained 1.5 mM MgCl2. The primers for DNA sequencing, amplification and annealing temperature are listed in S1 Table. The purified PCR products of five Meishan and five Large White pigs were cloned into the pMD18-T vector (TaKaRa, Japan), and sequenced commercially (Sangon, China). Then these sequences were aligned using Clustalw (http://www.ebi.ac.uk/Tools/msa/clustalw2/) to detect the SNPs. Direct sequencing of 568 bp PCR products containing the SNPs (g.1015 A>G and g.1018 T>C) were further conducted to infer the haplotypes. Haplotypes were inferred using Haploview software (Version 4.2) [38].
Vectors construction of promoter fragments carrying the four haplotypes
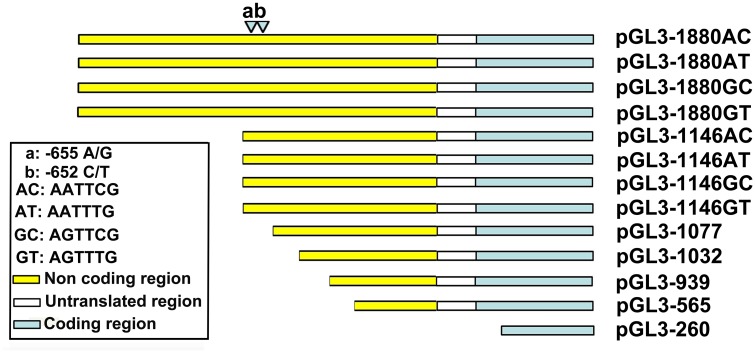
The luciferase reporter gene assays were performed using vector constructs containing the 5’ flanking region of PPARD gene: pGL3-1880GT (from − 1415 bp to + 464 bp), pGL3-1146GT (− 681 bp to + 464 bp), pGL3-1077 (− 612 bp to + 464 bp), pGL3-1032 (− 567 bp to + 464 bp), pGL3-939 (− 474 bp to + 464 bp), pGL3-565 (− 100 bp to + 464 bp) and pGL3-260 (+ 205 bp to + 464 bp) (relative to the transcription start site) (Fig 1). The 5’ terminal primer oligonucleotides contained KpnIand SacIenzymes sites. The amplified fragments were inserted into the pGL3-Basic vector (Promega, USA). Five deletion fragments of the PPARD promoter were generated by PCR using the pGL3-1880GT vector as a template. All primers are listed in S2 Table.
Fig 1. Schematic representation of the deletions of PPARD promoter linked with the luciferase gene in vectors.
The nucleotides were numbered relative to the transcription start site that was assigned as + 1.
Using the pGL3-1880GT and pGL3-1146GT vectors as templates, two sets of promoter luciferase report gene vectors were constructed by site directed mutagenesis respectively. The vectors included haplotype AC (pGL3-1880AC and pGL3-1146AC), AT (pGL3-1880AT and pGL3-1146AT), GT (pGL3-1880GT and pGL3-1146GT) and GC (pGL3-1880GC and pGL3-1146GC).
The eukaryotic overexpression vector of TCF7L2 (pCDNA3.1-TCF7L2) was constructed. The primer (F: 5’ GGGGTACCGCCACCATGCCGCAGCTGAACGG 3’, R: 5’ GCTCTAGACTATTCTAAAGACTTGGTGACCAGG 3’) was designed according to the GenBank database sequence (NM001142922.1) and used to obtain the CDS (coding sequence) of the swine TCF7L2 gene. The CDS was digested with restriction enzyme KpnI(forward primer) and XbaI(reverse primer), and then ligated into pcDNA3.1 using T4 DNA Ligase (Takara, Japan) to generate pcDNA3.1-TCF7L2 vector.
Dual luciferase assay
C2C12, 3T3-L1 and PK cells were grown in Dulbecco's Modified Eagle's Medium (DMEM) (Hyclone, USA) supplemented with 10% (v/v) fetal bovine serum (Gibco, Australia), maintained in a 5% CO2 incubator at 37°C. Twenty-four hours before transfection, cells were plated onto 24-well plates or 6-well plates. When cells reached 80% confluence, they were transfected with promoter constructs (1 μg) or eukaryotic expression vectors (4 μg) using lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions.
After transfection for 24 h, cells were lysed in 100 μl of lysis buffer, and firefly and renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). The luciferase activity was measured using PerkinElmer 2030 Multilabel Reader (PerkinElmer, USA). To normalize the transfection efficiency, the pRL-TK plasmid vector (Promega, USA) was co-transfected with the reporter construct as described above. The experiments were performed in three replicates for each construct and the data were the average of three replicates.
Quantitative real-time PCR
For tissue expression pattern analysis, nine tissues including heart, liver, spleen, lung, kidney, stomach, intestines, longissimus dorsi (LD), and backfat were sampled from three France Large White pigs. For differential gene expression analysis between different genotypes, muscle and adipose tissues were sampled from three France Large White pigs with genotypes AG and GG, respectively. RNA samples from different tissues of the France Large White pigs were isolated using the TRIzol reagent (Invitrogen, USA). Reverse transcription of RNA (1 μg) was performed using random primers (Invitrogen, USA) and Moloney Murine Leukemia Virus (M-MLV) Reverse Transcriptase (Invitrogen, USA) according to the manufacturer’s instructions. The primers used to amplify the cDNA corresponding to PPARD, TCF7L2, and β-actin genes are listed in S3 Table. Quantitative RT-PCR was performed on a LightCycler 480 II Real-Time (Roche, Switzerland) using the FastStart DNA Master SYBR Green I reagent (Roche, Switzerland). The gene expression results were normalized with regard to the expression of the β-actin. The Ct (2-ΔΔCt) method was used to analyze the relative gene expression [39].
Western blotting
Tissues and cells were lysed in RIPA buffer according to manufacturer’s instruction (Beyotime, China). Protein lysates were heated at 95°C 5min in 5 × Sodium Dodecyl Sulfate (SDS) sample buffer separated by 12% SDS-PAGE (20 μg each lane), then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, USA) using Mini Trans-Blot Cell (Bio-Rad, USA). After being blocked with 5% non-fat milk for 1.5 h, the membranes were incubated with primary antibodies overnight at 4°C. After washed three times, the membranes were hybridized with secondary antibody for 1 h at 37°C, and washed three times. The targeted proteins were detected using the ImageQuant LAS 4000 mini (GE, USA) according to the manufacturer instructions. Primary antibodies were specific for PPARD (Abcam, USA, ab23673; 1:1000 dilution), TCF7L2 (Abcam, USA, ab32873; 1:1000 dilution), and β-actin (Boster, China, BM0627; 1:1000 dilution). Secondary antibodies were goat anti-mouse IgG-HRP (Santa Cruz, USA, sc-2005; 1:3000 dilution) and goat anti-rabbit IgG-HRP (Santa Cruz, USA, sc-2004; 1:3000 dilution).
