Abstract
Bacteria have developed several strategies to communicate and compete with one another in complex environments. One important mechanism of inter-bacterial competition is contact-dependent growth inhibition (CDI), in which some Gram-negative bacteria use CdiB/CdiA two-partner secretion proteins to suppress the growth of neighboring target cells. CdiB is an Omp85 outer-membrane protein that exports and assembles CdiA exoproteins onto the inhibitor-cell surface. CdiA binds to receptors on susceptible bacteria and subsequently delivers its C-terminal toxin domain (CdiA-CT) into the target cell. CDI systems also encode CdiI immunity proteins, which specifically bind to the CdiA-CT and neutralize its toxin activity, thereby protecting CDI+ cells from auto-inhibition. Remarkably, CdiA-CT sequences are highly variable between bacteria, as are the corresponding CdiI immunity proteins. Variations in CDI toxin/immunity proteins suggest that these systems function in bacterial self/nonself recognition and thereby play an important role in microbial communities. In this review, we discuss recent advances in the biochemistry, structural biology and physiology of CDI.
Keywords: biofilms, self/nonself recognition, toxin/immunity proteins, type V secretion
Graphical abstract
Introduction
Bacteria generally live in mixed species communities, forming intricate networks in nearly all environments1. These bacterial networks play important host-associated roles in insects, plants and animals2. Recent studies show that microbial communities influence host physiology and behavior, affecting sexual preference, host development, endocrine signaling, brain/nervous system diseases, immunity and other complex traits3; 4; 5; 6; 7; 8. Within each niche, bacteria communicate and compete with one another for colonization and survival. Much is known about the secreted factors that govern inter-cellular communication through quorum sensing9; 10 and the soluble siderophores used in the competition for iron acquisition11; 12; 13. Similarly, the functions of diffusible antibiotics and bacteriocins, which directly inhibit or kill surrounding cells, have been examined in detail over the past 90 years14. By contrast, we know less about inter-cellular communication/competition systems that rely on direct physical contact. Here, we discuss recent insights into bacterial contact-dependent growth inhibition (CDI) – a mechanism by which growth inhibitory toxin domains are translocated into neighboring target bacteria following direct cell-to-cell contact.
The discovery of contact-dependent growth inhibition (CDI)
CDI was discovered in 2005 during analysis of Escherichia coli strain EC93, which was identified as a dominant isolate from rat intestine15. As with many natural isolates, E. coli EC93 has a significant growth advantage over domesticated E. coli strains and reduces E. coli K-12 viability several thousand-fold after a few hours of co-culture. However, unlike previously characterized inhibitory strains, E. coli EC93 does not secrete diffusible microcin or colicin toxins. Instead, EC93 cells must make direct contact with target cells to inhibit growth15. This new mode of bacterial competition was termed "contact-dependent growth inhibition" or CDI15. The CDI locus was isolated from a cosmid library of E. coli EC93 genomic DNA and found to contain three genes, cdiB, cdiA and cdiI (Fig. 1a), which are sufficient to convert E. coli K-12 into a CDI+ inhibitor strain15. The cdiB and cdiA genes encode a two-partner secretion (TPS) system. TPS proteins constitute a sub-family of type V secretion systems and include a number of important adhesins that are used by bacterial pathogens to adhere to eukaryotic host cells16; 17; 18; 19; 20. As indicated by its name, TPS systems are characterized by two proteins, generically known as TpsB (CdiB) and TpsA (CdiA), that are secreted across the Gram-negative envelope. TpsB partners are outer-membrane proteins of the Omp85 family, and export TpsA exoprotein cargos through the central lumen of their β-barrels21. In many instances, the TpsA partner is thought to remain tethered non-covalently to TpsB, thereby projecting the exoprotein outward from the surface of the secreting bacterium (Fig. 1b). CdiA and other TpsA exoproteins are often quite large and probably form long β-helical filaments that extend several hundred angstroms from the cell surface22; 23. Therefore, the E. coli EC93 CdiB/CdiA proteins represent a specialized TpsB/TpsA pair used to bind and inhibit the growth of neighboring E. coli target cells (Fig. 1c). The cdiI gene encodes a small hydrophobic peptide that acts as an immunity protein to protect E. coli EC93 cells from auto-inhibition. The CdiI protein is also sufficient to confer immunity to E. coli K-12 target strains, allowing them to grow unabated in co-culture with E. coli EC9315; 24. Together, these findings led to the hypothesis that CDI is used to compete with other bacteria for environmental resources.
Fig. 1. Contact-dependent growth inhibition (CDI).
a) CDI+ bacteria carry cdiBAI gene clusters that encode CdiB-CdiA two-partner secretion proteins and CdiI immunity proteins. b) Model for the CdiB/CdiA complex. CdiB is represented by the crystal structure of B. pertussis FhaC (PDB: 3NJT), and CdiA is modeled as concatenated β-helices from E. coli Ag43 (PDB: 4KH3). c) CdiA binds to receptors on neighboring bacteria and delivers its C-terminal toxin domain (red star) into the target cell. If target cells lack immunity (left pathway), then their growth is inhibited. In contrast, CdiI neutralizes the toxin in isogenic CDI+ bacteria, preventing growth inhibition (right pathway).
When CDI was first reported, there were only a handful of known TpsA proteins with significant homology to CdiAEC93 15. As more bacterial genomes were sequenced, it became apparent that cdi gene clusters are found in several α-, β- and γ-proteobacteria25. Probable cdi loci are also found in Fusobacteria and the Negativicutes, which are a class of didermal Firmicutes. Comparative analysis of CdiA proteins from a given species typically shows significant sequence variability over the C-terminal region (CdiA-CT). Moreover, most of the predicted cdiI immunity genes downstream of cdiA do not share significant sequence identity with one another. Together, these observations suggest that CDI growth inhibition activity resides in the CdiA-CT region and that many different toxins are carried by CdiA effectors. Both hypotheses are supported by biochemical analyses of CdiA-CT/CdiI pairs. The CdiA-CTEC93 toxin from E. coli EC93 appears to form pores in membranes, because it dissipates the proton-motive force and reduces ATP synthesis24. Most other characterized CDI toxins have nuclease activities that are blocked by their cognate CdiI immunity proteins25; 26; 27; 28. Thus, CdiA effectors can be armed with an array of C-terminal toxin domains, each associated with a specific CdiI immunity protein to prevent self intoxication. Because immunity proteins do not protect against non-cognate toxins, CDI is thought to mediate inter-strain competition and contribute to self/non-self recognition.
