Abstract
To examine the T cell receptor structure in the absence of B cells, the TCR β CDR3 was sequenced from DNA of 15 X-linked agammaglobulinemia (XLA) subjects and 18 male controls, using the Illumina HiSeq platform and the ImmunoSEQ analyzer. V gene usage and the V–J combinations, derived from both productive and nonproductive sequences, were significantly different between XLA samples and controls. Although the CDR3 length was similar for XLA and control samples, the CDR3 region of the XLA T cell receptor contained significantly fewer deletions and insertions in V, D, and J gene segments, differences intrinsic to the V(D)J recombination process and not due to peripheral T cell selection. XLA CDR3s demonstrated fewer charged amino acid residues, more sharing of CDR3 sequences, and almost completely lacked a population of highly modified Vβ gene segments found in control DNA, suggesting both a skewed and contracted T cell repertoire in XLA.
Keywords: XLA, T cell receptor, High throughput sequencing, Junctional diversity, Amino acid sequence
1. Introduction
With the increasing use of B cell depleting therapies in human autoimmune diseases, the impact of loss of B cells on the T cell repertoire has been of broad interest [1]. Examining the T cell repertoire of subjects with X-linked agammaglobulinemia (XLA), who lack B cell development, has revealed that broad alterations of the T cell compartment can be observed in these subjects, including decreased numbers of peripheral blood memory CD 45RA cells, effector memory CD4+ T cells, and follicular helper T cells [2–4]. These changes could potentially be due to the abnormal germinal center formation characteristic of this immune defect [5]. As seen in subjects treated with anti-CD20 monoclonal B cell depleting antibody [6], circulating Th17 cells also appear diminished in subjects with XLA [7], further suggesting that loss of B cells affects the T cell compartment. Testing cellular immunity in XLA has, however, generally demonstrated preserved global T cell responses, with normal proliferation to influenza vaccine, tetanus toxoid, and non-specific mitogens [8–10], and with development of T cell memory after hepatitis B vaccination [3]. In contrast, impaired delayed type hypersensitivity (DTH) reactions, impaired anti-meningococcal T cell immunity [11], and, in an older study, skewing to a Th1 cytokine response [12] have been observed in XLA. To examine the T cell repertoire in XLA more closely, in this study we have used high throughput sequencing to examine the structure of the XLA T cell receptor as compared to age appropriate normal male subjects. Our results show that relative to controls, subjects with XLA have skewed V–J gene usage, fewer deletions and insertions, differential use of amino acids, increased CD3 hydrophilicity, and lack of a population of T cell receptors with more extensive nucleotide deletions from V genes.
2. Methods
2.1. Demographics and clinical characteristics
Fifteen male children and adults with X-linked agammaglobulinemia defined by congenital agammaglobulinemia and a mutation in the Btk gene and/or family history of congenital agammaglobulinemia were recruited for this study (Table 1). Two subjects were brothers, and two sets of cousins were included. Eighteen banked control male PBMC DNA samples were obtained from the Department of Genetics and Genomics at the Icahn School of Medicine and used as controls. The age ranges for XLA subjects (2 years to 54 years) and controls (3 years to 42 years) were not significantly different (p-value = 0.12). All XLA patients were receiving replacement immunoglobulin at time of study participation; none were ill or on immunomodulatory or immunosuppressive medications at the time of blood collection. This study was approved by the Institutional Review Board of Mount Sinai Hospital, and written informed consent was obtained from all patients or their parents.
Table 1.
Clinical information.