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts from C2C12 myoblasts, 3T3-L1 cells and pig longissimus muscle were prepared by Nuclear Protein Extraction Kit (BestBio, China). The protein concentration was determined by Bicinchoninic Acid assay (Beyotime, China). The double-stranded DNA probes of four haplotypes were prepared by annealing the desired sense and anti-sense oligonucleotides which were 5’ end-labeled with biotin. The DNA binding activity of protein was detected by chemiluminescent EMSA Kit (Beyotime, China). The protein-DNA complexes were analyzed by electrophoresis in 6.0% polyacrylamide gels at 100 V for 1 h in 0.5 TBE running buffer (44.58 mM Tris Base, 44.58 mM Boric Acid, 1.25 mM Na2EDTAH2O), and then were transferred to a nylon membrane. The dried nylon was visualized using the ECL (Bio-Rad, USA). The bands were detected using the the ImageQuant LAS 4000 mini (GE, USA) according to the manufacturer’s instruction.
DNA pull-down
The probes for DNA pull-down assay were the same as EMSA assay. The probes were conjugated with M-280 Streptavidin Dynabeads (Invitrogen, USA) in binding buffer (10 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM MgCl2, 1 mM EDTA, pH 8.0, 1 mM Na3VO4, 5 mM DTT, 5% glycerol, 0.3% NP-40) for 40 min at room temperature. Non-denaturing total proteins were extracted from 3T3-L1cells using Western and IP Cell lysis Buffer (Sangon Biotech, China). The proteins (400 μg) were incubated with unconjugated Dynabeads for 2 h at 4°C using Dynal MPC-S magnetic particle concentrator (Dynal Biotech, Norway), and the non-specific binding proteins were removed. The supernatant (supernatantI) was collected for binding reaction. The probes were incubated with the Dynabeads for 5 h at 4°C, and then the Dynabeads-probes complexes were washed three times with lysis buffer and incubated with supernatant I for 2 h at 4°C in the presence of 25 μg/ml poly (dI:dC) which could prevent the nonspecific binding of protein with DNA. The precipitates (Dynabeads-probes-protein) were washed with lysis buffer, and then were eluted in SDS sample buffer. The supernatantII was collected for β-actin detection. The precipitates and supernatant were assayed by western blotting.
SNP genotyping and statistical analysis
The SNP (g.1015A>G) was genotyped by PCR-RFLP using EcoRI restriction enzyme. 8.5 μl PCR products were digested with 5 U EcoRI restriction enzyme (Fermentas, Canada) for 6 h at 37°C, and then were separated by electrophoresis on a 1.5% agarose gel with ethidium bromide in 1× TAE buffer.
The association of SNP genotypes with fat deposition and carcass traits in Large White × Meishan F2 pigs, France, and American Large White pigs were conducted with the general linear model (GLM) procedure of SAS version 8.0 (SAS Institute, Cary, NC, 2000). Both additive and dominance effects were estimated using the REG procedure of SAS version 8.0. The data are presented as mean ± S.D., and statistical significance level was set at p < 0.05.
The model of association analysis in 274 Large White × Meishan F2 pigs was as following:
Where, T ijkl is the observed values of a given trait; μ is the overall mean; S i is effect of sex (i = 1 for male or 2 for female); Y j is the effect of year (j = 1 for year 2000 or 2 for year 2003); G k is the effect of genotype (k = AA, AG and GG); F l is the effect of family (l = 37); b ijkl is the regression coefficient of the slaughter weight for carcass traits, X ijkl is the slaughter weight; e ijkl is the random residual.
The model of association analysis in 710 France Large White pigs and 409 American Large White pigs was as following:
Where, T ijkl is the corrected backfat thickness at 100 kg live weight; G k is the effect of genotype (k = AA, AG and GG); S i is the effect of sex (i = 1 for male or 2 for female); F j is effect of family (j = 17 for France Large White and j = 20 for American Large White); B l is the effect of batch (l = 20 for France Large White and l = 8 for American Large White); e ijkl is the random residual.
Results
Identification of two SNPs in the upstream of the 5’ regulatory region of PPARD gene
Two SNPs, g.1015A>G and g.1018 T>C (GenBank accession no. GU565976.1) were detected at the upstream − 655 bp (A/G) and − 652 bp (T/C) (relative to the transcription start site + 1) of the 5’regulatory region of PPARD gene (Fig 1). The corresponding variation ID of two SNPs in the SNP database (http://www.ncbi.nlm.nih.gov/SNP) was rs80912557 and rs80964726, respectively. The haplotypes of two SNPs were inferred in 90 pigs randomly selected from seven breeds including Bamei, Erhualian, Huainan, Duroc, American Larger White, France Larger White, and Large White × Meishan F2 pigs. In these pig populations, the two SNPs were in strong linkage disequilibrium (D = 0.925, R2 = 0.633) and the frequency of haplotypes GT, GC, AC and AT was 52.9%, 8.9%, 36.7% and 1.6%, respectively. In American Large White pigs, the frequency of haplotypes GT, GC, AC and AT was 36.7%, 6.7%, 57.7%, and 0%, respectively. In France Large White pigs, the frequency of haplotypes GT, GC, AC and AT was 86.7%, 10.0%, 3.3%, and 0%, respectively. In Large White × Meishan F2 pigs, the frequency of haplotypes GT, GC, AC and AT was 32.1%, 25.9%, 42.9%, and 0%, respectively. The results suggested that the allele A at rs80912557 of the PPARD was mostly linked with allele C at rs80964726, and allele G at rs80912557 was linked with allele T or C at rs80964726 in the pig population studied.