Architecture of CdiA effector proteins
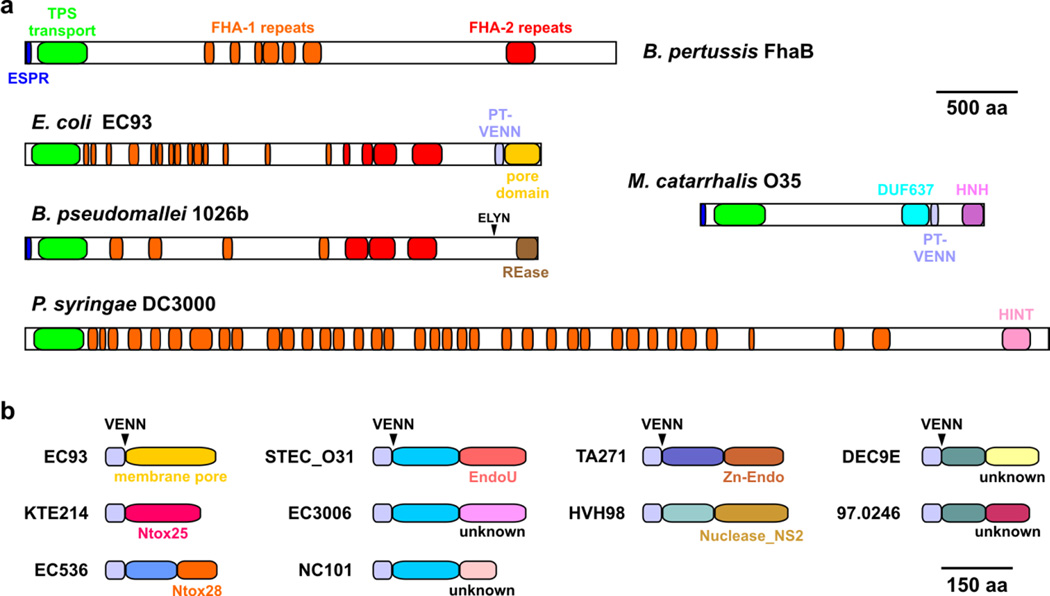
CdiA effectors exhibit considerable diversity in sequence and size depending on bacterial species and strain. The smallest recognizable CdiA protein is from Moraxella catarrhalis O35E (MhaB1, Uniprot: A5JFL2) at ~182 kDa29, and the largest is a ~636 kDa protein encoded by the PSPTO_3229 locus of Pseudomonas syringae DC3000 (Fig. 2a). Despite this heterogeneity, most CdiA proteins have an architecture similar to that of the filamentous hemagglutinin (FHA/FhaB) adhesins of Bordetella species (Fig. 2a). These proteins share N-terminal elements that guide secretion across the cell envelope. All CdiA proteins carry an N-terminal signal sequence for Sec-dependent secretion, though many have an unusual extended signal peptide region (ESPR, Pfam: PF13018) in common with FHA/FhaB (Fig. 2a)30; 31. The N-terminal region also contains the TPS transport domain (also known as the hemagglutinin activation domain, PF05860), which is required for export across the outer membrane. CdiB recognizes the TPS transport domain as it emerges into the periplasm and transports CdiA through the central pore in its β-barrel domain21; 32; 33. CdiA proteins also share FHA-1 (PF05594) and FHA-2 (PF13332) peptide repeats with FHA/FhaB (Fig. 2a). FHA peptide repeats are degenerate 20-residue sequences characterized by alternating polar and non-polar residues34. These sequences are predicted to form right-handed parallel β-helices with a pitch of 4.8 Å per repeat22; 35. Given this conformation, CdiA proteins could extend 40 to 140 nm from the surface of CDI+ bacteria. Domain composition is much more variable over the C-terminal region of CdiA proteins. CdiA proteins from the Pseudomonadaceae and Moraxellaceae often contain the predicted α-helical DUF637 domain of unknown function (PF04830); and those from the Enterobacteriaceae, Moraxellaceae, Pasteurellaceae and Neisseria meningitidis strains usually contain the pretoxin-VENN domain (PF04829), which demarcates the variable CdiA-CT toxin region (Fig. 2a). Although CdiA proteins from other species lack the pretoxin-VENN domain, the CdiA-CT is identifiable as a sequence-variable region at the extreme C-terminus. For example, Burkholderia CdiA proteins show abrupt divergence of the C-terminal region after a conserved (E/Q)LYN peptide motif28; 36. CdiA-CT regions also vary in domain organization. The CdiA-CTEC93 pore-forming toxin appears to be a composed of a single domain, as is the predicted Ntox25 RNase toxin from E. coli strain KTE214 (Fig. 2b)37. Many other CdiA-CTs are composed of two domains. The C-terminus of MhaB1 from M. catarrhalis O35E contains an unannotated domain between the pretoxin-VENN and HNH nuclease domains, and the CdiA-CT region from P. syringae DC3000 contains a predicted hedgehog-intein domain (PF07591) followed by a C-terminal domain of unknown function (Fig. 2a). Analysis of CdiA-CTs from E. coli strains shows that the two constituent domains can be rearranged in new combinations. For example, the CdiA-CT regions from E. coli strains STEC_O31, 3006 and NC101 all share the same N-terminal domain, but have different C-terminal domains (Fig. 2b). The same is true of CdiA-CTs from E. coli strains DEC9E and 97.0246, which contain related N-terminal domains, but divergent C-terminal domains (Fig. 2b). The C-terminal domain of the CdiA-CT region contains the actual toxin activity and these domains are typically nucleases. For example, the C-terminal domain of CdiA-CTSTECO31 is a predicted RNase related to eukaryotic EndoU RNA processing enzymes37; 38 (Fig. 2b), and the C-terminal domains of other experimentally characterized CdiA-CTs have toxic nuclease activities26; 28; 39; 40. Recent genetic studies suggest that the N-terminal domain mediates transport of the tethered nuclease toxin into target bacteria41. Perhaps the most remarkable feature of the CdiA-CT region is its modularity. CdiA-CT sequences can be exchanged between CdiA proteins to generate chimeric effectors. Heterologous CdiA-CTs from Yersinia pestis, Dickeya dadantii, Enterobacter cloacae and Photorhabdus luminescens are all functional when fused at the VENN sequence of E. coli CdiA proteins25; 26; 41; 42. Similarly, functional chimeras have been generated by fusing different Burkholderia CdiA-CT sequences at the (E/Q)LYN motif of CdiAIIBp1026b 28. This modularity allows cdi loci to acquire new toxins through horizontal gene transfer and recombination. In fact, CdiA-CT related toxins are commonly found at the C-terminus of Rhs, LXG and MafB proteins, which represent different classes of secreted effectors that also transfer toxin domains between bacteria37; 40; 43; 44; 45. Together, these observations indicate that different bacterial competition systems share the same pool of toxin/immunity genes through horizontal exchange.
Fig. 2. Domain structure of CdiA.
a) Architectures of putative CdiA proteins. Predicted domain structures are presented for CdiA proteins from E. coli EC93 (Uniprot: Q3YL96), M. catarrhalis O35E (A5JFL2), B. pseudomallei 1026b (I1WVY3) and P. syringae DC3000 (Q880E1). The domain structure of B. pertussis FhaB is also presented for comparison. The extended signal peptide region (ESPR) and TPS transport domain are required for TpsA/CdiA secretion. FHA-1 (PF05594) and FHA-2 (PF13332) peptide repeats were first identified in FhaB and are predicted to form β-helical structures34; 35. In many species, the pretoxin-VENN domain (PF04829) demarcates the variable C-terminal (CT) region, which contains the CDI toxin activity. DUF637 (PF04830) is a domain of unknown function found in a subset of CdiA proteins. HINT indicates the pretoxin-HINT domain (hedgehog intein; PF07591), and HNH indicates a predicted colicin DNase domain (PF12639). The restriction endonuclease (REase) domain at the C-terminus of CdiABp1026b was determined though X-ray crystallography27. b) Domain structures of CdiA-CT regions from different E. coli CdiA proteins. CdiA-CTEC93 and CdiA-CTKTE214 are formed from single domains, but many other CdiA-CTs are composed of two domains. The extreme C-terminal domain usually contains nuclease activity, whereas the function of the variable N-terminal domain is not known. E. coli strains are indicated as superscripts and domains are color-coded to indicate sequence variation. The position of the common VENN peptide motif is indicated.