| Patient | Clinical information | Year of birth | T cells %# (normal range 55–89%) | CD4% (normal range 33–65%) | CD8% (normal range 10–41%) | B cells% (normal range 5–15%) | |
|---|---|---|---|---|---|---|---|
| 1 | Pneumonia; brother died of ECHO virus | Base pair substitution C to T codon 582 | 1956 | NA | NA | NA | <1 |
| 2 | History of respiratory infections | Missense mutation exon 15 | 1976 | 84 | 57 | 25 | 0 |
| 3 | History of respiratory infections; sinusitis; 2 affected nephews | ND; positive family history | 1980 | 90 | 57 | 30 | 0 |
| 4 | History of respiratory infections; joint pain | ND; positive family history | 1971 | 83 | 64 | 19 | <1 |
| 5 | History of respiratory infections† | Exon 11 18-bp insertion | 1982 | NA | NA | NA | <1 |
| 6 | Pneumonia in childhood‡ | Missense mutation in the pleckstrin homology domain | 1987 | NA | NA | NA | <1 |
| 7 | Pseudomonas aeruginosa bacteremia age 2† | Exon 11 18-bp insertion | 1993 | NA | NA | NA | <1 |
| 8 | Pneumonia in childhood | 2 kb deletion that removes exon 6 and 7 | 1996 | 93 | 60 | 30 | <1 |
| 9 | History of respiratory infections‡ | Missense mutation in the pleckstrin homology domain | 1997 | 89 | 69 | 24 | 0 |
| 10 | Pneumonia in childhood† | Nonsense mutation exon 13 | 1999 | 92 | 55 | 32 | 1 |
| 11 | Pneumocystis carnii pneumonia age 1† | Nonsense mutation exon 13 | 2006 | 94 | 69 | 24 | 0 |
| 12 | Respiratory infections | Frameshift mutation leading to stop codon | 2007 | 93 | 71 | 19 | <1 |
| 13 | Acute bacterial meningitis age 11 months | Nonsense mutation exon 13 | 2008 | 98 | 72 | 24 | <1 |
| 14 | Bacterial pneumonia age 2 | Missense mutation exon 6; leads to stop codon | 2008 | 92 | 65 | 25 | <1 |
| 15 | Upper respiratory infections age 1; RSV | ND; positive family history; 3 relatives | 2010 | 95 | 71 | 32 | <1 |
*, †, and ‡ are related patients.
NA = not available.
normal lymphocyte ranges are for subjects over age 10.
2.2. TCR β sequencing and analysis
PBMC DNA was prepared as described recently [13]. Equivalent amounts of control and CVID DNA were used for sequencing. A 60 bp sequence of the rearranged TCRβ CDR3 region was amplified and sequenced for all samples using the immunoSEQ™ assay, a high-throughput multiplex PCR assay for the re-arranged DNA of T cells (Adaptive Biotechnologies Inc.). Average number of sequence for patients was 95,129.47 (range of 18,049–185,067), and for controls 35,777.72 (range of 10,796–53956). PCR bias was controlled using synthetic templates [14]. For each unique nucleotide sequence, the V, D, and J gene usage, n-nucleotide insertions, base deletions, copy number, and frequency were determined. The predicted amino acid sequence of the productive sequences was determined. The mean CDR3 Kyte–Doolittle Hydropathy index was interpolated from this sequence.
2.3. Statistical methods
Statistical analyses and graphing were performed using the R statistical programming language (version 2.15.2) and GraphPad Prism (version 5.01). Normality was determined using histograms and the Shapiro–Wilk test. Unpaired t-tests were used for comparison of normally distributed numerical data, while nonparametric data were assessed with Wilcoxon tests. Pearson coefficients were calculated to test correlation. A p-value of <0.05 was regarded as significant. V, D, or J family and gene usage was compared using a two-way anova with Tukey HSD multiple comparisons test. Clustering analysis of V genes was performed using the hclust function in R using scaled data and with an algorithm using Manhattan distances and complete clustering.
3. Results
3.1. XLA patients have a unique pattern of V gene usage
The T cell receptor repertoire is determined by the V, D and J genes used and by subsequent recombination events that include nucleotide insertions and deletions. During V(D)J recombination, if the recombination event results in a sequence containing a stop codon or a frame-shift mutation, a second locus is rearranged. If this rearrangement results in a productive sequence the cell carries the productive and the previously rearranged non-productive sequence in its genome. Sequencing genomic DNA allowed us to examine both loci. We examined the distribution of individual V genes, J genes and, more specifically, V–J combinations in XLA as compared to controls. When assessing productive sequences, V gene usage and V–J combination use, but not J gene usage, were significantly different between XLA and normal controls (Fig. 1a,b). Anova with Tukey HSD correction for multiple comparisons; V–J combination p-value = 0.005, V genes p-value = 0.003, J genes p-value = 0.385). Similar differences were also seen when examining the VJ use of nonproductive sequences (anova with Tukey HSD correction for multiple comparisons; V–J combination p-value of <0.0001). Non-biased clustering was performed on the basis of V gene usage. As seen in Fig. 1c, patients clustered independently of the control subjects.
Fig. 1.