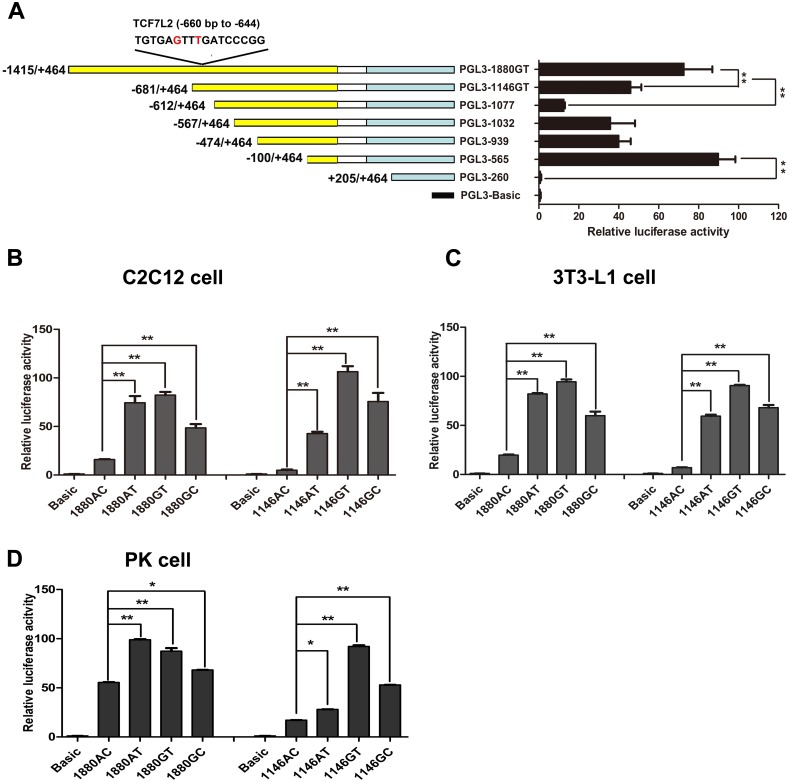
The SNPs in the promoter of PPARD gene significantly affected its transcriptional activity
The bioinformatics prediction of transcription factor binding sites showed that five transcription factors could bind to the segment containing the two SNPs, and the prediction scores of the binding capacity of transcription factors with different haplotype promoter fragments are listed in S4 Table. According to the bioinformatics prediction of transcription factor binding sites by TESS and Promoter 2.0 software, we constructed a series of deletion fragments by fusing different fragments of the 5’ flanking region to luciferase reporter vector pGL3-basic (Fig 1). All constructs were transfected into C2C12 myoblasts and their promoter activities were examined. The luciferase activity of the promoter fragments PGL3-260 (+ 205 bp to + 464 bp) was the lowest in all the luciferase activities (Fig 2A), suggesting that this region between nucleotides − 100 bp to + 205 bp was the core promoter region. The transcription activity of the promoter fragments PGL3-1077 (− 612 bp to + 464 bp) was lower than that of the other two promoter fragments PGL3-1880GT (− 1415 bp to + 464 bp), and PGL3-1146GT (− 681 bp to + 464 bp). The results showed that the promoter fragments from position − 1415 bp to − 612 bp contained the potential positive regulatory elements. To determine the effects of the SNPs on the promoter transcriptional activities, we designed and studied two sets of promoter luciferase report gene vectors carrying the four haplotypes constructed by site-directed mutagenesis using pGL3-1146GT and pGL3-1880GT as templates. The promoter vectors were transfected into C2C12, 3T3-L1, and PK cells, respectively. In C2C12 myoblasts, the transcriptional activity driven by pGL3-1146AC decreased 8.5-fold, 12.9-fold, and 9.7-fold compared with that driven by pGL3-1146AT, pGL3-1146GT and pGL3-1146GC, respectively (Fig 2B). The same trends were found in 3T3-L1 cells and PK cells. The luciferase expression level driven by pGL3-1880AC was also the lowest among the four haplotype constructs (Fig 2C and 2D). The results indicated that the SNPs in the promoter of PPARD gene significantly affected its transcription activities. The transcription activities of PPARD gene carrying haplotype AC were the lowest and these of haplotype GT the highest.
Fig 2. Transcriptional activities analysis of deletion constructs of PPARD promoter carrying the four haplotypes.
(A) Transcriptional activities of a series of deletion fragments determined by luciferase assay in C2C12 myoblasts. Left panel, schematic representation of the deleted fragments linked with the luciferase gene in the pGL3 vector. The nucleotides were numbered relative to the transcription start site that was assigned as + 1. Right panel, the relative activities of a series of deletion fragments of pGL3-1880GT vector determined by luciferase assay. (B)(C)(D) The relative activities of a series of deletion fragments of PPARD promoter carrying the different haplotypes in C2C12, 3T3-L1, and PK cells. Error bar represents mean ± S.D. (three independent replicates per group). Asterisk (*) and (**) represent the significance level at P < 0.05 and P < 0.01, respectively (The same below).
Potential transcription factors binding to the promoter fragments carrying the four haplotypes
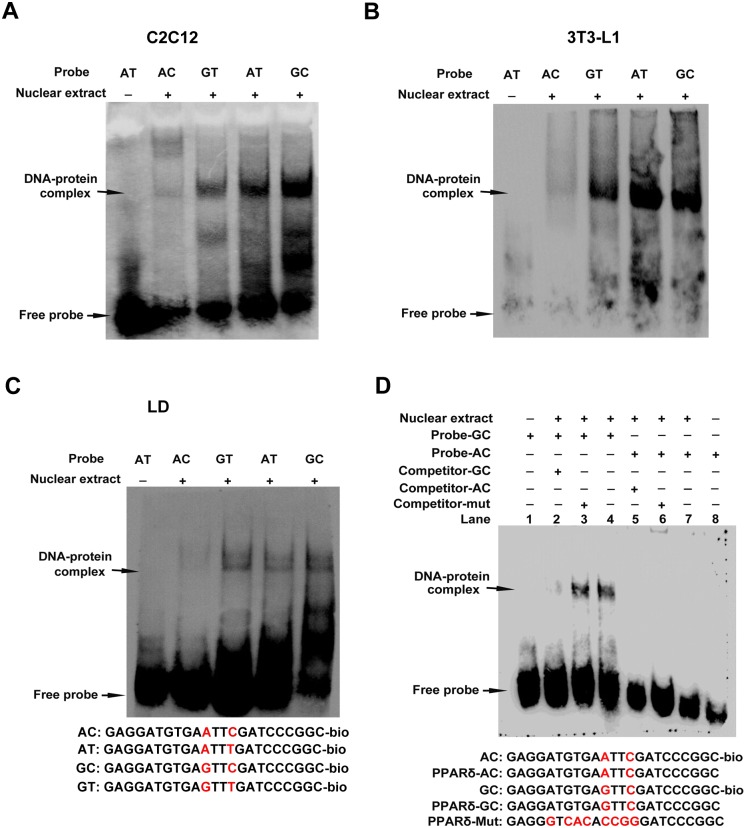
We used genomatrix (http://www.genomatrix.de/) to investigate the potential transcription factors in the promoter region harboring the SNPs identified. The results showed that there were potential ETS1, MEL1, Foxh1, POU2F1, and TCF7L2 binding sites in the promoter region, and the binding capacities of Foxh1, POU2F1 and TCF7L2 to the promoter changed when the SNPs were introduced to the DNA sequences (S4 Table). EMSA was performed to detect the binding capacities of different haplotypes to the nuclear extracts in C2C12 myoblasts, 3T3-L1 cells and pig LD muscle. The results of EMSA showed that the binding capacity of haplotype AC to the nuclear extracts was the weakest in C2C12 myoblasts, 3T3-L1 cells and pig LD muscle (Fig 3A, 3B and 3C), confirming that there are potential transcription factors differentially binding to the promoter fragments carrying the different haplotypes. EMSA was further conducted in C2C12 myoblasts with the probes of haplotypes AC and GC which showed the largest difference in the binding capacities (Fig 3D). The incubation of nuclear extracts of C2C12 myoblasts with probe GC formed a DNA-protein complex. The complex became weaker with 1 × cold probes in the mixture. The complex did not change in the mutation cold probe reaction. By contrast, the incubation of nuclear extracts of C2C12 myoblasts with probe AC did not form the DNA-protein complex. These results demonstrated that transcription factors can bind to the DNA sequence with haplotype AT, GT and GC, but not to haplotype AC.