CdiA-CT activities and structures
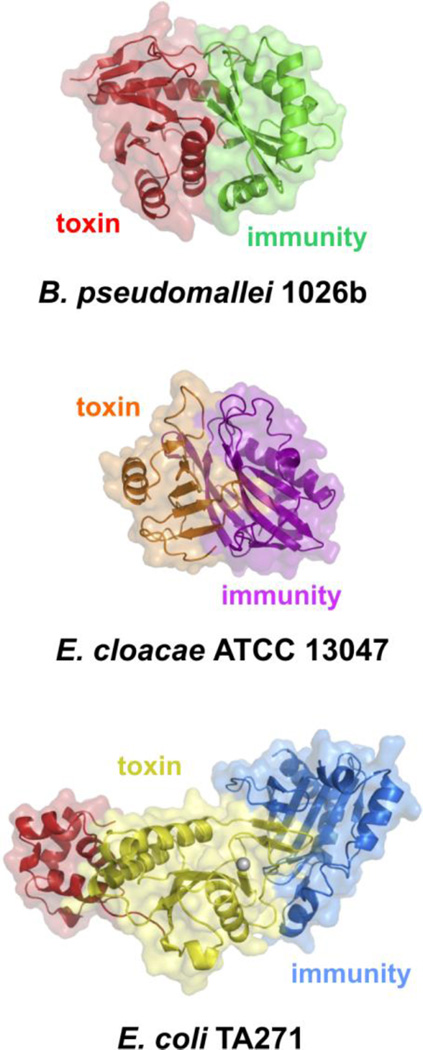
The first CDI toxin to be characterized was the pore-forming CdiA-CTEC93 from E. coli EC9324. DNase and RNase toxins were subsequently identified through sequence homology to bacteriocin nuclease domains. CdiA-CTs encoded by Dda3937_02098 of Dickeya dadantii 3937 and BPSS2053 of Burkholderia pseudomallei K96243 share significant sequence identity with the C-terminal nuclease domains from pyocin S3 and colicin E5, respectively25. Aravind and colleagues have conducted comprehensive sequence analyses of prokaryotic toxin/immunity pairs and predict that CdiA proteins also carry toxin domains with RNA deaminase and peptidase activities37; 38. However, despite these combined experimental and informatic analyses, the activities of most CDI toxins remain unknown. For example, there are at least 20 distinct CdiA-CT sequence types present in E. coli strains, yet only five of these toxin domains currently have Pfam designations (Table 1). Biochemical screens have uncovered the activities of some toxins like CdiA-CTEC869 from E. coli EC869 (ECH7EC869_5848), which cleaves near the 3´-end of tRNAGln (Table 1) and CdiA-CTBp1655 from Burkholderia pseudomallei 1655 (DP51_5554), which cleaves tRNA T-loops28; 46. Structural studies have informed the activities of other CDI toxins. Crystal structures of toxin/immunity protein complexes from B. pseudomallei 1026b (CdiA-CT/CdiIIIBp1026b) and E. coli TA271 (CdiA-CT/CdiITA271) show that each CdiA-CT contains a C-terminal nuclease domain with a type IIS restriction endonuclease fold27 (Fig. 3). Though these nuclease domains are similar in structure, they only share ~18% sequence identity and have distinct substrate specificities. CdiA-CTTA271 is a Zn2+-dependent DNase, whereas CdiA-CTIIBp1026b is an RNase that preferentially cleaves the aminoacyl acceptor stem of tRNAAla molecules27; 28. The CdiA-CTTA271 and CdiA-CTIIBp1026b nuclease domains also interact with their immunity proteins in distinct manners. The Bp1026b toxin and immunity proteins interact through complementary shape and electrostatics, with the immunity protein binding directly over the toxin active site. In contrast, the CdiA-CT/CdiITA271 complex is formed through an unusual β-augmentation interaction in which the toxin extends a β-hairpin into a binding pocket within the immunity proteins to complete a six-stranded anti-parallel sheet27. The nuclease active site remains exposed in the inactive CdiA-CT/CdiITA271 complex, and it is remains unclear how toxin activity is neutralized by the immunity protein.
Table 1.
E. coli CdiA-CT toxin families.
| Class | Strain | Genbank ID | Activity | Pfam designation |
|---|---|---|---|---|
| 1 | E. coli DEC9E | EHW54111.1 | - | - |
| 2 | E. coli 97.0246 | EIG93024.1 | - | - |
| 3 | E. coli EC1738 | EIP59427.1 | - | - |
| 4 | E. coli STEC_O31 | EJK94116.1 | RNase | EndoU (PF14436) |
| 5 | E. coli NC101 | EFM55009.1 | - | - |
| 6 | E. coli HVH 98 | ESK02023.1 | DNase | Endonuclease_NS_2 (PF13930) |
| 7 | E. coli O32:H37 | EIF16908.1 | - | - |
| 8 | E. coli 96.154 | EIH97650.1 | - | DUF4258 (PF14076) |
| 9 | E. coli 3006 | EKI34460.1 | - | - |
| 10 | E. coli EC93 | AAZ57198.1 | pore-forming | - |
| 11 | E. coli 536 | ABG72516.1 | tRNA anticodon nuclease | Ntox28 (PF15605) |
| 12 | E. coli EC869 | EDU89581.1 | cleaves tRNAGln/tRNAAsn | - |
| 13 | E. coli TA271 | EGI36612.1 | DNase | - |
| 14 | E. coli 0.1288 | EKJ55728.1 | - | - |
| 15 | E. coli B088 | EFE64162.1 | - | - |
| 16 | E. coli KTE214 | ELD41871.1 | RNase | Ntox25 (PF15530) |
| 17 | E. coli M605 | EGI12780.1 | - | - |
| 18 | E. coli B799 | EIG47241.1 | - | - |
| 19 | E. coli KTE75 | ELE47736.1 | - | - |
| 20 | E. coli 3-267-03_S3_C2 | KDU01818.1 | - | - |
Fig. 3. Crystal structures of selected CdiA-CT/CdiI complexes.
Toxin/immunity protein complexes from B. pseudomallei 1026 (PDB: 4G6V), E. cloacae ATCC 13047 (PDB: 4NTQ) and E. coli TA271 (PDB: 4G6U) are presented26; 27. All CdiA-CT structures contain only the C-terminal nuclease domain, with the exception of CdiA-CTTA271, for which a portion of the N-terminal domain (shown in red) has been resolved.
Though the nuclease domains of CdiA-CTIIBp1026b and CdiA-CTTA271 share the same fold, other CDI toxins do not (Fig. 3). The C-terminal Ntox21 domain of CdiA-CTECL from Enterobacter cloacae ATCC 13047 has a BECR (barnase/EndoU/colicin E3/RelE) fold and cleaves 16S rRNA between residues A1493 and G1494 (E. coli numbering)26; 37. This activity is identical to that of colicin E3 and the structures of the two nuclease domains superimpose with rmsd of 2.1 Å. However, the CdiA-CTECL active site is distinct from that of colicin E3, suggesting the toxins do not share the same catalytic mechanism. Moreover, CdiIECL shares no sequence or structural homology with the ImmE3 immunity protein. CdiIECL is structurally similar to the Whirly family of single-stranded DNA binding proteins26; 47, but was recently proposed to have evolved from an ancestral SUKH family immunity protein38; 48. Intriguingly, the CdiIo2MC58-1 immunity protein encoded by locus NMB0503 of Neisseria meningiditis MC58 also shares some structural homology with Whirly proteins49. The cognate toxin for this latter immunity protein is a predicted EndoU RNase37; 49. Because EndoU and BECR toxin folds are related37, it appears that the Ntox21 and EndoU nuclease families evolved from a common ancestor that was associated with an SUKH immunity protein.
