Abstract
Most aphids are highly specialized on one or two related plant species and generalist species often include sympatric populations adapted to different host plants. Our aim was to test the hypothesis of the existence of host specialized lineages of the aphid Melanaphis sacchari in Reunion Island. To this end, we investigated the genetic diversity of the aphid and its association with host plants by analyzing the effect of wild sorghum Sorghum bicolor subsp. verticilliflorum or sugarcane as host plants on the genetic structuring of populations and by performing laboratory host transfer experiments to detect trade-offs in host use. Genotyping of 31 samples with 10 microsatellite loci enabled identification of 13 multilocus genotypes (MLG). Three of these, Ms11, Ms16 and Ms15, were the most frequent ones. The genetic structure of the populations was linked to the host plants. Ms11 and Ms16 were significantly more frequently observed on sugarcane, while Ms15 was almost exclusively collected in colonies on wild sorghum. Laboratory transfer experiments demonstrated the existence of fitness trade-offs. An Ms11 isofemale lineage performed better on sugarcane than on sorghum, whereas an Ms15 lineage developed very poorly on sugarcane, and two Ms16 lineages showed no significant difference in performances between both hosts. Both field and laboratory results support the existence of host plant specialization in M. sacchari in Reunion Island, despite low genetic differentiation. This study illustrates the ability of asexual aphid lineages to rapidly undergo adaptive changes including shifting from one host plant to another.
Introduction
Most aphid genera or families are host specialized and generalist species represent only 1% of total aphid species. Some specialized aphid species are specific to a single plant species, while other genera or families are associated with a single plant genus or family [1]. Moreover, even in aphid species considered as polyphagous, sympatric populations adapted to different host plants may coexist. The existence of such host races [2] has been demonstrated in Aphis gossypii Glover [3]; Acyrtocyphon pisum Harris [4], Myzus persicae (Sulzer) [5] or Schizaphis graminum (Rondani) [6], for example. Trade-offs in fitness across hosts are regarded as one of the factors that favor such ecological specialization, according to the ‘Jack of all trades-master of none’ theory, which states that genotypes displaying highly performance in a given environment will perform poorly in other environments [7,8].
The aphid Melanaphis sacchari (Zehnter), is distributed worldwide, with apomictic parthenogenetic reproduction, and is mainly restricted to Poaceae hosts [9]. It is a major pest of sorghum [10] and a major virus vector in sugarcane [11]. Both sugarcane (Saccharum spp.) and sorghum (Sorghum spp.) are members of the Poaceae family and the Andropogoneae tribe. In the USA, a recent outbreak of M. sacchari has been causing serious economic damage to sorghum in the southern states since 2013 [12]. This recent change in its pest status occurred although M. sacchari has been reported in Florida since 1922 and has been a common pest of sugarcane since the late 1970s, but has never caused such outbreaks on sorghum [12–14]. This change in its pest status on sorghum raised the hypothesis [15] that these outbreaks could be due to the emergence of a new variant of sugarcane aphid that has a high preference for sorghum or to the introduction of the species Melanaphis sorghi (Theobald). Melanaphis sorghi, which has been regarded as a synonym of M. sacchari by Remaudière and Remaudière [16], is considered as a separate species by Blackman and Eastop [9], and could prefer sorghum to sugarcane [17].
The question of host plant specialization in M. sacchari has never been deeply investigated. In Brazil, Lopes da Silva et al. [18] showed that a clonal lineage derived from a female collected on sugarcane exhibited higher demographic parameters on sorghum than on sugarcane. In a previous study [19], we analyzed the worldwide genetic diversity of M. sacchari. Five multilocus lineages (MLL), or parthenogenetic lineages, were identified, each lineage grouping several multilocus genotypes (MLG) differing from each other by stepwise mutations. The distribution of the five MLLs revealed a strong geographic structuring: MLL-A in West and East Africa, MLL-B in Australia, MLL-D in the USA, MLL-E in China, and MLL-C in a wide region covering South America, the Caribbean, East Africa and the Indian Ocean. That study was carried out on samples collected on sugarcane and on three wild or cultivated Sorghum species. When considering the MLL–host plant association, we did not detect any effect of the host plant on genetic diversity. However, our study was carried out at a large spatial scale and considering MLL as homogeneous genetic entities, we were therefore unable to check the absence of host plant specialization at a finer spatial and genetic scale.
In the present study, to test the hypothesis of the existence of host specialized lineages in M. sacchari, we characterized its genetic diversity and its association with host plants in Reunion Island by (i) analyzing the effect of the host plant, wild sorghum or sugarcane, on the genetic structuring of populations and (ii) by performing laboratory host transfer experiment to detect trade-offs in host use.