Fig 3. Nuclear extracts differentially bind to the promoter fragments carrying the four haplotypes.
(A) (B) (C) EMSA results showing the binding capacities of four haplotypes to the nuclear extracts in C2C12 myoblasts, 3T3-L1 cells, and porcine LD muscle. (D) The EMSA results showing the binding capacities of nuclear extracts to the promoter fragments with haplotypes AC and GC, Lane 1 and Lane 8 were negative control; Lane 2–4 and 5–7 in turn were sample reactions, mutation competitive reactions, and cold competitive reactions for AC and GC reaction groups, respectively. The probes were incubated with nuclear extracts in the absence or presence of 1-fold excess of various competitor probes (mutant or non-labeled probe). The specific DNA-protein complex bands were indicated by arrows. The sequences of various probes were shown under the panel.
The PPARD gene SNPs showed different interactions with transcription factor TCF7L2
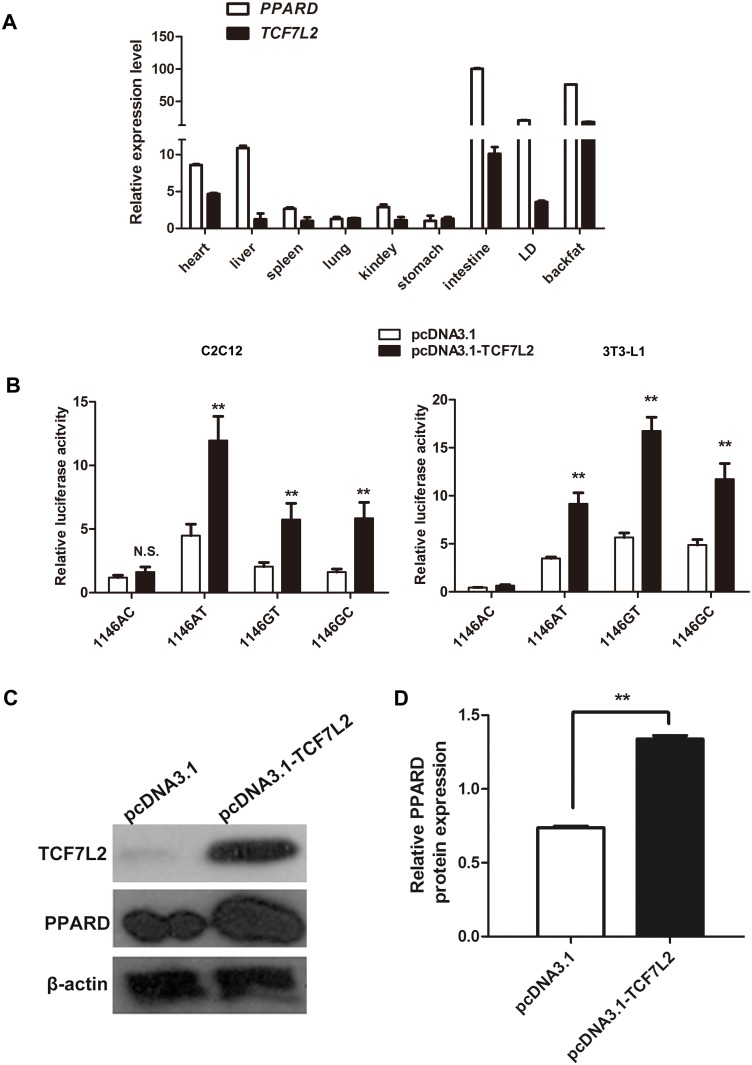
The binding motif of transcription factor TCF7L2 contains AC/GA/TTCAAAG [40], which is similar to the reverse sequence of the region harboring the haplotypes. In the present study, a classic motif of TCF7L2 was not identified in PPARD promoter, but a variant TCF7L2 binding sequences ccgggATCAAACtcaca was located at − 660/− 644 bp. TCF7L2 could bind to the promoter of PPARD and increase its expression in human colorectal cancer (CRC) cells [41]. Therefore, we hypothesized that SNPs alters transcriptional activities of the promoter through affecting the binding capacities of TCF7L2 to its binding sites. We initially studied the tissue expression patterns of TCF7L2 and PPARD genes in ten tissues including heart, liver, spleen, lung, kindey, brain, stomach, intestine, LD, and backfat of the four-month-old Large White pigs. The tissue expression profiles of two genes were very similar. They were all broadly expressed in the ten tissues, and highly expressed in liver and adipose which are active in fat synthesis (Fig 4A). To investigate the effects of TCF7L2 overexpression on the promoter transcriptional activities of PPARD gene, we constructed the eukaryotic overexpression vector of the porcine TCF7L2 (pCDNA3.1-TCF7L2) and co-transfected with the promoter fragments carrying the four haplotypes into C2C12 myoblasts and 3T3-L1 cells. TCF7L2 overexpression significantly improved the transcriptional activities of the promoter fragments with haplotypes AT, GT, and GC, whereas there were not significant effects with haplotype AC (Fig 4B). The protein expression levels of PPARD were also up-regulated by TCF7L2 overexpression in C2C12 myoblasts (Fig 4C and 4D). The binding capacities of TCF7L2 to the promoter fragments carrying four haplotypes were further analyzed by DNA pulldown in 3T3-L1 cells. The binding capacities of the fragments carrying haplotypes AT, GT and GC to TCF7L2 were significantly stronger than those of haplotype AC, and the binding capacity of haplotypes GT was the strongest. These results were in agreement with the results of the luciferase assay (Fig 5).
Fig 4. The SNPs in the 5’ regulatory region of PPARD gene affected transcriptional activation of TCF7L2.
(A) The relative mRNA expression profiles of the porcine PPARD and TCF7L2 gene in nine different tissues. (B) Transcriptional activities of four haplotypes determined by luciferase assay before and after TCF7L2 overexpression in C2C12 myoblasts and 3T3-L1 cells. (C) The protein expression levels of TCF7L2 and PPARD before and after TCF7L2 overexpression in C2C12 myoblasts. C2C12 myoblasts were transfected with pcDNA3.1-TCF7L2 vector and empty vector, and then the total protein extracted after 48 h transfection. The protein levels of TCF7L2 and PPARD genes were analyzed by western blotting. (D) Relative PPARD protein expression levels represented by ratio of detected protein to β-actin protein expression level after TCF7L2 overexpression.