Genomic organization and orphan toxin/immunity modules
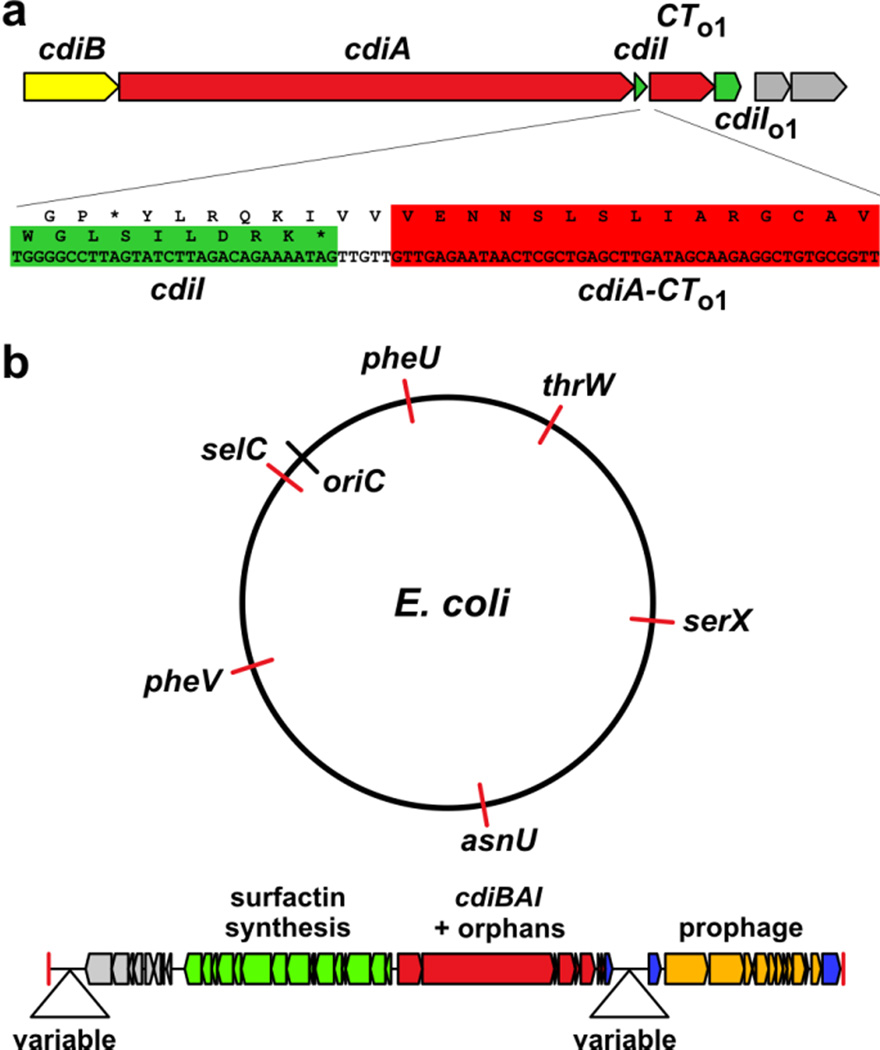
The simplest cdi loci contain only three genes: cdiB, cdiA and cdiI (Fig. 1a), but many systems are linked to additional toxin/immunity protein coding sequences. In E. coli EC93, the cdiI immunity gene is closely followed by sequences that encode a toxin/immunity protein pair that closely resembles CdiA-CT/CdiI from uropathogenic E. coli 53640. The toxin open reading frame (ORF) lacks an initiation codon and begins with Val of the VENN motif (Fig. 4a). These toxin/immunity gene pairs have been termed "orphans" because they resemble the displaced 3´-ends of cdiA cistrons with their associated immunity genes. Orphan regions usually contain predicted transposase and integrase genes as well as insertion sequence (IS) elements40. These observations suggest that orphan regions are hotspots for horizontal gene flow. In this model, new toxin/immunity pairs are collected and eventually undergo recombination with the full-length cdiA gene to deploy a new toxin type on the cell surface. Moreover, CDI systems are typically encoded on genomic and pathogenicity islands, indicating that entire cdi loci are transferred horizontally between bacteria. Many uropathogenic E. coli strains carry a cdi gene cluster on pathogenicity associated island-2 (PAI-2)50. Examination of E. coli strains from the Broad Institute UTI genome sequencing initiatives has revealed another family of genomic islands that contain cdi genes flanked by a putative surfactin biosynthesis operon and prophage sequences (Fig. 4b). These islands are inserted at six different tRNA genes in over 50 E. coli strains (Fig. 4b). Moreover, the cdiA genes on these latter islands encode 11 different CdiA-CT toxins, indicating that smaller scale horizontal gene transfer events can abruptly replace cdiA-CT/cdiI sequences. The cdi genes in B. pseudomallei strains are also found on genomic islands. The cdi genes of B. pseudomallei K96243 are within genomic island 16 (GI-16), and the three cdi clusters of B. pseudomallei 1106a are found in GI-5a.1, GI-11.1 and GI-16.151; 52.
Fig. 4. Genomic organization and horizontal gene transfer.
a) The cdi gene cluster from E. coli EC93 is depicted with the cdiA-CT/cdiIo1 orphan gene pair. The genes rendered in gray encode a predicted IS3-family transposase. The cdiA-CTo1 toxin coding sequence is outlined in red and lacks an initiating methionine codon. b) E. coli cdi genes are found on genomic islands. A family of cdi gene containing islands is inserted at several different tRNA genes in E. coli strains.
In contrast to the cdiBAI gene organization common in γ-proteobacteria and Neisseria, the cluster is arranged in an alternative cdiAIB order in other β-proteobacteria. The latter arrangement dominates in Burkholderia, Commodus and Variovorax, whereas Ralstonia species have both types of loci. The toxin and immunity protein coding sequences remain closely linked, but the cdiAIB organization appears to preclude the accumulation of extensive orphan gene pair arrays. B. pseudomallei serves as a good model for cdi genetic organization in the Burkholderiales because more than a hundred isolates have been sequenced and each strain carries at least one cdi locus28. B. pseudomallei strains contain 10 distinct cdi locus types, each characterized by a unique cdiA-CT/cdiI sequence28; 36, though the type VII and VIII loci encode closely related RNA deaminase toxin/immunity protein pairs37. Most B. pseudomallei clusters contain additional small ORFs between cdiI and cdiB, and many of these sequences encode putative or known immunity proteins (Fig. 5). For example, the intervening gene in the type locus of B. pseudomallei K96243 has weak homology to a predicted CdiI immunity protein from Serratia sp. DD3 (SRDD_21370), and the ORFs from strains NTCC 13179 (class IV) and 576 (class VI) systems are distantly related to the CdiI protein from Enterobacter aerogenes ATCC 13048 (EAE_10270) (Fig. 5). B. pseudomallei type V loci contain a copy of the type IV cdiI immunity gene together with 43 codons of the upstream type IV cdiA-CT coding sequence (Fig. 5). This orphan cdiA-CT/cdiI fragment is flanked by 125-bp direct repeats, strongly suggesting that the immunity gene was transferred horizontally into the type locus. Another interesting example is the type IX locus, which contains two cdiI immunity genes that share 42% sequence identity with each other. These observations indicate that the cdiAIB genetic organization still allows limited transfer and retention of orphan immunity genes.