Material and Methods
Insect samples
Here, an ‘individual’ refers to one individual aphid and a ‘sample’ refers a several individuals collected from the same host plant species at a given location on the same date. The complete set of individuals (S1 Table) comprised 31 samples collected from two host plants (Table 1): sugarcane (i.e. commercial cultivars derived from inter-specific crosses between S. officinarum and S. spontaneum) and wild sorghum Sorghum bicolor (L.) Moench subsp. verticilliflorum (Steud.) de Wet ex Wiersema & J. Dahlb. On Reunion Island, sugarcane is the dominant crop, cultivated on more than 50% of agricultural lands, while wild sorghum is a common ruderal weed. Sampling was carried out from 2007 to 2010. Collects were carried out during the austral summer (hot and wet season), from October to June. The geographic coordinates of sampling localities are listed in S1 Table. Most of sampling localities, except five of them (Run8, Run17, Run18, Run28, Run30), were situated within the sugarcane growing area. No specific permission was required to sample aphids in these locations. Sampling did not involve endangered or protected species. Eight of these samples (Run1 to Run8) were already included in our previous study [19].
Table 1. Characteristics of samples.
| Sample | Locality | Date of sampling | Host plant |
|---|---|---|---|
| Run1 | Sainte Marie | June, 2007 | sugarcane |
| Run2 | Etang Salé | April, 2008 | sugarcane |
| Run3 | Saint-Pierre | October, 2007 | sugarcane |
| Run4 | Savanna | April, 2008 | sugarcane |
| Run5 | Saint André | April, 2008 | sugarcane |
| Run6 | Entre Deux | March, 2009 | wild sorghum |
| Run7 | Bras Panon | January, 2010 | wild sorghum |
| Run8 | Salazie | March, 2010 | wild sorghum |
| Run9 | Sainte Marie | October, 2007 | sugarcane |
| Run10 | Sainte Marie | October, 2007 | sugarcane |
| Run11 | Saint Leu | April, 2008 | sugarcane |
| Run12 | Petite Île | April, 2008 | sugarcane |
| Run13 | Saint Philippe | April, 2008 | sugarcane |
| Run14 | Piton Sainte Rose | April, 2008 | sugarcane |
| Run15 | Entre Deux | June, 2008 | wild sorghum |
| Run16 | La Saline les Hauts | January, 2010 | sugarcane |
| Run17 | Salazie | March, 2010 | sugarcane |
| Run18 | Grande Chaloupe | January, 2010 | wild sorghum |
| Run19 | Saint Pierre | February, 2010 | sugarcane |
| Run20 | Savanna | January, 2010 | wild sorghum |
| Run21 | Saint André | January, 2010 | wild sorghum |
| Run22 | Piton Sainte Rose | February, 2010 | wild sorghum |
| Run23 | Saint Gilles les Hauts | February, 2010 | wild sorghum |
| Run24 | Sainte Anne | February, 2010 | wild sorghum |
| Run25 | Saint Pierre | February, 2010 | wild sorghum |
| Run26 | Petite Île | March, 2010 | wild sorghum |
| Run27 | Etang Salé | March, 2010 | wild sorghum |
| Run28 | Cilaos | April, 2010 | wild sorghum |
| Run29 | Saint Philippe | April, 2010 | wild sorghum |
| Run30 | Cilaos | April, 2010 | wild sorghum |
| Run31 | Sainte Anne | April, 2009 | sugarcane |
Only a few aphids were collected on each plant sampled to avoid collecting several individuals from the same colony. The number of plants to be sampled was not fixed and varied according to the abundance of aphid colonies. Aphids were immediately placed in 95% ethanol in Eppendorf tubes, and kept frozen at -80°C until processed.
DNA extraction, genotyping and sequencing
DNA extraction and genotyping
DNA was extracted using the protocol of Sunnucks and Hales [20].
Ten microsatellite loci (Table 2) were selected among the 14 previously developed by our team for M. sacchari [21]. PCR reactions were performed with labelled primers and multiplexed as described by Nibouche et al. [19]. Genotyping was carried out using an ABI PRISM 3110 and alleles were identified at each locus by comparison with the size standard using GeneMapper version 2.5 software (Applied Biosystems).
Table 2. Observed microsatellite multilocus genotypes (MLG): allele size (bp) at each locus and distribution according to the host plant.