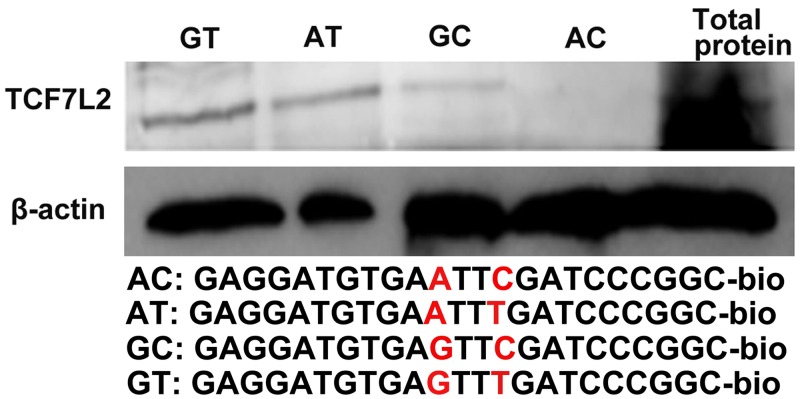
Fig 5. DNA pull-down results showing the interaction of TCF7L2 with the four haplotypes in 3T3-L1 cell in vivo.
3T3-L1 cells were seeded in 10-cm dishes for 48 h, harvested and pull-downed by antibodies. Lane GT, AT, GC, and AC: immunoprecipitation results with anti-TCF7L2 monoclonal antibody. Lane total protein: the analysis of total cell lysates before immunoprecipitation to verify expression of TCF7L2. The sequences of four probes were shown under the panel.
The SNP g.1015A>G was significantly associated with fat deposition traits in three pig populations
We used EcoRIPCR-RFLP to genotype the polymorphism g.1015A>G. Digestion of the PCR fragments with EcoRIrestriction enzyme generated one fragment (genotype GG, 568bp), two fragments (genotype AA, 314 bp + 254 bp) or three fragments (genotype AG, 568 + 314 + 254 bp) (S1 Fig). Allele frequency distributions of EcoRI-RFLP polymorphism in thirteen pig populations are shown in Table 1. This polymorphic SNP was segregated in the tested pig populations, most of which had high frequencies of allele G at this site. The association analysis of the genotypes with the fat deposition and carcass traits were conducted in three pig populations including the Large White × Meishan F2 pigs, France, and American Large White pigs. There were significant differences in the fat deposition and carcass traits among different genotypes in Large White × Meishan F2 populations. Pigs carrying genotype GG had higher BFT, RIBBF, LFP and IFR, but lower CL than the AA or AG animals (Table 2). Furthermore, significant additive effects were also observed for these traits (Table 2). The significant associations of g.1015 A>G with the corrected live backfat thickness were also found in France and American Large White pigs (Table 3). Taken together, we concluded that this functional SNP was significantly associated with fat deposition traits.
Table 1. Allele distribution of g.1015 A>G polymorphism in thirteen pig populations.
| Allele frequency | ||||
|---|---|---|---|---|
| Population | Location | number | G | A |
| Bamei | Gansu province | 12 | 0.71 | 0.29 |
| Erhualian | Jiangsu province | 34 | 0.68 | 0.32 |
| Exihei | Hubei province | 16 | 0.69 | 0.31 |
| Huainan | Henan province | 27 | 0.78 | 0.22 |
| Jianli | Hubei province | 19 | 0.79 | 0.21 |
| Meishan | Shanghai | 44 | 0.52 | 0.48 |
| Wannan | Anhui province | 19 | 0.63 | 0.37 |
| Yangxin | Hubei province | 19 | 0.47 | 0.53 |
| English Large White | Hubei province | 31 | 0.82 | 0.18 |
| American Large White | Zhejiang province | 409 | 0.68 | 0.32 |
| France Large White | Zhejiang province | 710 | 0.89 | 0.11 |
| Landrace | Hubei province | 34 | 0.80 | 0.20 |
| Large White × Meishan F2 pigs | Hubei province | 274 | 0.56 | 0.44 |
Table 2. Association results of g.1015 A>G polymorphism with carcass traits in Large White × Meishan F2 pig population.
| Genotype | Additive effect | Dominant effect | |||
|---|---|---|---|---|---|
| Traits | AA | AG | GG | a | d |
| BFT1 (cm) | 3.44±0.11a | 3.48±0.05a | 3.76±0.08b | 0.16±0.07* | 0.04±0.03 |
| 67RIBBF (cm) | 2.70±0.09a | 2.72±0.04a | 2.94±0.07b | 0.12±0.05* | 0.04±0.03 |
| ABT (cm) | 2.35±0.08a | 2.43±0.04a | 2.62±0.06b | 0.13±0.05* | 0.02±0.02 |
| BFT2 (cm) | 1.91±0.08c | 2.04±0.04ac | 2.17±0.06ab | 0.13±0.05* | -0.002±0.02 |
| BFT3 (cm) | 1.71±0.1 | 1.83±0.05 | 1.94±0.07 | 0.11±0.06 | 0.001±0.03 |
| CL1 (cm) | 92.40±0.67a | 91.10±0.34a | 89.80±0.50b | -1.23±0.40 | -0.1±0.20 |
| CL2 (cm) | 78.20±0.5A | 77.40±0.28A | 75.60±0.40B | -1.27±0.34* | -0.26±0.18 |
| RN | 14.70±0.10 | 14.68±0.10 | 14.69±0.20 | -0.007±0.06 | 0.01±0.03 |
| LFP (kg) | 0.66±0.03c | 0.71±0.03ac | 0.77±0.02ab | 0.05±0.02* | 0.007±0.01 |
| IFR (%) | 1.10±0.05ac | 1.06±0.02c | 1.18±0.04ab | 0.04±0.03 | 0.03±0.02 |
BFT1: backfat thickness at shoulder; ABT: average backfat thickness; BFT2: backfat thickness at thorax-waist; BFT3: backfat thickness at buttock; 67RIBBF: backfat thickness between 6th and 7th ribs; RN: rib numbers; CL1: Carcass body length 1; CL2: Carcass body length 2; LFP: Leaf fat percentage; IFR: Internal fat rate. Data were shown as means ± S.D. (standard deviation). Different superscript small letters in one row indicate significance level at P<0.05; Different superscript large letters in one row indicate significance level at P<0.01; Asterisk (*) represents significance level at P<0.05.
Table 3. Association results of g.1015 A>G polymorphism with the corrected backfat thickness at 100 kg body weight in France and American Large White pigs.