Fig. 5. Burkholderia pseudomallei cdi loci.
Organization of the 10 cdi sequence types found in strains of B. pseudomallei. The cdiA-CT to cdiB regions are shown from representative strains, with cdiA genes identified by their ordered locus tags. Immunity genes are shown in green, bcpO in brown and predicted orphan immunity genes in light blue. The direct repeats surrounding the orphan cdiI immunity gene within the type locus are shown as arrows.
Several Burkholderia cdi loci, including the B. pseudomallei type VII, VIII, IX and X clusters contain an unusual intervening gene that is not related to known immunity genes (Fig. 5). Cotter and colleagues have termed this gene, bcpO (Burkholderia competition protein O), and have characterized its deletion phenotype in Burkholderia thailandensis36. BcpO is a predicted lipoprotein required for the full inhibition effect of the B. thailandensis CDI system. Closely related homologs of bcpO are associated with cdi loci in other Burkholderia species including B. gladioli, B. glumae and B. phymatum. There are also genes encoding more distantly related lipoproteins in B. phytofirmans, B. ambifaria, B. cenocepacia and Pseudomonas species, though these latter homologues are not linked to cdi gene clusters. The precise function of BcpO remains unclear, but given its presumed localization in the outer membrane, perhaps this lipoprotein collaborates with CdiB to export CdiA onto the inhibitor-cell surface. Many other CDI+ bacteria contain yet another accessory gene that encodes a predicted hemolysin activator (HlyC) or RTX toxin acyltransferase. This gene is found between cdiB and cdiA in strains of E. coli (e.g. ECIG_03035 locus of E. coli M605), Enterobacter cloacae, Ralstonia solanacearum, Pseudomonas fluorescens and other bacteria. The HlyC gene product is related to fatty acyl transferases that modify lysyl residues within hemolysin53; 54. The role of HlyC acyltransferases in CDI has not been explored, but presumably this enzyme modifies either CdiB or CdiA to promote their association with membranes.
Regulation of cdi gene expresssion
The expression of most CDI systems is tightly regulated, with the notable exception of E. coli EC93, which constitutively expresses one of its two cdi gene clusters15. The cdi loci of soft-rot pathogens appear to be expressed only when the bacteria are grown on plant hosts. Collmer and colleagues first characterized cdiIEC16 from Erwinia (Dickeya) chrysanthemi EC16 as a virulence gene (virA) because its disruption significantly reduced virulence on plant hosts55. Now that the EC16 hecBA-virA cluster is recognized as cdiBAIEC16 25; 26, an alternative explanation for these findings is that the locus is only expressed during infection. Thus, the immunity gene is not required under laboratory growth conditions when cdiA is not expressed, but the bacteria either auto-inhibit or inhibit neighboring sibling cells when inoculated onto the host. Similar findings have been obtained with a related plant pathogen, D. dadantii 3937, which expresses its cdi gene cluster on chicory but not in laboratory media25. Cotter and colleagues have shown differential cdi expression within regions of an individual colony36, suggesting stochastic activation of the gene cluster. Greenberg and colleagues have shown further that cdi expression in B. thailandensis E264 is induced in response to acylhomoserine signaling56. However, quorum sensing does not regulate the two cdiA genes (BP1026B_I2481 and BP1026B_II2207) in closely related B. pseudomallei 1026b57. Given the energy required to synthesize large CdiA exoproteins and their exposure to immune surveillance systems, most bacteria probably regulate expression to ensure that cdi genes are only expressed at high cell densities when cell-cell contact is favored.
Recognition of target cells and toxin delivery
CDI requires direct interaction between inhibitor and target bacteria, suggesting that CdiA proteins recognize receptors on the surface of target cells. Aoki et al. used genetic selections to identify the receptor for CdiAEC93, reasoning that disruption of the receptor gene would confer resistance to CDI (CDIR)58. This approach led to the isolation of the E. coli bamA101 mutant, which carries a transposon insertion that decreases bamA expression approximately five-fold. BamA is an essential outer membrane β-barrel protein that forms the core of the β-barrel assembly machine (BAM) complex. The BAM complex is required for the biogenesis of all β-barrel outer-membrane proteins (OMP) in Gram-negative bacteria and eukaryotic plastids59; 60; 61; 62; 63. Several observations indicate that BamA is the receptor for CdiAEC93. First, other components of the BAM complex are not required for CDI, and a biogenesis inactive version of BamA lacking its POTRA-3 domain still complements the bamA101 mutation with respect to CDI sensitivity58. Second, antibodies to BamA block the binding of CDIEC93 inhibitor cells to target bacteria and also protect target cells from growth inhibition58. Finally, allelic exchange of E. coli bamA with the bamA genes of other enterobacterial species renders E. coli cells completely resistant to CDIEC93 46; 64; 65. The latter BamA-replacement strategy facilitated localization of the CdiAEC93 binding site to extracellular loops L6 and L7 of BamA64. These loop sequences vary considerably between different species, restricting the CDIEC93 target-cell range to E. coli. Because most cell-surface epitopes are variable due to strong positive selection, these observations suggest that other CdiA proteins probably have narrow target-cell ranges. However, there are two examples where CDI acts across species. Enterobacter cloacae ATCC 13047 can inhibit E. coli K-12 derivatives using its CDI system26, and the CDIIIBp1026b system from B. pseudomallei 1026b inhibits closely related Burkholderia thailandensis E26428; 66. The receptors for these latter CdiA proteins have not been identified, but there is some evidence that lipopolysaccharide (LPS) may serve as a receptor for CdiAIIBp1026b. Disruption of the BTH_I0986 locus in B. thailandensis target bacteria confers resistance to CDIIIBp1026b 66. The BTH_I0986 gene encodes a predicted LPS glycosyltransferase, and ΔBTH_I0986 mutants have altered LPS and show decreased binding to inhibitor cells that express CdiAIIBp1026 66. Thus, it seems likely that CdiA effectors exploit several different cell-surface receptors to identify susceptible target cells.
Many CDI toxins are nucleases and therefore must translocate into the target-cell cytoplasm to reach their substrates. CdiA-CT fragments have been detected inside target bacteria directly using immunofluorescence microscopy42 and indirectly by monitoring nuclease activities26; 46. Thus, C-terminal toxin domains must be transported across both outer- and inner-membranes of target bacteria. Recent work shows that CDI toxin translocation requires the proton gradient across the inner membrane of target bacteria46. This gradient generates an electrochemical potential called the proton motive force (pmf), which is used to power diverse processes in bacteria including the import of small molecules and flagellum rotation. Colicins also require the pmf to enter bacteria, using pmf-energized Tol or Ton systems for transport across the outer membrane14. However, tol and ton mutants are not resistant to CDI, and there is evidence that the pmf is not required for CdiA-CT translocation across the target-cell outer membrane46. CdiA proteins are easily removed from the surface of inhibitor cells with extracellular protease; but the CdiA-CT toxin is protected from protease when inhibitor cells are bound to target bacteria in the presence of uncoupling agents, which dissipate the pmf. Under these conditions, much of the CdiA filament is still degraded by extracellular protease, converting the inhibitor-target cell aggregates into a suspension of individual cells. Remarkably, toxin activity is detectable within target cells shortly after the cells are washed to remove the uncoupling agent. This result suggests that the CdiA-CT is transferred into the periplasm of target bacteria, protecting it from extracellular protease. Further, toxin can apparently dwell in the target-cell periplasm for several minutes and remain competent to resume translocation once the pmf is reestablished. Together, these findings indicate that the pmf is required for the final translocation step across the target-cell inner membrane.