Altered V–J gene usage in XLA. Circos plot representation of VJ usage for a) XLA and b) controls (http://circos.ca). V genes (green to purple) and J gene (red to yellow) usage is shown. Samples are color coded by name. Width of arc represents mean copy number. Width of chords represents contribution of each VJ combination. c) A dissimilarity matrix was generated using Canberra distance and non-biased clustering (Complete method) was performed based on the VJ gene combination usage. Controls (blue) and XLA patients (red) segregated almost entirely from each other. Length of branches represent distance. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Decreased V, D, and J deletions and n-nucleotide insertions in XLA
We next examined the structure of the CDR3 region of the XLA TCRs. XLA patients had significantly fewer total deletions from V, D, and J genes (XLA 14.95 ± 0.15; controls 15.84 ± 0.21; p = 0.002; Fig. 2a). There were fewer reciprocal insertions of non-templated nucleotides (XLA 7.998 ± 0.1254; 8.768 ± 0.1736; p = 0.002; Fig. 2b). CDR3 length was comparable between the groups (XLA 38.07 ± 0.05167; Controls 38.06 ± 0.06563; p = 0.91). Perhaps as a result of fewer deletions and insertions, we observed fewer non-productive sequences.
Fig. 2.

XLA patients have fewer CDR3 deletions and insertions. The mean number of deletions from V, D and J genes (a, c) and V–D and D–J insertions (b, d) in productive (a, b) and nonproductive sequences (c, d) for XLA (red square) and controls (blue filled circle). a, c) The sum of the numbers of deletions from Vβ, Dβ, and Jβ sequences were calculated for each sequence. The mean number of deletions (in bases) for each patient (red square) or control (blue filled circle) is shown (y-axis). The horizontal bar represents the mean of each group and the whiskers represent the standard error of the mean. b, d) The sum of the number of insertions between Vβ and Dβ, and Dβ and Jβ sequences was calculated for each sequence. The mean number of insertions (in bases) for each patient (red square) or control (blue filled circle) is shown (y-axis). The horizontal bar represents the mean of each group and the whiskers represent the standard error of the mean. p-values of t-test are indicated. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
This skewing towards fewer deletions during recombination could conceivably be the result of a thymic or an extra-thymic selection event. To dissect the influence of initial recombination and subsequent selection, we examined non-productive sequences (the recombined sequences that resulted in stop codons or frameshift mutations) that are not expressed on the cell surface, and are thus exempt from peripheral selection. As for the productive sequences, the non-productive sequences also had fewer deletions (XLA 15.03 ± 0.1517; 16.13 ± 0.2131; p = 0.0003; Fig. 2c) and insertions (XLA 10.15 ± 0.1209; Controls 11.00 ± 0.1941; p = 0.0013; Fig. 2d). As such, this decrease in n-nucleotide addition and in base deletion is intrinsic to XLA T cell V(D)J recombination and is not a result of selection.
In a previous study we noted that DNA of peripheral blood lymphocytes of normal subjects contains a population of highly modified T cell receptors, characterized by greater than 12 deletions from the Vβ gene segment [13]. We examined the XLA receptor repertoire for this population and observed that very few clones with >12 deletions from the Vβ gene were present in these XLA samples (normalized number of clones 1.9 ± 1.9), unlike normal controls (normalized number of clones 28.7 ± 1.6, p < 0.0001 Fig. 3). Only three of the 15 XLA patients had any detectable sequences with >12 deletions. There is evidence that the coding end can influence the number of base deletions from the Vβ gene [15]. However, in these 3 patients there was no evidence that any of the V-genes were selectively responsible for the fewer numbers of highly modified sequences observed.
Fig. 3.