| Genotype | Additive effect | Dominant effect | |||
|---|---|---|---|---|---|
| Population | AA | AG | GG | a | d |
| American Large White | 9.13±0.21Aa | 9.59±0.09b | 9.85±0.09Bc | 0.36±0.11 | -0.06±0.07 |
| France Large White | 8.53±0.10A | 8.87±0.05B | |||
The SNPs could significantly affect the expression of PPARD gene in vivo
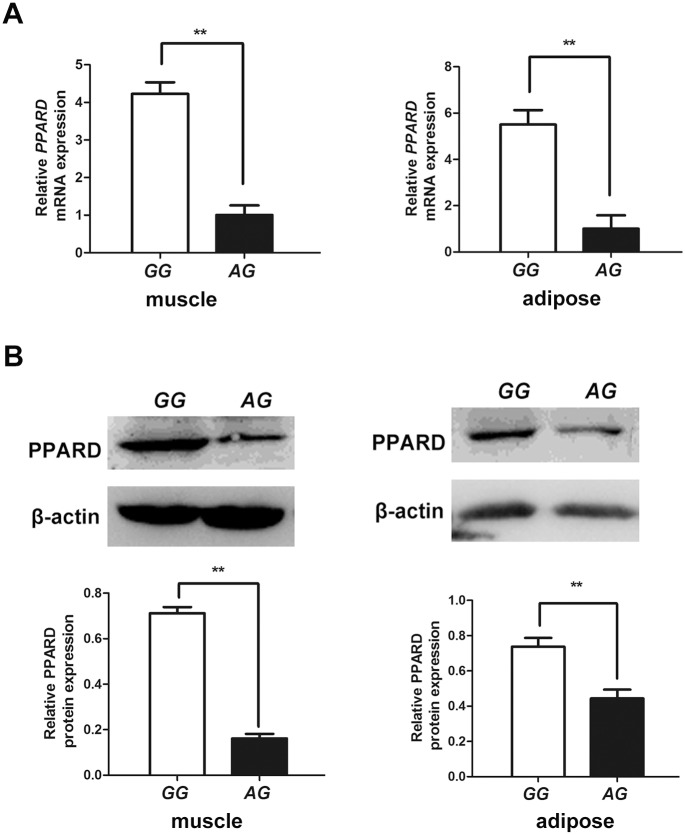
To estimate whether the SNP g.1015A>G could affect the gene expression in vivo, we selected France Large White pigs with genotypes AG and GG to compare the expression of PPARD gene in adipose and muscle tissues. The expression of PPARD gene between two genotypes was significantly different at both mRNA and protein level. Pigs with genotype GG had significant higher expression level than that of pigs with genotype AG (Fig 6).
Fig 6. The SNPs significantly affect the expression of PPARD gene in vivo.
(A) The relative mRNA expression levels of PPARD gene in muscle and adipose between genotype AG and GG. (B) The protein expression levels of PPARD gene in muscle and adipose between genotype AG and GG.
Discussion
Functional mutations in the regulatory region have effects on the gene expression and phenotypic variation. PPARD is a promising candidate gene for pig carcass traits [27, 33, 42]. Currently, there is not report about any functional SNPs in the 5’ regulatory region of the porcine PPARD gene. In this study, we identified the functional SNPs in the 5’ regulatory region which directly affected the promoter transcriptional activities. The transcriptional activity of the PPARD promoter fragment carrying haplotype AC was the lowest, and haplotype AC had the lowest binding capacities with TCF7L2. TCF7L2 is a member of the T cell factor (TCF) family of transcription factors, and involved in the control of cell growth and signaling of wingless-type MMTV integration site family (Wnt) receptors [43]. TCF7L2 interacts with β-catenin in the nucleus and regulates the expression of Wnt target genes by acting as transcriptional activator or repressor [44–47]. TCF4-binding sites are located at long distances from transcription start sites, and defined as evolutionarily conserved A-C/G-A/T-T-C-A-A-A-G motifs [40]. In our study, three TCF7L2 binding sites were predicted in the promoter region between nucleotides −1636 bp and + 464 bp, and one of the three sites was located within the promoter sequence harboring the two SNPs. TCF7L2 can bind to the promoter of PPARD gene and alter gene expression in human CRC cells [41]. We also confirmed that the expression of PPARD gene was up-regulated by TCF7L2 overexpression in C2C12 cells. Besides, the porcine TCF7L2 gene improved the transcriptional activities of the PPARD promoter fragments carrying the haplotypes AT, GT, and GC. However no significant effects on the haplotype AC were found, because there was no interaction between promoter fragments carrying haplotype AC and TCF7L2 protein.
PPARD gene has been implicated in lipid metabolism via several signaling pathways including Wnt/FZD and Wnt/β-catenin pathways. On the one hand, PPARD promote the accumulation of cholesterol and serum high density lipoprotein (HDL), and decrease the level of triglyceride in human macrophages by lipogenesis pathways such as Wnt/FZD and Wnt/β-catenin pathway [48, 49]. On the other hand, it reduces body weight, lipid droplet number and size, and up-regulates gene expression related to fatty acid oxidation [34]. In human population, three SNPs in the PPARD affect (lifestyle intervention) LI-induced changes in overall adiposity hepatic fat storage [50]. However, the molecular mechanism by which genetic variants within upstream regulatory region of PPARD gene affect fat deposition traits still remain unclear. In this study, the allele A was mostly linked with allele C at two loci in all the pig breeds studied, and no haplotype AT was found in American Large White pigs, France Large White pigs and Large White × Meishan F2 pigs studied. We also found that the functional variant (g. 1015A>G) had significant association with fat deposition traits in three pig populations and allele A was associated with lower fat deposition traits as well as lower gene expression of PPARD in vitro and in vivo. Previous studies showed that PPARD and CDKN1A gene in preadipocytes had lower expression levels in heterozygote MSQTL7/LWQTL7 (the heterozygous individuals carrying Meishan and Large White SSC7 QTL alleles) pigs than in homozygous LWQTL7/LWQTL7 (the homozygous individuals carrying Large White SSC7 QTL alleles) pigs at 28 and 150 days ages [51], which was in agreement with our results. Taken together, it can be inferred that haplotype AC inhibited the promoter transcriptional activity through decreasing the binding capacities of TCF7L2 to its binding sites, thereby repressed the PPARD expression and resulted in decreased fat deposition traits. Therefore, the expression change of PPARD gene may be an important cause of the major QTL effects for fat deposition.
In pig breeding schemes, the lean percentage in carcass can be indirectly selected by live backfat thickness because there is significant negative correlation between these two traits. In this study, there were significant differences in fat deposition traits between different genotypes. Pigs with genotypes AA or AG had significant lower fat deposition trait values compared with those of genotype GG in the pig populations studied. Considering the effects of allele A in this study, enhancing the allele frequency of allele A will lead to the reduction of backfat thickness in the pig populations of which the allele was not fixed at this locus. However, selection of allele A also has adverse effects on intramuscular fat and meat quality, as intramuscular fat and backfat deposition are negatively correlated.
In conclusion, our results provided a new evidence that the newly identified functional SNPs in the 5’ regulatory region of PPARD genes have significant effects on the fat deposition and carcass traits. Further work will be necessary to confirm the effects of this functional SNP in more pig populations and investigate the molecular mechanism regarding the roles of SNPs in phenotype variation.
Supporting Information
Lane Marker: DNA molecular marker DL2000 (TaKaRa, Dalian, China); Lane 1, 2, and 6: genotype GG, 568 bp; Lane 3, 4: genotype AG, 568 bp + 314 bp + 254 bp; Lane 5: genotype AA, 314 bp + 254 bp.