Contact-dependent growth inhibition in Gram-positive bacteria
CdiB/CdiA proteins have evolved to transport cargo exoproteins across the Gram-negative outer membrane and therefore are not found in Gram-positive bacteria, which are surrounded by a single membrane and thick peptidoglycan layer. However, there is an analogous toxin-delivery system commonly found in Bacillus and Listeria species. Wall-associated protein A (WapA) was first identified in Bacillus subtilis as an abundant secreted protein that associates non-covalently with the peptidoglycan wall67. WapA exhibits a number of features in parallel with CdiA effector proteins. Both proteins are cell-surface associated and very large. WapA from B. subtilis 168 has a predicted molecular mass of 258 kDa, though the full-length chain is difficult to detect and cleavage products are far more abundant67; 68. WapA also contains peptide-repeat regions, but the repeats are similar to YD-peptide/Rhs sequences rather than the FHA repeats of CdiA. Most importantly, the C-terminal region of WapA is highly variable between different strains of B. subtilis and these domains have toxic nuclease activities44. All of the characterized WapA-CT toxins have tRNase activities. WapA-CT168 from B. subtilis 168 cleaves four nucleotides from the 3´-end of tRNA molecules, whereas the toxins from B. subtilis natto and B. subtilis T-UB-10 specifically cleave near the anticodon loops of tRNAGlu and tRNASer, respectively44. Like cdi loci, each wapA gene is closely linked to a downstream immunity gene that is responsible for protecting the cell from WapA-CT toxin activity. wapAI genes are found in many different strains of B. subtilis, B. cereus, B. thringiensis, B. anthracis, B. amyloliquefaciens, Geobacillus sp., Anoxybacillus sp. and Listeria monocytogenes, suggesting that these systems play important roles in self/nonself recognition in Gram-positive bacteria. Because this growth inhibition appears to require direct contact between inhibitor and target cells44, WapA presumably binds to cell-surface receptors and translocates its C-terminal nuclease domains into the cytoplasm.
CDI and cooperative behavior
Although much of the work on CDI has focused on inter-cellular competition, these systems also contribute to cooperative group activities. Collmer and colleagues provided the first evidence for this function before the growth inhibition activity of CdiA proteins was appreciated. They found that HecA (CdiAEC16) is required for the pathogenesis of E. chrysanthemi EC16 on plant hosts69. hecA mutants have defects in adherence to plant epidermal cells and do not auto-aggregate like wild-type cells. Similar findings have been reported for Neisseria meningitidis, Xylella fastidiosa and Xanthomonas axonopodis70; 71; 72; 73; 74, suggesting that inter-cellular adhesion between isogenic cells contributes to host colonization and infection. These latter reports also show that cdiA mutants are defective for biofilm formation, a finding that has been confirmed and extended in B. thailandensis E264 and E. coli EC93 (Fig. 6)36; 65; 75; 76. Using Burkholderia thailandensis E264 as a model, Cotter and colleagues have shown that cdiA and cdiB mutants have profound biofilm defects36. Intriguingly, point mutations that ablate the nuclease activity of the CdiA-CTE264 toxin domain also interfere with biofilm formation76. Although toxin activity is required for biofilm formation, cell killing is not, suggesting that the exchange of the CdiA-CTE264 between isogenic (and immune) cells could influence gene expression to promote biofilms76. Of course, the cell-cell adhesion activity of CdiA proteins also promotes the biofilm lifestyle. Recent work with E. coli EC93 shows that CdiA binding to the BamA receptor is important for the formation of mature biofilms. However, EC93 cells that express a heterologous bamA gene, and therefore lack a suitable receptor for CdiAEC93, still form biofilm structures65. CdiAEC93 contains an additional adhesin domain that probably mediates homotypic interactions and facilitates auto-aggregation independently of the BamA receptor65. Together, these reports show that CDI promotes collective behaviors directly through cell-cell adhesion, but also indirectly through uncharacterized signaling pathways. Thus, CDI promotes the establishment of kin communities, where isogenic cells collaborate to form biofilms, and the growth inhibition activity of these systems excludes non-identical bacteria from participating in the group behavior75.
Fig. 6. E. coli EC93 ΔcdiA mutants are defective for biofilm formation.
Biofilms of wild-type (cdiA+) and ΔcdiA strains of E. coli EC93 after 24 h incubation in a flow-cell. Inset scale bars equal 40 µm. The left panels show three-dimensional reconstructions of biofilm structures. Images courtesy of Loni Townsley and Fitnat Yildiz (UC Santa Cruz).
Highlights.
Contact-dependent growth inhibition (CDI) systems mediate inter-bacterial competition.
CdiA effector proteins carry a variety of C-terminal toxin domains.
CDI toxins are specifically neutralized by cognate immunity proteins.
CDI systems also contribute to community structure by promoting biofilm formation.