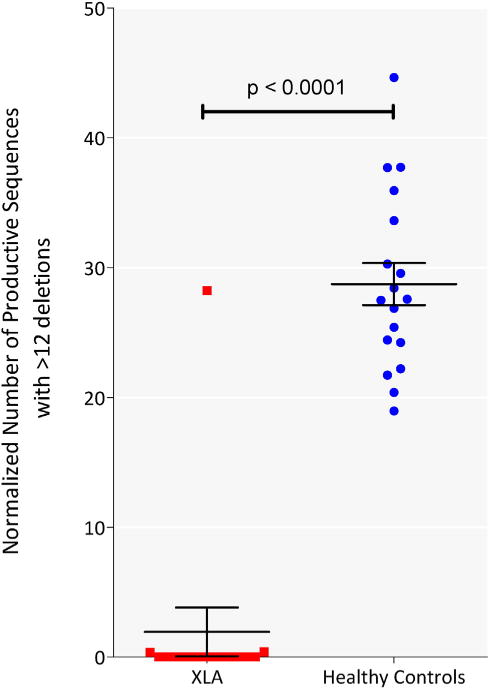
XLA patients lack a population of TCRs characterized by >12 V gene deletions. The number of productive sequences with more than 12 deletions from the V gene was determined and normalized to the total number of sequences for each patient (red) or control (blue), expressed as number of sequences per 10,000 productive sequences. Horizontal line represents mean and whiskers represent standard error of mean. The t-test p-values is indicated. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Altered amino acid sequence in XLA leads to more hydrophilic TCRs
The alterations in the V(D)J recombination led us to probe the effect of these changes on the amino acid sequences of the TCRs. We assessed the amino acid composition of the TCRβ beyond the conserved part of the Vβ gene (ending in the amino acids CASS), and found that the amino acid sequence was significantly different between XLA subjects and controls (anova with Tukey HSD correction for multiple comparisons p-value = 0.004). The receptor function of the TCR is dependent on the charged residues and hydrophobicity. To determine if the sequence changes may have functional effects we specifically examined the percentages of charged residues (histidine, arginine, aspartate, glutamate, lysine) in the productive TCR clones in XLA samples and controls. XLA subjects had fewer of these charged residues (13.96 ± 0.057%) compared to controls (14.17 ± 0.048%, p = 0.007 Fig. 4a; t-test). On comparing the mean hydrophobicity of XLA samples (mean hydropathy index −0.33 ± 0.019) and controls (mean hydropathy index −0.35 ± 0.018), we found that XLA sequences were significantly more hydrophilic (t-test p-value = 0.01, Fig. 4b).
Fig. 4.

Amino acid usage was significantly different between XLA and controls. a) The percent charged amino acids (histidine, arginine, aspartate, glutamate, lysine) in each productive sequence was determined and the mean for each control (blue circle) and XLA (red square) subject determined. b) Hydrophobicity was estimated by the Kyte–Doolittle index for each sequence and the mean for each control (blue circles) and XLA subject (red squares). Horizontal line represents mean and whiskers represent standard error of mean. t-Test p-values are indicated. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Investigations of clonality and clone sharing in XLA T cells
Several primary immune deficiencies are associated with an increase in T cell clonality, including severe combined immune deficiency, MHC class II defects, and, most recently, CVID [13,16–20] There was, however, no difference in clonality in T cell sequences between XLA patients and control subjects (Fig. 5).
Fig. 5.
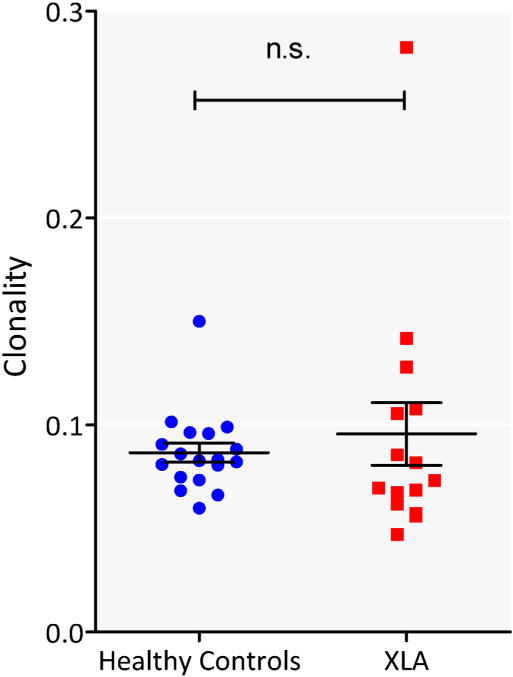
No difference in clonality. Clonality of XLA (red square) and control DNA (blue filled circle) is shown. Horizontal line represents mean and whiskers represent standard error of mean. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Prior studies have demonstrated that a high degree of TCR sequence sharing exists between any two normal persons, irrespective of HLA match [21]. To compare sharing of clones amongst XLA patients and amongst controls, we randomly resampled the DNA from subjects from each cohort separately, 12 at a time, for 100 rounds of comparisons, to determine how many individuals in each cohort shared a TCR, as defined by amino acid sequence. As seen in Fig. 6, XLA patients shared a larger number of sequences compared to the controls. We examined the highly-shared sequences that were present in at least half the subjects or controls to determine the pair-wise overlap in the amino acid repertoire. The results showed that the percent of overlap between XLA subjects was higher than controls (XLA mean overlap = 0.034% ± 0.012%; controls, 0.013% ± 0.004%), a highly significant difference (p < 0.0001). The V and J gene usages also differed between sequences shared by patients and controls (χ2 p-value of <0.0001 and <0.0001 respectively). The proportional distribution of V and J genes in the shared population mirrored their distribution in the total population of productive sequences, suggesting that sharing was not attributable to a few V–J gene combinations that were