| MLG | CIR-Ms-G08 | CIR-Ms-G403 | CIR-Ms-B03 | CIR-Ms-C08* | CIR-Ms-G01 | CIR-Ms-E01* | CIR-Ms-G12 | CIR-Ms-G02* | CIR-Ms-E03* | CIR-Ms-D02* | Number of individuals | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Saccharum sp. | Sorghum sp. | |||||||||||
| Ms11 | 233 / 233 | 251 / 259 | 213 / 213 | 197 / 199 | 185 / 210 | 247 / 247 | 212 / 216 | 199 / 199 | 186 / 193 | 228 / 232 | 120 | 20 |
| Ms12 | 233 / 233 | 251 / 259 | 213 / 213 | 195 / 197 | 185 / 210 | 247 / 247 | 212 / 216 | 199 / 205 | 186 / 193 | 228 / 232 | 24 | 5 |
| Ms13 | 233 / 233 | 251 / 259 | 213 / 213 | 197 / 199 | 185 / 210 | 247 / 247 | 212 / 216 | 199 / 201 | 186 / 201 | 228 / 232 | 18 | |
| Ms14 | 233 / 233 | 251 / 259 | 213 / 213 | 195 / 197 | 185 / 210 | 247 / 249 | 212 / 216 | 199 / 203 | 186 / 193 | 228 / 232 | 5 | 1 |
| Ms15 | 233 / 233 | 251 / 259 | 213 / 213 | 197 / 199 | 185 / 210 | 247 / 247 | 212 / 216 | 199 / 201 | 186 / 193 | 228 / 232 | 2 | 253 |
| Ms16 | 233 / 233 | 251 / 259 | 213 / 213 | 195 / 197 | 185 / 210 | 247 / 247 | 212 / 216 | 199 / 203 | 186 / 193 | 228 / 232 | 233 | 99 |
| Ms17 | 233 / 233 | 251 / 259 | 213 / 213 | 197 / 199 | 185 / 210 | 247 / 247 | 212 / 216 | 199 / 201 | 186 / 193 | 224 / 228 | 2 | |
| Ms18 | 233 / 233 | 251 / 259 | 213 / 213 | 197 / 201 | 185 / 210 | 247 / 247 | 212 / 216 | 199 / 201 | 186 / 193 | 228 / 232 | 1 | |
| Ms19 | 233 / 233 | 251 / 259 | 213 / 213 | 195 / 197 | 185 / 210 | 247 / 247 | 212 / 216 | 199 / 203 | 186 / 193 | 220 / 228 | 1 | |
| Ms20 | 233 / 233 | 251 / 259 | 213 / 213 | 195 / 197 | 185 / 210 | 247 / 247 | 212 / 216 | 199 / 203 | 186 / 193 | 228 / 230 | 23 | |
| Ms21 | 233 / 233 | 251 / 259 | 213 / 213 | 195 / 197 | 185 / 210 | 247 / 247 | 212 / 216 | 199 / 203 | 186 / 193 | 228 / 234 | 29 | |
| Ms22 | 233 / 233 | 251 / 259 | 213 / 213 | 197 / 199 | 185 / 210 | 247 / 247 | 212 / 216 | 199 / 199 | 186 / 193 | 220 / 232 | 6 | 1 |
| Ms23 | 233 / 233 | 251 / 259 | 213 / 213 | 197 / 199 | 185 / 210 | 247 / 247 | 212 / 216 | 199 / 199 | 188 / 193 | 228 / 232 | 5 | |
* polymorphic loci
Sequencing
A portion of the mitochondrial COI gene was amplified and sequenced in a total of 26 aphids. The individuals were chosen among the samples to represent at least one individual for each microsatellite multilocus genotype (MLG, see below). Two MLGs, Ms18 and Ms19, were not represented because we collected only one individual of each and their PCR amplification failed. COI fragments were amplified using the LCO1490 and HCO2198 primers designed by Folmer et al. [22]. PCR was carried out using the protocol of Kim and Lee [23]. PCR products were purified and sequenced by a subcontractor (Cogenics, Takeley, Essex, U.K.), and a consensus sequence of 658 pb was defined. Sequence alignments were performed using Geneious software version 5.6.6 [24].
Host transfer experiment
To confirm the existence of host specialization, the development of several M. sacchari isofemale lineages belonging to different MLG was monitored on sugarcane and S. verticilliflorum plantlets in the laboratory. Four isofemale lineages were compared. An Ms11 lineage founded from individuals collected on sugarcane, an Ms15 from S. verticilliflorum, an Ms16 from sugarcane and an Ms16 from S. verticilliflorum. These are referred to as Ms11sugarcane, Ms15sorghum, Ms16sugarcane and Ms16sorghum respectively.
The aphids used to inoculate the plants in this experiment were obtained from laboratory clonal isofemale lineages started from females collected on sugarcane at two locations and on S. verticilliflorum at two locations. The aphids were reared in the laboratory on the same host plant as the plant on which they were collected. We used the rearing method on detached leaves described by Abu Ahmad et al. [25], at 28°C under a L12:D12 light dark regime and 80% relative humidity. Detached sugarcane leaves from cultivar MQ 76/53 were collected in the field. Detached leaves of S. verticilliflorum were collected from potted plants grown in the greenhouse. Several isofemale lineages were produced from individuals collected in the field. After establishment in the laboratory, one individual was collected from each lineage and was genotyped with the microsatellite markers as described above. The MLG of each lineage was therefore identified and only one of each lineage required (i.e. Ms11sugarcane, Ms15sorghum, Ms16sugarcane and Ms16sorghum) was retained for the following experiment.
The experiment was performed on potted plantlets in a no-choice trial, using the experimental design described by Fartek et al. [26, 27]. The potted sugarcane plantlets were grown from stem cuttings collected in the field. Plantlets were grown in pots filled with potting soil at 25°C with a 12 hour light dark regime, until they reached the three leaf stage. The sugarcane cultivar used was ‘R570’, which is one of the two most widely grown cultivars in Reunion Island and has intermediate susceptibility to M. sacchari [27]. The S. verticilliflorum plantlets were grown from seeds collected in natural populations. Plantlets at the three leaf stage were obtained with the cultivation method described above for sugarcane.