Acknowledgments
We thank Loni Townsley and Fitnat Yildiz for providing images of CDI-dependent biofilms. CDI research in the Goulding, Low and Hayes laboratories is supported by grant U01 GM102318 (D.A.L. & C.S.H.) from the National Institutes of Health. J.L.E.W. was supported by National Science Foundation Graduate Research Fellowship DGE-1144085, and Z.C.R. was supported by the Tri-Counties Blood Bank Postdoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. Microbial diversity in the deep sea and the underexplored "rare biosphere". Proc Natl Acad Sci U S A. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engel P, Moran NA. The gut microbiota of insects - diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 3.Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2010;107:20051–20056. doi: 10.1073/pnas.1009906107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol. 2013;14:668–675. doi: 10.1038/ni.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt C. Mental health: thinking from the gut. Nature. 2015;518:S12–S15. doi: 10.1038/518S13a. [DOI] [PubMed] [Google Scholar]
- 6.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, Patterson PH, Mazmanian SK. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, Alam A, Gates CL, Wu H, Swanson PA, Lambeth JD, Denning PW, Neish AS. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 2013;32:3017–3028. doi: 10.1038/emboj.2013.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Mello C, Ronaghan N, Zaheer R, Dicay M, Le T, MacNaughton WK, Surrette MG, Swain MG. Probiotics Improve Inflammation-Associated Sickness Behavior by Altering Communication between the Peripheral Immune System and the Brain. J Neurosci. 2015;35:10821–10830. doi: 10.1523/JNEUROSCI.0575-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2 doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saha R, Saha N, Donofrio RS, Bestervelt LL. Microbial siderophores: a mini review. J Basic Microbiol. 2013;53:303–317. doi: 10.1002/jobm.201100552. [DOI] [PubMed] [Google Scholar]
- 12.Ahmed E, Holmstrom SJ. Siderophores in environmental research: roles and applications. Microb Biotechnol. 2014;7:196–208. doi: 10.1111/1751-7915.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buckling A, Harrison F, Vos M, Brockhurst MA, Gardner A, West SA, Griffin A. Siderophore-mediated cooperation and virulence in Pseudomonas aeruginosa. FEMS Microbiol Ecol. 2007;62:135–141. doi: 10.1111/j.1574-6941.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- 14.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- 16.Cotter PA, Yuk MH, Mattoo S, Akerley BJ, Boschwitz J, Relman DA, Miller JF. Filamentous hemagglutinin of Bordetella bronchiseptica is required for efficient establishment of tracheal colonization. Infect Immun. 1998;66:5921–5929. doi: 10.1128/iai.66.12.5921-5929.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishibashi Y, Claus S, Relman DA. Bordetella pertussis filamentous hemagglutinin interacts with a leukocyte signal transduction complex and stimulates bacterial adherence to monocyte CR3 (CD11b/CD18) J Exp Med. 1994;180:1225–1233. doi: 10.1084/jem.180.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Relman D, Tuomanen E, Falkow S, Golenbock DT, Saukkonen K, Wright SD. Recognition of a bacterial adhesion by an integrin: macrophage CR3 (alpha M beta 2, CD11b/CD18) binds filamentous hemagglutinin of Bordetella pertussis. Cell. 1990;61:1375–1382. doi: 10.1016/0092-8674(90)90701-f. [DOI] [PubMed] [Google Scholar]
- 19.Noel GJ, Barenkamp SJ, St Geme JW, 3rd, Haining WN, Mosser DM. High-molecular-weight surface-exposed proteins of Haemophilus influenzae mediate binding to macrophages. J Infect Dis. 1994;169:425–429. doi: 10.1093/infdis/169.2.425. [DOI] [PubMed] [Google Scholar]
- 20.Noel GJ, Love DC, Mosser DM. High-molecular-weight proteins of nontypeable Haemophilus influenzae mediate bacterial adhesion to cellular proteoglycans. Infect Immun. 1994;62:4028–4033. doi: 10.1128/iai.62.9.4028-4033.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baud C, Guerin J, Petit E, Lesne E, Dupre E, Locht C, Jacob-Dubuisson F. Translocation path of a substrate protein through its Omp85 transporter. Nat Commun. 2014;5:5271. doi: 10.1038/ncomms6271. [DOI] [PubMed] [Google Scholar]
- 22.Makhov AM, Hannah JH, Brennan MJ, Trus BL, Kocsis E, Conway JF, Wingfield PT, Simon MN, Steven AC. Filamentous hemagglutinin of Bordetella pertussis A bacterial adhesin formed as a 50-nm monomeric rigid rod based on a 19-residue repeat motif rich in beta strands and turns. J Mol Biol. 1994;241:110–124. doi: 10.1006/jmbi.1994.1478. [DOI] [PubMed] [Google Scholar]
- 23.Clantin B, Delattre AS, Rucktooa P, Saint N, Meli AC, Locht C, Jacob-Dubuisson F, Villeret V. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science. 2007;317:957–961. doi: 10.1126/science.1143860. [DOI] [PubMed] [Google Scholar]
- 24.Aoki SK, Webb JS, Braaten BA, Low DA. Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli. J Bacteriol. 2009;191:1777–1786. doi: 10.1128/JB.01437-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, Low DA. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beck CM, Morse RP, Cunningham DA, Iniguez A, Low DA, Goulding CW, Hayes CS. CdiA from Enterobacter cloacae delivers a toxic ribosomal RNase into target bacteria. Structure. 2014;22:707–718. doi: 10.1016/j.str.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morse RP, Nikolakakis KC, Willett JL, Gerrick E, Low DA, Hayes CS, Goulding CW. Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems. Proc Natl Acad Sci U S A. 2012;109:21480–21485. doi: 10.1073/pnas.1216238110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikolakakis K, Amber S, Wilbur JS, Diner EJ, Aoki SK, Poole SJ, Tuanyok A, Keim PS, Peacock S, Hayes CS, Low DA. The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems. Mol Microbiol. 2012;84:516–529. doi: 10.1111/j.1365-2958.2012.08039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balder R, Hassel J, Lipski S, Lafontaine ER. Moraxella catarrhalis strain O35E expresses two filamentous hemagglutinin-like proteins that mediate adherence to human epithelial cells. Infect Immun. 2007;75:2765–2775. doi: 10.1128/IAI.00079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desvaux M, Cooper LM, Filenko NA, Scott-Tucker A, Turner SM, Cole JA, Henderson IR. The unusual extended signal peptide region of the type V secretion system is phylogenetically restricted. FEMS Microbiol Lett. 2006;264:22–30. doi: 10.1111/j.1574-6968.2006.00425.x. [DOI] [PubMed] [Google Scholar]
- 31.Desvaux M, Scott-Tucker A, Turner SM, Cooper LM, Huber D, Nataro JP, Henderson IR. A conserved extended signal peptide region directs posttranslational protein translocation via a novel mechanism. Microbiology. 2007;153:59–70. doi: 10.1099/mic.0.29091-0. [DOI] [PubMed] [Google Scholar]
- 32.Delattre AS, Saint N, Clantin B, Willery E, Lippens G, Locht C, Villeret V, Jacob-Dubuisson F. Substrate recognition by the POTRA domains of TpsB transporter FhaC. Mol Microbiol. 2011;81:99–112. doi: 10.1111/j.1365-2958.2011.07680.x. [DOI] [PubMed] [Google Scholar]
- 33.Norell D, Heuck A, Tran-Thi TA, Gotzke H, Jacob-Dubuisson F, Clausen T, Daley DO, Braun V, Muller M, Fan E. Versatile in vitro system to study translocation and functional integration of bacterial outer membrane proteins. Nat Commun. 2014;5:5396. doi: 10.1038/ncomms6396. [DOI] [PubMed] [Google Scholar]
- 34.Relman DA, Domenighini M, Tuomanen E, Rappuoli R, Falkow S. Filamentous hemagglutinin of Bordetella pertussis: nucleotide sequence and crucial role in adherence. Proc Natl Acad Sci U S A. 1989;86:2637–2641. doi: 10.1073/pnas.86.8.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kajava AV, Cheng N, Cleaver R, Kessel M, Simon MN, Willery E, Jacob-Dubuisson F, Locht C, Steven AC. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol Microbiol. 2001;42:279–292. doi: 10.1046/j.1365-2958.2001.02598.x. [DOI] [PubMed] [Google Scholar]