over represented. While 135 amino acid sequences were shared amongst any 9 or more of the 18 controls, 1966 sequences were shared by any 7 or more XLA subjects. These sequences also occupied a larger fraction of the repertoire (1.66% ± 0.06%) of these subjects as compared to controls (0.21% ± 0.01%, p < 0.0001). However, the number of V genes contributing to each amino acid sequence was not different between the two groups (p = 0.173). As the 15 subjects with XLA contain three sets of two related subjects, we separated the related and unrelated XLA subjects and determined the number of sequences shared by the related pairs or randomly selected pairs of the unrelated XLA subjects or controls. However, there was no significant difference in the sharing of TCR sequences between related or unrelated XLA subjects (p = 0.55), and the pairwise sharing in related or unrelated XLA subject was still significantly greater than controls (p < 0.0001 for both comparisons).
Fig. 6.

Increased sequence sharing in XLA patients. Iterative resampling of XLA (red squares) and control samples (blue filled circles) was performed for 12 individuals at a time. The amino acid sequences of productive clones were compared separately amongst patients or healthy controls. The number of individuals that share each clone in a sample of DNA was determined (x-axis). The mean number of clones (normalized to number of sequences) shared amongst 2 to 12 individuals was then determined (y-axis log10 scale). Error bars represent the 95% confidence interval. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
The role of T cells in B cell development and differentiation is well described; however the importance of B cells in T cell development has not been as well defined. This is the first report that provides a detailed look at the TCR repertoire that develops in the absence of mature B cells in humans. As B cells are present in the normal thymus, bearing MHC class II capable of presenting diverse peptides, these cells are likely to play a role in co-stimulation and the generation of T cell diversity. On this basis, older work showed that mice lacking B cells had a significant decrease in CD4 and CD8 T cells [22]. More recently, several studies have demonstrated that XLA subjects had a reduced T cell memory compartment, including fewer CD4 + CD45RO + and CD4 + CD45ROCXCR5 + T cells [2,23]. While these reports suggest alterations in T cell populations in XLA, the molecular structure of the TCR repertoire in XLA, which develops in the absence of B cells, has not been examined.
Examining the V gene repertoire in XLA we found that for both productive and non-productive sequences, V gene usage, and the V–J combinations were significantly different between XLA and normal controls. In addition, while the CDR3 length was similar for XLA and control samples, the CDR3 region of the XLA T cell receptor contained significantly fewer deletions in V, D, and J gene segments. CDR3 lengths are tightly constrained [24], therefore there was reciprocal decrease in the number of insertions. As this was true for both productive and non-productive clones, these differences appear intrinsic to the V(D)J recombination process, and are not likely to be due to peripheral T cell selection. Sequencing of XLA CDR3s also demonstrated a reduced use of charged residues, more sharing of CDR3 sequences, and almost entirely lack the population of highly modified Vβ gene segments found in control DNA, suggesting both a skewed and contracted T cell repertoire in XLA. While populations of T cell subsets could not be separately examined here (and in fact data on T cell subsets have not been published for healthy controls thus no comparisons are possible), a relative reduction of CD4 + CD45RO + T cells in XLA could potentially contribute to the alterations in CDR3 structure noted here. The alteration in T cell populations may, in turn, result from the well-known lack of germinal centers in peripheral lymph tissues in XLA [5], or, alternatively, from the loss of btk signals, important in the innate immunity such as toll like responses of monocyte and dendritic signaling pathways [25]. Any of these reasons might skew the resulting XLA TCR structure as described here.