Each host-plant × lineage modality was isolated from the others, but aphids could move from one plant to the next one within each host-plant × lineage modality. Ten apterous nymphs, aged 3–4 days (i.e., approximately 2nd or 3rd instar), were placed on each plantlet with a camel hair brush, and the number of aphids on the plants was scored 10 days after infestation. Three replications were carried out, set up successively on May 20, May 31, and June 14, 2011. Each replication used 12 sugarcane plantlets and 12 sorghum plantlets per isofemale lineage. Experiments were carried out at 25°C with a 12 hour light dark regime. In these conditions, the mean duration of one M. sacchari generation (adult to adult) is between 9 to 11 days and the mean longevity of individuals is between 11 and 18 days [26]. Consequently, 10 days after infestation the plantlets were colonized both by F1 progenies and by some of the initial individuals that had survived over the course of the experiment.
Data analysis
Clonal diversity analysis
Single combinations of alleles were retrieved from genotyping data and arranged as unique multilocus genotypes (MLGs). Given the clonal reproduction of M. sacchari, we assumed that the different occurrences of the same MLG in a sample were the result of local clonal reproduction. We therefore kept only one representative of each MLG in each of the 31 samples for genetic and diversity analysis.
Using GENCLONE software [28], we computed a matrix of pairwise genetic distance between MLGs as the number of allelic differences between MLGs. Examination of the distribution of these distances enabled us to define a threshold below which MLGs were considered to belong to the same multilocus lineage (MLL), i.e. genotypes that differed slightly due to mutation or scoring errors according to Arnaud‐Haond et al. [29]. The same matrix of pairwise distances was also used to construct a minimum spanning network using HAPSTAR software [30]. On the set of identical loci within each MLL, we computed psex, the probability that the repeated MLGs originated from distinct sexual reproductive events. A psex value lower than 0.01 supported the hypothesis that MLGs originated from the same MLL [29]. To describe clonal diversity, we computed the clonal richness index as RMLG = (G-1)/(N-1), where G is the number of MLGs detected, and N is the number of samples [31].
Prevalence of MLGs in sugarcane or sorghum
Prevalence of a MLG was defined as the proportion of samples in which at least one individual was detected. For each of the three most prevalent MLGs, i.e. Ms11, Ms15 and Ms16, a logistic model was used to analyze differences in prevalence between sorghum and sugarcane using SAS GLIMMIX procedure [32]. Because the sampling location and the host plant might be confounding effects (i.e. only one host was sampled in each location), we carried out a spatial analysis with a mixed logistic model with a G-side spherical spatial covariance structure [33]. Laplace approximation of the likelihood was used to allow model comparisons. However, the spatial correlation models could not estimate positive spatial covariance components and did not provided an improved fit (based on the AIC criterion) compared with the simple logistic model. For this reason, the results of these spatial analyses were not further taken into account. To compare the level of specialization of the three most prevalent MLGs, their prevalence ratio was computed as the ratio of the prevalence in their most preferred host to the prevalence in the least preferred host. The estimation of the prevalence ratios with their 95% confidence interval was performed with a Poisson regression with a robust error variance using the SAS GENMOD procedure [34–35].
Host transfer experiment
Count data from the laboratory experiment were analyzed with a generalized mixed linear model using a negative binomial distribution. The negative binomial distribution was preferred over the Poisson distribution because a strong overdispersion was obvious with the Poisson distribution model, which yielded a scaled Pearson statistic of 13.83, far from the expected 1.0 value. The lineage, the host plant, the lineage × host plant interaction and the replication (i.e. the three successive experiments) were considered as fixed effects. The 12 plantlets were considered as repeated observations and taken into account by a random lineage × host plant × replication effect. Analyses were performed using the SAS GLIMMIX procedure [32].
Results
Genetic and clonal diversity
A total of 855 aphids were genotyped with the 10 microsatellite markers. After retaining a single representative of each MLG in each of the 31 samples, the final dataset comprised 64 individuals (S1 Table).
Among the 10 microsatellite loci, five were polymorphic (Table 2). These five loci defined 13 MLGs. The most frequently observed MLG were Ms11 (16% of the 855 individuals), Ms15 (30%) and Ms16 (39%). The global clonal richness was low, RMLG = 0.40. All pairwise numbers of distinct alleles between MLG were lower than four, and the distribution was unimodal (S1 Fig). The value of psex was <0.001, which confirmed that the 13 MLGs belonged to the same multilocus lineage (MLL), i.e. genotypes that differed slightly due to step mutations or scoring errors (Fig 1).
Fig 1. Minimum spanning network of Melanaphis sacchari microsatellite distances computed as the number of allele differences between MLGs.
Each node represents one step in the network, i.e. a distance of one allele. The numbers in the circles refer to the numbers given to MLGs in Table 1.
Among the 26 COI sequences obtained (GenBank accessions listed in S1 Table), a single haplotype was observed.
Host plant specialization
The number of sorghum or sugarcane samples hosting each MLG is summarized in Table 3. Three MLGs (Ms11, Ms15 and Ms16) were frequent enough to allow a statistical comparison of their prevalence in sorghum vs. sugarcane samples (Fig 2). All the results of the tests were significant. Ms11 (F = 5.15; df = 1, 29; P = 0.0308) and Ms16 (F = 5.27 df = 1, 29; P = 0.0291) showed higher prevalence in sugarcane samples than in sorghum samples. Conversely, Ms15 showed a significantly higher prevalence in sugarcane than in sorghum (F = 11.38; df = 1, 29; P = 0.0021). The estimated prevalence ratios [95% CI] in favor of sugarcane for Ms11 and Ms16 were respectively 4.27 [1.07; 16.96] and 1.87 [1.12; 3.10]. For Ms15, the prevalence ratio in favor of sorghum was 12.19 [1.80; 80.14]. The large confidence intervals due to the small number of samples (31), prevented us from ranking the prevalence ratios to rank the specialization of the three MLGs.