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
Acknowledgments
We thank Dr. Jinzeng Yang from University of Hawaii and Prof. Chunyan Mu for their kind assistance in manuscript writing.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The study was financially supported by the Key National High Technology Development Project of China (Grant No. 2011AA100301), the National Key Technology Support Program of China (Grant No. 2014BAD20B01), the Agricultural Innovation Fund of Hubei Province (Grant No. 2007-620), the Fundamental Research Funds for the Central Universities (Grant No. 2014PY038) and the Key Project of Science and Technology of Zhejiang Province (Grant No. 2014C02010). The last funder is Tianpeng Group. The funder provided support in the form of salaries for authors [ZZ, YM, HM, LW, GW], but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Evans RM, Barish GD, Wang YX. PPARs and the complex journey to obesity. Nat Med. 2004; 10: 355–361. [DOI] [PubMed] [Google Scholar]
- 2. Pesant M, Sueur S, Dutartre P, Tallandier M, Grimaldi PA, Rochette L, et al. Peroxisome proliferator-activated receptor delta (PPARdelta) activation protects H9c2 cardiomyoblasts from oxidative stress-induced apoptosis. Cardiovasc Res. 2006; 69: 440–9. [DOI] [PubMed] [Google Scholar]
- 3. Chen L, Yang G. PPARs Integrate the mammalian clock and energy metabolism. PPAR Res. 2014; 2014: 653017 10.1155/2014/653017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neher MD, Weckbach S, Huber-Lang MS, Stahel PF. New insights into the role of peroxisome proliferator-activated receptors in regulating the inflammatory response after tissue injury. PPAR Res. 2012; 2012: 728461 10.1155/2012/728461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim DJ, Bility MT, Billin AN, Willson TM, Gonzalez FJ. Peters JM. PPARbeta/delta selectively induces differentiation and inhibits cell proliferation. Cell Death Differ. 2006; 13: 53–60. [DOI] [PubMed] [Google Scholar]
- 6. Kliewer SA, Forman BM, Blumberg B, Ong ES, Borgmeyer U, Mangelsdorf DJ, et al. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A. 1994; 91: 7355–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Higashiyama H, Billin AN, Okamoto Y, Kinoshita M, Asano S. Expression profiling of peroxisome proliferator-activated receptor-delta (PPAR-delta) in mouse tissues using tissue microarray. Histochem Cell Biol. 2007; 127: 485–94. [DOI] [PubMed] [Google Scholar]
- 8. Barish GD, Narkar VA, Evans RM. PPAR delta: a dagger in the heart of the metabolic syndrome. J Clin Invest. 2006; 116: 590–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu G, Li X, Li Y, Tang X, Xu J, Li R, et al. PPARδ agonist GW501516 inhibits PDGF-stimulated pulmonary arterial smooth muscle cell function related to pathological vascular remodeling. Biomed Res Int. 2013; 2013: 903947 10.1155/2013/903947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ali FY, Egan K, FitzGerald GA, Desvergne B, Wahli W, Bishop-Bailey D, et al. Role of prostacyclin versus peroxisome proliferator-activated receptor beta receptors in prostacyclin sensing by lung fibroblasts. Am J Respir Cell Mol Biol, 2006; 34: 242–6. [DOI] [PubMed] [Google Scholar]
- 11. Teunissen BE, Smeets PJ, Willemsen PH, De Windt LJ, Van der Vusse GJ, Van Bilsen M. Activation of PPARdelta inhibits cardiac fibroblast proliferation and the transdifferentiation into myofibroblasts. Cardiovasc Res. 2007; 75: 519–29. [DOI] [PubMed] [Google Scholar]
- 12. Zhang H, Pi R, Li R, Wang P, Tang F, Zhou S, et al. PPARbeta/delta activation inhibits angiotensin II-induced collagen type I expression in rat cardiac fibroblasts. Arch Biochem Biophys. 2007; 460: 25–32. [DOI] [PubMed] [Google Scholar]
- 13. Grimaldi PA. The roles of PPARs in adipocyte differentiation. Prog Lipid Res. 2001; 40: 269–281. [DOI] [PubMed] [Google Scholar]
- 14. Bastie C, Luquet S, Holst D, Jehl-Pietri C, Grimaldi PA. Alterations of peroxisome proliferator-activated receptor delta activity affect fatty acid-controlled adipose differentiatio. J Biol Chem. 2000; 275: 38768–73. [DOI] [PubMed] [Google Scholar]
- 15. Barak Y, Liao D, He W, Ong ES, Nelson MC, Olefsky JM, et al. Effects of peroxisome proliferator-activated receptor delta on placentation, adiposity, and colorectal cancer. Proc Natl Acad Sci U S A. 2002; 99: 303–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peters JM, Lee SS, Li W, Ward JM, Gavrilova O, Everett C, et al. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor beta(delta). Mol Cell Biol. 2000; 20: 5119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doerks T, Copley RR, Schultz J, Ponting CP, Bork P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002; 12: 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burch LR, Zhou K, Donnelly LA, Doney AS, Brady J, Goddard C, et al. A single nucleotide polymorphism on exon-4 of the gene encoding PPARdelta is associated with reduced height in adults and children. J Clin Endocrinol Metab. 2009; 94: 2587–93. 10.1210/jc.2009-0392 [DOI] [PubMed] [Google Scholar]
- 19. Andrulionyte L, Peltola P, Chiasson JL, Laakso M, STOP-NIDDM Study Group. Single nucleotide polymorphisms of PPARD in combination with the Gly482Ser substitution of PGC-1A and the Pro12Ala substitution of PPARG2 predict the conversion from impaired glucose tolerance to type 2 diabetes: the STOP-NIDDM trial. Diabetes 2006; 55: 2148–52. [DOI] [PubMed] [Google Scholar]
- 20. Grarup N, Albrechtsen A, Ek J, Borch-Johnsen K, Jørgensen T, Schmitz O, et al. Variation in the peroxisome proliferator-activated receptor delta gene in relation to common metabolic traits in 7,495 middle-aged white people. Diabetologia 2007; 50: 1201–8. [DOI] [PubMed] [Google Scholar]
- 21. Burch LR, Donnelly LA, Doney AS, Brady J, Tommasi AM, Whitley AL, et al. Peroxisome proliferator-activated receptor-delta genotype influences metabolic phenotype and may influence lipid response to statin therapy in humans: a genetics of diabetes audit and research Tayside study. J Clin Endocrinol Metab. 2010; 95: 1830–7. 10.1210/jc.2009-1201 [DOI] [PubMed] [Google Scholar]
- 22. Xu ZY, Yang H, Xiong YZ, Deng CY, Li FE, Lei MG, et al. Identification of three novel SNPs and association with carcass traits in porcine TNNI1 and TNNI2. Mol Biol Rep. 2010; 37: 3609–13. 10.1007/s11033-010-0010-9 [DOI] [PubMed] [Google Scholar]