As previously reported for other healthy controls, the sequences of control CDR3 DNA revealed a degree of TCR clonal sharing, believed to be due to the emergence of public T cell responses to common pathogens such as viruses [21]. However, for unclear reasons, subjects with XLA exhibited significantly increased clonal sharing as compared to normal subjects. Though some of these individuals were related, excluding them from analyses did not change this result, suggesting that this is disease specific. This may be due to the decreased junctional diversity observed in XLA, with fewer insertions or deletions of n- and p- nucleotides. While other primary immune deficiencies that involve loss of T cell immunity have been shown to have increased clonality in the T cell compartment [13, 16–20], this was not observed In the XLA subjects examined here.
The clinical outcome of a contracted T cell repertoire in XLA is unclear. T cell immunity has been investigated in various studies; while proliferation, T cell vaccine responses, and cytokine production appear globally normal [3,8,9], a few studies have demonstrated selected T cell defects in XLA [4,11]. With immune globulin replacement, subjects with XLA are generally healthy [26], but unusual infections [27,28], unexpected inflammatory complications [29], and clonal CD8 + T cell lymphomas have been reported [30,31], suggesting the possibility of discrete cellular defects. Suggesting that the loss of B cells in general effects the T cell compartment, subjects with rheumatoid arthritis treated with rituximab have a sustained loss of CD4 + T cells and a decreased CD8 + CD45RO +/RA + ratio [32,33].
These studies demonstrate that B cells are likely to be involved in the development and maintenance of normal T cell populations in humans and that loss of these cells, based on genetic defects or temporary pharmacologic elimination, may alter and possibly constrict the available T cell repertoire.
Acknowledgments
We would like to thank Lin Radigan for her technical help without which this study would not have been possible. We would like to thank Harlan Robins and Adaptive Biotech Inc. for their generous support for sequencing and informatics. We would like to acknowledge the invaluable assistance of Lisa J. Edelmann PhD and Mount Sinai Genetic testing Laboratory who provided the control DNA used in this work. This work was supported in part through the computational resources and staff expertise provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai.
This work was supported by grants from the National Institutes of Health (AI 101093, AI-086037, AI-48693, T32-GM007280), The Jeffrey Modell Foundation, and the David S Gottesman Immunology Chair.
References
- 1.Lund FE, Randall TD. Effector and regulatory B cells: modulators of CD4+ T cell immunity. Nat Rev Immunol. 2010;10:236–247. doi: 10.1038/nri2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martini H, Enright V, Perro M, Workman S, Birmelin J, Giorda E, Quinti I, Lougaris V, Baronio M, Warnatz K, Grimbacher B. Importance of B cell co-stimulation in CD4(+) T cell differentiation: X-linked agammaglobulinaemia, a human model. Clin Exp Immunol. 2011;164:381–387. doi: 10.1111/j.1365-2249.2011.04377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paroli M, Accapezzato D, Francavilla V, Insalaco A, Plebani A, Balsano F, Barnaba V. Long-lasting memory-resting and memory-effector CD4+ T cells in human X-linked agammaglobulinemia. Blood. 2002;99:2131–2137. doi: 10.1182/blood.v99.6.2131. [DOI] [PubMed] [Google Scholar]
- 4.Crockard A, Boyd N, McNeill T, McCluskey D. CD4 lymphocyte subset abnormalities associated with impaired delayed cutaneous hypersensitivity reactions in patients with X-linked agammaglobulinaemia. Clin Exp Immunol. 1992;88:29–34. doi: 10.1111/j.1365-2249.1992.tb03034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conley ME. Early defects in B cell development. Curr Opin Allergy Clin Immunol. 2002;2:517–522. doi: 10.1097/00130832-200212000-00007. [DOI] [PubMed] [Google Scholar]