Table 3. Number of samples hosting at least one individual of each of the 13 MLGs on sugarcane or sorghum samples.
| MLG | Number of samples hosting each MLG | |
|---|---|---|
| sorghum | sugarcane | |
| Ms11 | 2 | 8 |
| Ms12 | 2 | 3 |
| Ms13 | 1 | 0 |
| Ms14 | 1 | 1 |
| Ms15 | 13 | 1 |
| Ms16 | 8 | 14 |
| Ms17 | 1 | 0 |
| Ms18 | 1 | 0 |
| Ms19 | 0 | 1 |
| Ms20 | 1 | 0 |
| Ms21 | 0 | 2 |
| Ms22 | 1 | 1 |
| Ms23 | 0 | 2 |
| Total number of samples | 16 | 15 |
Fig 2. Prevalence of the three most frequent MLGs on sorghum (white bars) and sugarcane (grey bars) samples.
Vertical lines represent standard errors of prevalences.
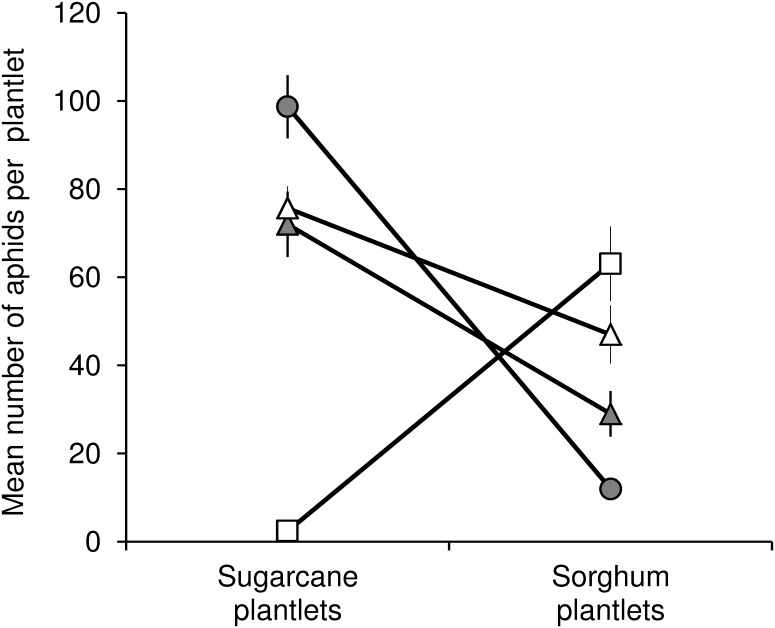
The results of the host transfer experiment are illustrated in Fig 3. At 240 hours post infestation, the lineage × host plant effect was significant (F = 19.31; df = 3, 14; P < 0.0001) and revealed a significant host specialization. The Ms11sugarcane aphid population size was significantly lower on sorghum than on sugarcane (t = 3.69; df = 14; P = 0.0024). Conversely, the Ms15sorghum population size was significantly higher on sorghum than on sugarcane (t = 6.22; df = 14; P < 0.0001). No significant difference in Ms16sorghum was observed between sugarcane and sorghum (t = 1.20; df = 14; P = 0.2488). The difference in Ms16sugarcane was close to the significance level but remained non-significant (t = 2.11; df = 14; P = 0.0532).
Fig 3. Laboratory comparison of fitness trade-offs in three MLGs.
Ms11 (circles), Ms15 (squares), or Ms16 (triangles). Host transfer experiments were carried out with four isofemale lineages derived from individuals collected either on sorghum (white symbols) or on sugarcane (grey symbols). The figure represents the mean number of aphids per sugarcane or sorghum plantlet 240 hours after 12 plantlets had each been infested with 10 individuals. The experiment was repeated three times. Vertical lines represent the standard errors of means.
Ms15sorghum appeared to be more specialized than Ms11sugarcane. The population of Ms15sorghum on sugarcane decreased 4.2-fold, from 10 aphids per plantlet at the beginning of the experiment to 2.36 ± 0.73 aphids per plantlet 240 hours after infestation. On sorghum, the Ms11sugarcane population remained stable over the course of the experiment, increasing slightly from 10 to 11.97 ± 1.94 aphids per plantlet. At 240 hours, the population size of Ms11sugarcane on sorghum was significantly higher than the population of Ms15sorghum on sugarcane (t = 3.48; df = 14; P = 0.0037), suggesting a higher level of specialization by Ms15 than by Ms11.
Fig 4 presents the results of the host transfer experiment illustrating the negative correlation of fitness on sugarcane and fitness on sorghum.
Fig 4. Illustration of the negative correlation of the performances on sorghum and sugarcane of four isofemale lineages in the laboratory host transfer experiments.
The lineages belong to three MLGs: Ms11 (circles), Ms15 (squares), or Ms16 (triangles). Vertical and horizontal bars represent the standard errors of means.