- 23. Zuo B, Xiong YZ, Su YH, Deng CY, Lei MG, Zheng R, et al. Mapping of quantitative trait loci on porcine chromosome 7 using combined data analysis. Asian-Aust J Anim Sci. 2004; 17: 1350–1354. [Google Scholar]
- 24. Su YH, Xiong YZ, Zhang Q, Jiang SW, Lei MG, Yu L, et al. Mapping quantitative trait loci for fat deposition in carcass in pigs. Acta Genetica Sinica. 2002; 29: 681–684. [PubMed] [Google Scholar]
- 25. Zuo B, Xiong YZ, Su YH, Deng CY, Zheng R, Lei MG, et al. Mapping and genetic effect analysis of swine quantitative trait loci using 24 microsatellites. Acta Veterinaria et Zootechnica Sinica 2003; 34: 139–146. [Google Scholar]
- 26. Bidanel JP, Milan D, Iannuccelli N, Amigues Y, Boscher MY, Bourgeois F, et al. Detection of quantitative trait loci forgrowth and fatness in pigs. Genet Sel Evol. 2001; 33: 289–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Koning DJ, Janss LL, Rattink AP, van Oers PA, de Vries BJ, Groenen MA, et al. Detection of quantitative trait loci for backfat thickness and intramuscular fat content in pigs. Genetics 1999; 152: 1679–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rattink AP, De Koning DJ, Faivre M, Harlizius B, van Arendonk JA, Groenen MA. Fine mapping and imprinting analysis for fatness trait QTLs in pigs. Mamm Genome 2000; 11: 656–61. [DOI] [PubMed] [Google Scholar]
- 29. Demeure O, Sanchez MP, Riquet J, Iannuccelli N, Demars J, Fève K, et al. Exclusion of the swine leukocyte antigens as candidate region and reduction of the position interval for the Sus scrofa chromosome 7 QTL affecting growth and fatness. J Anim Sci. 2005; 83: 1979–87. [DOI] [PubMed] [Google Scholar]
- 30. Qiao R, Gao J, Zhang Z, Li L, Xie X, Fan Y, et al. Genome-wide association analyses reveal significant loci and strong candidate genes for growth and fatness traits in two pig populations. Genet Sel Evol. 2015; 47:17 10.1186/s12711-015-0089-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ai H, Ren J, Zhang Z, Ma J, Guo Y, Yang B, et al. Detection of quantitative trait loci for growth- and fatness-related traits in a large-scale White Duroc × Erhualian intercross pig population. Anim Genet. 2012; 43:383–91. 10.1111/j.1365-2052.2011.02282.x [DOI] [PubMed] [Google Scholar]
- 32. Huang WH, Ma ZX, Xu ZY, Xiong YZ, Zuo B. Detection of novel SNPs and mapping of the fatness QTL on pig chromosome 7q1.1–1.4 region. Genet Mol Res. 2011; 10: 3090–7. 10.4238/2011.December.14.2 [DOI] [PubMed] [Google Scholar]
- 33. Meidtner K, Schwarzenbacher H, Scharfe M, Severitt S, Blöcker H, Fries R. Haplotypes of the porcine peroxisome proliferator-activated receptor delta gene are associated with backfat thickness. BMC Genet. 2009; 10: 76 10.1186/1471-2156-10-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang YX, Lee CH, Tiep S, Yu RT, Ham J, Kang H, et al. Peroxisome-proliferator-activatedreceptor delta activates fat metabolism to prevent obesity. Cell 2003, 113: 159–170. [DOI] [PubMed] [Google Scholar]
- 35. Ren J, Duan Y, Qiao R, Yao F, Zhang Z, Yang B, et al. A missense mutation PPARD causes a major QTL effect on ear size in pigs, PLoS Genet. 2011; 7: e1002043 10.1371/journal.pgen.1002043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duan Y, Brenig B, Wu X, Ren J, Huang L. The G32E functional variant reduces activity of PPARD by nuclear export and post-translational modification in pigs. PLoS One. 2013; 8:e75925 10.1371/journal.pone.0075925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiong YZ, Deng CY. Principle and method of swine testing. 1999; Chinese Agricultural Press. [Google Scholar]
- 38. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–5. [DOI] [PubMed] [Google Scholar]
- 39. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 40. Hatzis P, van der Flier LG, van Driel MA, Guryev V, Nielsen F, Denissov S, et al. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol Cell Biol. 2008; 28: 2732–44. 10.1128/MCB.02175-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. He TC, Chan TA, Vogelstein B, Kinzler KW. PPARdelta is an APC regulated target of nonsteroidal anti-inflammatory drugs. Cell 1999; 99: 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tanaka M, Suzuki K, Morozumi T, Kobayashi E, Matsumoto T, Domukai M, et al. Genomic structure and gene order of swine chromosome 7q1.1—>q1.2. Anim Genet. 2006; 37: 10–6. [DOI] [PubMed] [Google Scholar]
- 43. Jin T, Liu L. The Wnt signaling pathway effector TCF7L2 and type 2 diabetes mellitus. Mol Endocrinol. 2008; 22: 2383–92. 10.1210/me.2008-0135 [DOI] [PubMed] [Google Scholar]
- 44. Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009; 10: 468–477. 10.1038/nrm2717 [DOI] [PubMed] [Google Scholar]
- 45. MacDonald BT, Tamai K, He X. Wnt/beta-Catenin Signaling: Components, Mechanisms, and Diseases. Dev Cell. 2009; 17: 9–26. 10.1016/j.devcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Archbold HC, Yang YX, Chen L, Cadigan KM. How do they do Wnt they do?: regulation of transcription by the Wnt/β-catenin pathway. Acta Physiol. 2012; 204: 74–109. [DOI] [PubMed] [Google Scholar]
- 47. Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature 2005; 434: 843–850. [DOI] [PubMed] [Google Scholar]
- 48. Leibowitz MD. Activation of PPARdelta alters lipid metabolism in db/db mice. FEBS Lett. 2000; 473: 33–336. [DOI] [PubMed] [Google Scholar]
- 49. Oliver WR Jr, Shenk JL, Snaith MR, Russell CS, Plunket KD, Bodkin NL, et al. A selective peroxisome proliferator-activated receptors delta agonist promoters reverase cholesterol transport. Proc Natl Acad Sci U S A. 2001; 98: 5306–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thamer C, Machann J, Stefan N, Schäfer SA, Machicao F, Staiger H, et al. Variations in PPARD Determine the Change in Body Composition during Lifestyle Intervention: A Whole-Body Magnetic Resonance Study. J Clin Endocrinol Metab. 2008; 93: 1497–1500. 10.1210/jc.2007-1209 [DOI] [PubMed] [Google Scholar]
- 51. Gondret F, Riquet J, Tacher S, Demars J, Sanchez MP, Billon Y, et al. Towards candidate genes affecting body fatness at the SSC7 QTL by expression analyses. J Anim Breed Genet. 2012; 129: 316–24. 10.1111/j.1439-0388.2011.00965.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lane Marker: DNA molecular marker DL2000 (TaKaRa, Dalian, China); Lane 1, 2, and 6: genotype GG, 568 bp; Lane 3, 4: genotype AG, 568 bp + 314 bp + 254 bp; Lane 5: genotype AA, 314 bp + 254 bp.
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.