- 6.van de Veerdonk FL, Lauwerys B, Marijnissen RJ, Timmermans K, Di Padova F, Koenders MI, Gutierrez-Roelens I, Durez P, Netea MG, van der Meer JW, van den Berg WB, Joosten LA. The anti-CD20 antibody rituximab reduces the Th17 cell response. Arthritis Rheum. 2011;63:1507–1516. doi: 10.1002/art.30314. [DOI] [PubMed] [Google Scholar]
- 7.Barbosa RR, Silva SP, Silva SL, Melo AC, Pedro E, Barbosa MP, Pereira-Santos MC, Victorino RM, Sousa AE. Primary B-cell deficiencies reveal a link between human IL-17-producing CD4 T-cell homeostasis and B-cell differentiation. PLoS One. 2011;6:e22848. doi: 10.1371/journal.pone.0022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plebani A, Fischer M, Meini A, Duse M, Thon V, Eibl M. T cell activity and cytokine production in X-linked agammaglobulinemia: implications for vaccination strategies. Int Arch Allergy Immunol. 1997;114:90–93. doi: 10.1159/000237649. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Wu Y, Lam KT, Lee P, Tu W, Lau YL. Dendritic and T cell response to influenza is normal in the patients with X-linked agammaglobulinemia. J Clin Immunol. 2012;32:421–429. doi: 10.1007/s10875-011-9639-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amedei A, Romagnani C, Benagiano M, Azzurri A, Fomia F, Torrente F, Plebani A, D’Elios M, Del Prete G. Preferential Th1 profile of T helper cell responses in X-linked (bruton’s) agammaglobulinemia. Eur J Immunol. 2001;31:1927–1934. doi: 10.1002/1521-4141(200106)31:6<1927::aid-immu1927>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 11.Morales-Aza B, Glennie S, Garcez T, Davenport V, Johnston S, Williams N, Heyderman R. Impaired maintenance of naturally acquired T-cell memory to the meningococcus in patients with B-cell immunodeficiency. Blood. 2009;113:4206–4212. doi: 10.1182/blood-2008-08-171587. [DOI] [PubMed] [Google Scholar]
- 12.Smith CIE, Baskin B, Humiregreiff P, Zhou JN, Olsson PG, Maniar HS, Kjellen P, Lambris JD, Christensson B, Hammarstrom L, Bentley D, Vetrie D, Islam KB, Vorechovsky I, Sideras P. Expression of brutons agammaglobulinemia tyrosine kinase gene, BTK, is selectively down-regulated in T-lymphocytes and plasma-cells. J Immunol. 1994;152:557–565. [PubMed] [Google Scholar]
- 13.Ramesh M, Hamm D, Simchoni N, Cunningham-Rundles C. Clonal and constricted T cell repertoire in Common Variable Immune Deficiency. Clin Immunol. 2015 doi: 10.1016/j.clim.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, Steen MS, LaMadrid-Herrmannsfeldt MA, Williamson DW, Livingston RJ, Wu D, Wood BL, Rieder MJ, Robins H. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
- 15.Nadel B, Feeney AJ. Influence of coding-end sequence on coding-end processing in V(D)J recombination. J Immunol. 1995;155:4322–4329. [PubMed] [Google Scholar]
- 16.Lev A, Simon AJ, Broides A, Levi J, Garty BZ, Rosenthal E, Amariglio N, Rechavi G, Somech R. Thymic function in MHC class II-deficient patients. J Allergy Clin Immunol. 2013;131:831–839. doi: 10.1016/j.jaci.2012.10.040. [DOI] [PubMed] [Google Scholar]
- 17.Lev A, Simon AJ, Trakhtenbrot L, Goldstein I, Nagar M, Stepensky P, Rechavi G, Amariglio N, Somech R. Characterizing T cells in SCID patients presenting with reactive or residual T lymphocytes. Clin Dev Immunol. 2012;2012:261470. doi: 10.1155/2012/261470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minegishi Y, Akagi K, Nishikawa K, Okawa H, Yata J. Analysis of the CDR3 region of the rearranged IgH chain genes in patients with severe combined immunodeficiency and severe lymphopenia. J Immunol. 1996;156:4666–4671. [PubMed] [Google Scholar]