Discussion
In this study, the genetic diversity of M. sacchari in Reunion Island was represented by 13 MLGs which differed from one another by stepwise mutations. Three of the 13 MLGs, (Ms11, Ms16 and Ms15), were the most frequent. Among these 13 MLGs, only five (Ms11, Ms12, Ms15, Ms16, Ms21) had already been observed in our previous study [19]. The eight newly observed MLGs were rare and were only observed in one or two samples. This increase in the number of observed MLGs is the result of the increase in the number of samples, not to higher clonal diversity, as shown by the RMLG of 0.40, which is similar to the value observed previously at the world scale (0.37). The 13 MLGs observed in La Reunion belong to a unique multilocus lineage MLL-C, previously detected in South America, the Caribbean, East Africa and the Indian Ocean [19]. The COI haplotype is also the only one we observed previously in the individuals belonging to this MLL-C [19].
Both field and laboratory results support the hypothesis of host plant specialization in M. sacchari in Reunion Island. The genetic structure of the populations was linked to the host plants. Ms11 and Ms16 were significantly more frequently observed on sugarcane, while Ms15 was almost exclusively collected in colonies on the wild sorghum S. verticilliflorum. Laboratory transfer experiments demonstrated the existence of fitness trade-offs between the two host plants, and showed that fitness on sugarcane is negatively correlated to that on sorghum. Ms11 performed better on sugarcane than on sorghum, while the opposite was the case of Ms15. Ms16 was intermediate, its field prevalence was significantly higher on sugarcane than on sorghum, and its performance was intermediate on both hosts in the laboratory.
Although some authors demonstrated that conditioning or experience (i.e. the host plant on which aphids were reared before the experiment) does not interfere with the genetic based specialization [36], other studies demonstrated that conditioning or experience can alter the interpretation of host transfer experiments [37–38]. In our study, the four lineages were reared on the plant from which they originated and we did not include a conditioning step on another Poaceous host previous to the transfer experiments. Nevertheless, our results do not suggest a pronounced experience effect in the two Ms16 lineages collected and reared either on sorghum or on sugarcane in that no significant reduction in fitness was observed when they were transferred on the alternate host.
Host specialization has been demonstrated in several aphid species. In Aphis gossypii, Margaritopoulos et al. [39] reported morphometric differences between aphids originating from Compositae and those collected on Cucurbitaceae and Malvaceae. A study by Carletto et al. [3] demonstrated the existence of five A. gossypii host races specialized on Cucurbitaceae, cotton, eggplant, potato and chili or sweet pepper. In the Acyrthocyphon pisum complex, 11 well distinguished sympatric populations were also demonstrated on several Fabaceae species [4]. In Schizaphis graminum, analysis of COI revealed three clades whose host partitioning suggested the existence of host races [6]. Host races were also identified on several Poaceae hosts in Sitobion avenae [40–42]. In these four species, the existence of host races is associated with a strong genetic structuring revealed by DNA sequence variations in barcoding regions or by Bayesian clustering analysis of microsatellite data. In contrast, our study revealed strong host specialization despite low genetic differentiation. The Ms15 lineage was seen to be specialized on sorghum although it differed only by one allele in the CIR-Ms-G02 locus from the lineage Ms11, which developed better on sugarcane than on sorghum. This low genetic differentiation suggests that the appearance of the sorghum specialized host race Ms15 could be recent. On the other hand, Ms16 appears as intermediate, with a partial adaptation to sorghum, although Fig 1 shows that Ms16 is genetically closer to Ms11 than to Ms15, which could suggest that the partial adaptation of Ms16 to sorghum is independent from the adaptation of Ms15. Further experiments could aim at exploring the relationship between genetic diversity and host adaptation by characterizing the adaptation of the less frequent MLGs.
This study provides an example of the evolution of an ecological trait in a species with strictly parthenogenetic reproduction; such examples challenge the view that parthenogenesis is an evolutionary dead end and show that asexual aphid lineages can rapidly undergo adaptive changes including host plant shifts [43].
Supporting Information
(PDF)
(XLSX)
Acknowledgments
We are grateful to R. Tibère for technical assistance. B. Facon is acknowledged for his critical review of the manuscript. The authors greatly acknowledge the Plant Protection Platform (3P, IBISA).
Data Availability
All relevant data are within the paper and its Supporting Information files. COI data are available from the GenBank (https://www.ncbi.nlm.nih.gov/genbank/) database (accession numbers KJ083108, KJ083109, KJ083118-KJ083124, KJ083180-KJ083196).
Funding Statement
This work was funded by the European Union: European regional development fund (ERDF), by the Conseil Régional de la Réunion and by the Centre de coopération Internationale en Recherche Agronomique pour le Développement (CIRAD). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Peccoud J, Simon JC, von Dohlen C, Coeur d'Acier A, Plantegenest M, Vanlerberghe-Masutti F, et al. Evolutionary history of aphid-plant associations and their role in aphid diversification. C R Biol. 2010; 333: 474–487. 10.1016/j.crvi.2010.03.004 [DOI] [PubMed] [Google Scholar]
- 2. Drès M, Mallet J. Host races in plant–feeding insects and their importance in sympatric speciation. Philos Trans R Soc Lond B Biol Sci. 2002; 357: 471–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carletto J, Lombaert E, Chavigny P, Brévault T, Lapchin L, Vanlerberghe-Masutti F. Ecological specialization of the aphid Aphis gossypii Glover on cultivated host plants. Mol Ecol. 2009; 18: 2198–2212. 10.1111/j.1365-294X.2009.04190.x [DOI] [PubMed] [Google Scholar]
- 4. Peccoud J, Ollivier A, Plantegenest M, Simon J- C. A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc Natl Acad Sci U S A. 2009; 106: 7495–7500. 10.1073/pnas.0811117106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Margaritopoulos JT, Blackman RL, Tsitsipis JA, Sannino L. Co-existence of different host-adapted forms of the Myzus persicae group (Hemiptera: Aphididae) in southern Italy. Bull Entomol Res. 2003; 93: 131–135. [DOI] [PubMed] [Google Scholar]
- 6. Anstead J, Burd J, Shufran K. Mitochondrial DNA sequence divergence among Schizaphis graminum (Hemiptera: Aphididae) clones from cultivated and non-cultivated hosts: haplotype and host associations. Bull Entomol Res. 2002; 92: 17–24. [DOI] [PubMed] [Google Scholar]
- 7. Futuyma DJ, Moreno G. The evolution of ecological specialization. Annu Rev Ecol Evol Syst. 1988; 19: 207–233. [Google Scholar]
- 8. Via S. Ecological genetics and host adaptation in herbivorous insects: the experimental study of evolution in natural and agricultural systems. Annu Rev Entomol. 1990; 35: 421–446. [DOI] [PubMed] [Google Scholar]
- 9. Blackman RL, Eastop VF. Aphids on the world's herbaceous plants and shrubs. Chichester, UK: John Wiley & Sons Ltd; 2006. [Google Scholar]
- 10. Singh BU, Padmaja PG, Seetharama N. Biology and management of the sugarcane aphid, Melanaphis sacchari (Zehntner) (Homoptera: Aphididae), in sorghum: a review. Crop Prot. 2004; 23: 739–755. [Google Scholar]
- 11. Rott P, Mirkov TE, Schenck S, Girard JC. Recent advances in research on Sugarcane yellow leaf virus, the causal agent of sugarcane yellow leaf. Sugar Cane Int. 2008; 26: 18–27. [Google Scholar]
- 12. Armstrong SJ, Rooney WL, Peterson GC, Villenueva RT, Brewer MJ, Sekula-Ortiz D. Sugarcane Aphid (Hemiptera: Aphididae): host range and sorghum resistance including cross-resistance from greenbug sources. J Econ Entomol. 2015; 108: 576–582. 10.1093/jee/tou065 [DOI] [PubMed] [Google Scholar]
- 13. Mead FW. Sugarcane aphid (Melanaphis sacchari (Zehntner))—Florida—New. Cooperative Plant Pest Report. 1978; 3: 475. [Google Scholar]
- 14. White WH, Reagan TE, Hall DG. Melanaphis sacchari (Homoptera: Aphididae), a sugarcane pest new to Louisiana. Fla. Entomol. 2001; 84: 435–436. [Google Scholar]
- 15.Brewer M, Way M, Armstrong S, Biles S, Sekula D, Swart J, et al. Outbreak of sorghum/sugarcane aphid on sorghum: first detections, distribution, and notes on management. In: Castner E, editor. 25th Texas Plant Protection Conference Program; 2013 Dec 10–11; Bryan, TX. Conroe: Texas Plant Protection Association; 2013. Available: http://agrilife.org/ccag/files/2013/12/2013-Poster-TPPA-aphid.pdf
- 16. Remaudière G, Remaudière M. Catalogue of the world's Aphididae. Paris, France: INRA; 1997. [Google Scholar]
- 17.Blackman RL, Eastop VF, Brown PA. The biology and taxonomy of the aphids transmitting barley yellow dwarf virus. In: Burnett PA, editor. World Perspectives on Barley Yellow Dwarf Proceedings of the International Workshop; 1987 Jul 6–11; Udine, Italy. Mexico: CIMMYT; 1990. p. 197–214.