- 19.Sarzotti-Kelsoe M, Win CM, Parrott RE, Cooney M, Moser BK, Roberts JL, Sempowski GD, Buckley RH. Thymic output, T-cell diversity, and T-cell function in long-term human SCID chimeras. Blood. 2009;114:1445–1453. doi: 10.1182/blood-2009-01-199323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu X, Almeida JR, Darko S, van der Burg M, Deravin SS, Malech H, Gennery A, Chinn I, Markert ML, Douek DC, Milner JD. Human syndromes of immunodeficiency and dysregulation are characterized by distinct defects in T-cell receptor repertoire development. J Allergy Clin Immunol. 2014;133:1109–1115. e1114. doi: 10.1016/j.jaci.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robins H, Srivastava S, Campregher P, Turtle C, Andriesen J, Riddell S, Carlson C, Warren E. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010;2 doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.João C, Ogle B, Gay-Rabinstein C, Platt J, Cascalho M. B cell-dependent TCR diversification. J Immunol. 2004;172:4709–4716. doi: 10.4049/jimmunol.172.8.4709. [DOI] [PubMed] [Google Scholar]
- 23.Bateman EA, Ayers L, Sadler R, Lucas M, Roberts C, Woods A, Packwood K, Burden J, Harrison D, Kaenzig N, Lee M, Chapel HM, Ferry BL. T cell phenotypes in patients with common variable immunodeficiency disorders: associations with clinical phenotypes in comparison with other groups with recurrent infections. Clin Exp Immunol. 2012;170:202–211. doi: 10.1111/j.1365-2249.2012.04643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rock EP, Sibbald PR, Davis MM, Chien YH. CDR3 length in antigen-specific immune receptors. J Exp Med. 1994;179:323–328. doi: 10.1084/jem.179.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez-Herrera G, Vargas-Hernandez A, Gonzalez-Serrano ME, Berron-Ruiz L, Rodriguez-Alba JC, Espinosa-Rosales F, Santos-Argumedo L. Bruton’s tyrosine kinase—an integral protein of B cell development that also has an essential role in the innate immune system. J Leukoc Biol. 2014;95:243–250. doi: 10.1189/jlb.0513307. [DOI] [PubMed] [Google Scholar]
- 26.Howard V, Greene JM, Pahwa S, Winkelstein JA, Boyle JM, Kocak M, Conley ME. The health status and quality of life of adults with X-linked agammaglobulinemia. Clin Immunol. 2006;118:201–208. doi: 10.1016/j.clim.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Simons E, Spacek LA, Lederman HM, Winkelstein JA. Helicobacter cinaedi bacteremia presenting as macules in an afebrile patient with X-linked agammaglobulinemia. Infection. 2004;32:367–368. doi: 10.1007/s15010-004-3152-7. [DOI] [PubMed] [Google Scholar]
- 28.Turvey SE, Leo SH, Boos A, Deans GD, Prendiville J, Crawford RI, Senger C, Conley ME, Tilley P, Junker A, Janz L, Azana R, Hoang L, Morton TL. Successful approach to treatment of Helicobacter bilis infection in X-linked agammaglobulinemia. J Clin Immunol. 2012;32:1404–1408. doi: 10.1007/s10875-012-9750-8. [DOI] [PubMed] [Google Scholar]
- 29.Hernandez-Trujillo VP, Scalchunes C, Cunningham-Rundles C, Ochs HD, Bonilla FA, Paris K, Yel L, Sullivan KE. Autoimmunity and inflammation in X-linked agammaglobulinemia. J Clin Immunol. 2014;34:627–632. doi: 10.1007/s10875-014-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gualdi G, Lorenzi L, Arisi M, Maffeis M, Soresina A, Marocolo D, Plebani A, Calzavara-Pinton P, Facchetti F. Acral lympho-histiocytic dermatitis in X-linked agammaglobulinemia: a case report showing clonal CD8+ T cells with indolent clinical behaviour. J Eur Acad Dermatol Venereol ( 2014 doi: 10.1111/jdv.12839. [DOI] [PubMed] [Google Scholar]
- 31.Gammon B, Robson A, Deonizio J, Arkin L, Guitart J. CD8(+) granulomatous cutaneous T-cell lymphoma: a potential association with immunodeficiency. J Am Acad Dermatol. 2014;71:555–560. doi: 10.1016/j.jaad.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 32.Diaz-Torne C, Ortiz de Juana MA, Geli C, Canto E, Laiz A, Corominas H, Casademont J, de Llobet JM, Juarez C, Diaz-Lopez C, Vidal S. Rituximab-induced interleukin-15 reduction associated with clinical improvement in rheumatoid arthritis. Immunology. 2014;142:354–362. doi: 10.1111/imm.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melet J, Mulleman D, Goupille P, Ribourtout B, Watier H, Thibault G. Rituximab-induced T cell depletion in patients with rheumatoid arthritis: association with clinical response. Arthritis Rheum. 2013;65:2783–2790. doi: 10.1002/art.38107. [DOI] [PubMed] [Google Scholar]