- 18. Lopes da Silva M, Almeida Rocha D, Tayrane Bezerra da Silva K. Potential population growth of Melanaphis sacchari (Zethner) reared on sugarcane and sweet sorghum. Curr Agric Sci Technol. 2014; 20: 21–25. [Google Scholar]
- 19. Nibouche S, Fartek B, Mississipi S, Delatte H, Reynaud B, Costet L. Low genetic diversity in Melanaphis sacchari aphid populations at the worldwide scale. PLoS ONE. 2014; 9: e106067 10.1371/journal.pone.0106067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sunnucks P, Hales D. Numerous transposed sequences of mitochondrial cytochrome oxidase I-II in aphids of the genus Sitobion (Hemiptera: Aphididae). Mol Biol Evol. 1996; 13: 510–524. [DOI] [PubMed] [Google Scholar]
- 21. Andris M, Aradottir GI, Arnau G, Audzijonyte A, Bess EC, Bonadonna F, et al. Permanent genetic resources added to molecular ecology resources database 1 June 2010–31 July 2010. Mol Ecol Res. 2010; 10: 1106–1108. [DOI] [PubMed] [Google Scholar]
- 22. Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994; 3: 294–299. [PubMed] [Google Scholar]
- 23. Kim H, Lee S. A molecular phylogeny of the tribe Aphidini (Insecta: Hemiptera: Aphididae) based on the mitochondrial tRNA/COII, 12S/16S and the nuclear EF1α; genes. Syst Entomol. 2008; 33: 711–721. [Google Scholar]
- 24.Biomatters. Geneious version 5.6.6.; 2012
- 25. Abu Ahmad Y, Costet L, Daugrois J- H, Nibouche S, Letourmy P, Girard JC, et al. Variation in infection capacity and in virulence exists between genotypes of Sugarcane yellow leaf virus . Plant Dis. 2007; 91: 253–259. [DOI] [PubMed] [Google Scholar]
- 26. Fartek B, Nibouche S, Turpin P, Costet L, Reynaud B. Resistance to Melanaphis sacchari in the sugarcane cultivar R 365. Entomol Exp Appl. 2012; 144: 270–278. [Google Scholar]
- 27. Fartek B, Nibouche S, Atiama-Nurbel T, Reynaud B, Costet L. Genotypic variability of sugarcane resistance to the aphid Melanaphis sacchari, vector of the Sugarcane yellow leaf virus. Plant Breed. 2014; 133: 771–776. [Google Scholar]
- 28. Arnaud-Haond S, Belkhir K. GENCLONE: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol Ecol Notes. 2007; 7: 15–17. [Google Scholar]
- 29. Arnaud-Haond S, Duarte C, Alberto F, Serrão E. Standardizing methods to address clonality in population studies. Mol Ecol. 2007; 16: 5115–5139. [DOI] [PubMed] [Google Scholar]
- 30. Teacher AG, Griffiths DJ. HapStar: automated haplotype network layout and visualization. Mol Ecol Res. 2011; 11: 151–153. [DOI] [PubMed] [Google Scholar]
- 31. Dorken ME, Eckert CG. Severely reduced sexual reproduction in northern populations of a clonal plant, Decodon verticillatus (Lythraceae). J Ecol. 2001; 89: 339–350. [Google Scholar]
- 32. SAS Institute. SAS OnlineDoc® 9.2. Cary, NC: SAS Institute Inc; 2008. [Google Scholar]
- 33. Stroup WW. Generalized linear mixed models: modern concepts, methods and applications. Boca Raton, FL: CRC press; 2013. [Google Scholar]
- 34. Spiegelman D, Hertzmark E. Easy SAS calculations for risk or prevalence ratios and differences. Am J Epidemiol. 2005; 162: 199–200. [DOI] [PubMed] [Google Scholar]
- 35. Zou G. A modified poisson regression approach to prospective studies with binary data. Am J Epidemiol. 2004; 159: 702–706. [DOI] [PubMed] [Google Scholar]
- 36. Via S. Specialized host plant performance of Pea Aphid clones is not altered by experience. Ecology. 1991; 72: 1420–1427. [Google Scholar]
- 37. Montllor CB, Campbell BC, Mittler T. Natural and induced differences in probing behavior of two biotypes of the greenbug, Schizaphis graminum, in relation to resistance in sorghum. Entomol. Exp. Appl. 1983; 34: 99–106. [Google Scholar]
- 38. Ramírez CC, Niemeyer HM. The influence of previous experience and starvation on aphid feeding behavior. J. Insect Behav. 2000; 13: 699–709. [Google Scholar]
- 39. Margaritopoulos JT, Tzortzi M, Zarpas KD, Tsitsipis JA, Blackman RL. Morphological discrimination of Aphis gossypii (Hemiptera: Aphididae) populations feeding on Compositae. Bull Entomol Res. 2006; 96: 153–165. [DOI] [PubMed] [Google Scholar]
- 40. De Barro PJ, Sherratt TN, Carvalho GR, Nicol D, Iyengar A, MacLean N. Geographic and microgeographic genetic differentiation in two aphid species over southern England using the multilocus (GATA) 4 probe. Mol Ecol. 1995; 4: 375–384. [DOI] [PubMed] [Google Scholar]
- 41. Sunnucks P, Barro PJD, Lushai G, MacLean N, Hales D. Genetic structure of an aphid studied using microsatellites: cyclic parthenogenesis, differentiated lineages and host specialization. Mol Ecol. 1997; 6: 1059–1073. [DOI] [PubMed] [Google Scholar]
- 42. Weber G. On the ecological genetics of Sitobion avenae (F.) (Hemiptera, Aphididae). Z Angew Entomol. 1985; 100: 100–110. [Google Scholar]
- 43. Loxdale HD What’s in a Clone: The Rapid Evolution of Aphid Asexual Lineages in Relation to Geography, Host Plant Adaptation and Resistance to Pesticides In: Schön I, Martens K, van Dijk P, editors. Lost sex. Dordrecht: Springer; 2009. p. 535–557. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. COI data are available from the GenBank (https://www.ncbi.nlm.nih.gov/genbank/) database (accession numbers KJ083108, KJ083109, KJ083118-KJ083124, KJ083180-KJ083196).