- 36.Anderson MS, Garcia EC, Cotter PA. The Burkholderia bcpAIOB genes define unique classes of two-partner secretion and contact dependent growth inhibition systems. PLoS Genet. 2012;8:e1002877. doi: 10.1371/journal.pgen.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang D, Iyer LM, Aravind L. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 2011;39:4532–4552. doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beck CM, Diner EJ, Kim JJ, Low DA, Hayes CS. The F pilus mediates a novel pathway of CDI toxin import. Mol Microbiol. 2014;93:276–290. doi: 10.1111/mmi.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poole SJ, Diner EJ, Aoki SK, Braaten BA, t'Kint de Roodenbeke C, Low DA, Hayes CS. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet. 2011;7:e1002217. doi: 10.1371/journal.pgen.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willett JLE, Gucinski GC, Fatherree JP, Low DA, Hayes CS. Contact-dependent growth inhibition toxin exploit mulitple independent cell-entry pathways. Proc Natl Acad Sci U S A. 2015;112:11341–11346. doi: 10.1073/pnas.1512124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webb JS, Nikolakakis KC, Willett JL, Aoki SK, Hayes CS, Low DA. Delivery of CdiA nuclease toxins into target cells during contact-dependent growth inhibition. PLoS ONE. 2013;8:e57609. doi: 10.1371/journal.pone.0057609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holberger LE, Garza-Sanchez F, Lamoureux J, Low DA, Hayes CS. A novel family of toxin/antitoxin proteins in Bacillus species. FEBS Lett. 2012;586:132–136. doi: 10.1016/j.febslet.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koskiniemi S, Lamoureux JG, Nikolakakis KC, t'Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A. 2013;110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jamet A, Jousset AB, Euphrasie D, Mukorako P, Boucharlat A, Ducousso A, Charbit A, Nassif X. A new family of secreted toxins in pathogenic Neisseria species. PLoS Pathog. 2015;11:e1004592. doi: 10.1371/journal.ppat.1004592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruhe ZC, Nguyen JY, Beck CM, Low DA, Hayes CS. The proton-motive force is required for translocation of CDI toxins across the inner membrane of target bacteria. Mol Microbiol. 2014;94:466–481. doi: 10.1111/mmi.12779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desveaux D, Marechal A, Brisson N. Whirly transcription factors: defense gene regulation and beyond. Trends Plant Sci. 2005;10:95–102. doi: 10.1016/j.tplants.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 48.Zhang D, Iyer LM, Burroughs AM, Aravind L. Resilience of biochemical activity in protein domains in the face of structural divergence. Curr Opin Struct Biol. 2014;26:92–103. doi: 10.1016/j.sbi.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan K, Johnson PM, Stols L, Boubion B, Eschenfeldt W, Babnigg G, Hayes CS, Joachimiak A, Goulding CW. The structure of a contact-dependent growth-inhibition (CDI) immunity protein from Neisseria meningitidis MC58. Acta Crystallogr F Struct Biol Commun. 2015;71:702–709. doi: 10.1107/S2053230X15006585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobrindt U, Blum-Oehler G, Nagy G, Schneider G, Johann A, Gottschalk G, Hacker J. Genetic structure and distribution of four pathogenicity islands (PAI I(536) to PAI IV(536)) of uropathogenic Escherichia coli strain 536. Infect Immun. 2002;70:6365–6372. doi: 10.1128/IAI.70.11.6365-6372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tuanyok A, Leadem BR, Auerbach RK, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Mayo M, Wuthiekanun V, Brettin TS, Nierman WC, Peacock SJ, Currie BJ, Wagner DM, Keim P. Genomic islands from five strains of Burkholderia pseudomallei. BMC Genomics. 2008;9:566. doi: 10.1186/1471-2164-9-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tumapa S, Holden MT, Vesaratchavest M, Wuthiekanun V, Limmathurotsakul D, Chierakul W, Feil EJ, Currie BJ, Day NP, Nierman WC, Peacock SJ. Burkholderia pseudomallei genome plasticity associated with genomic island variation. BMC Genomics. 2008;9:190. doi: 10.1186/1471-2164-9-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trent MS, Worsham LM, Ernst-Fonberg ML. The biochemistry of hemolysin toxin activation: characterization of HlyC, an internal protein acyltransferase. Biochemistry. 1998;37:4644–4652. doi: 10.1021/bi971588y. [DOI] [PubMed] [Google Scholar]
- 54.Trent MS, Worsham LM, Ernst-Fonberg ML. HlyC, the internal protein acyltransferase that activates hemolysin toxin: the role of conserved tyrosine and arginine residues in enzymatic activity as probed by chemical modification and site-directed mutagenesis. Biochemistry. 1999;38:8831–8838. doi: 10.1021/bi990138y. [DOI] [PubMed] [Google Scholar]
- 55.Rojas CM, Ham JH, Schechter LM, Kim JF, Beer SV, Collmer A. The Erwinia chrysanthemi EC16 hrp/hrc gene cluster encodes an active Hrp type III secretion system that is flanked by virulence genes functionally unrelated to the Hrp system. Mol Plant Microbe Interact. 2004;17:644–653. doi: 10.1094/MPMI.2004.17.6.644. [DOI] [PubMed] [Google Scholar]
- 56.Majerczyk C, Brittnacher M, Jacobs M, Armour CD, Radey M, Schneider E, Phattarasokul S, Bunt R, Greenberg EP. Global analysis of the Burkholderia thailandensis quorum sensing-controlled regulon. J Bacteriol. 2014;196:1412–1424. doi: 10.1128/JB.01405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Majerczyk CD, Brittnacher MJ, Jacobs MA, Armour CD, Radey MC, Bunt R, Hayden HS, Bydalek R, Greenberg EP. Cross-species comparison of the Burkholderia pseudomallei, Burkholderia thailandensis and Burkholderia mallei quorum-sensing regulons. J Bacteriol. 2014;196:3862–3871. doi: 10.1128/JB.01974-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aoki SK, Malinverni JC, Jacoby K, Thomas B, Pamma R, Trinh BN, Remers S, Webb J, Braaten BA, Silhavy TJ, Low DA. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol. 2008;70:323–340. doi: 10.1111/j.1365-2958.2008.06404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 60.Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 62.Voulhoux R, Tommassen J. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res Microbiol. 2004;155:129–135. doi: 10.1016/j.resmic.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 63.Werner J, Misra R. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol Microbiol. 2005;57:1450–1459. doi: 10.1111/j.1365-2958.2005.04775.x. [DOI] [PubMed] [Google Scholar]
- 64.Ruhe ZC, Wallace AB, Low DA, Hayes CS. Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species. mBio. 2013;4:e00480–e00413. doi: 10.1128/mBio.00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruhe ZC, Townsley L, Wallace AB, King A, Van der Woude MW, Low DA, Yildiz FH, Hayes CS. CdiA promotes receptor-independent intercellular adhesion. Mol Microbiol. 2015 doi: 10.1111/mmi.13114. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koskiniemi S, Garza-Sanchez F, Edman N, Chaudhuri S, Poole SJ, Manoil C, Hayes CS, Low DA. Genetic analysis of the CDI pathway from Burkholderia pseudomallei 1026b. PLoS One. 2015;10:e0120265. doi: 10.1371/journal.pone.0120265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Foster SJ. Molecular analysis of three major wall-associated proteins of Bacillus subtilis 168: evidence for processing of the product of a gene encoding a 258 kDa precursor two-domain ligand-binding protein. Mol Microbiol. 1993;8:299–310. doi: 10.1111/j.1365-2958.1993.tb01574.x. [DOI] [PubMed] [Google Scholar]
- 68.Antelmann H, Yamamoto H, Sekiguchi J, Hecker M. Stabilization of cell wall proteins in Bacillus subtilis: a proteomic approach. Proteomics. 2002;2:591–602. doi: 10.1002/1615-9861(200205)2:5<591::AID-PROT591>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 69.Rojas CM, Ham JH, Deng WL, Doyle JJ, Collmer A. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc Natl Acad Sci U S A. 2002;99:13142–13147. doi: 10.1073/pnas.202358699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neil RB, Apicella MA. Role of HrpA in biofilm formation of Neisseria meningitidis and regulation of the hrpBAS transcripts. Infect Immun. 2009;77:2285–2293. doi: 10.1128/IAI.01502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tala A, Progida C, De Stefano M, Cogli L, Spinosa MR, Bucci C, Alifano P. The HrpB-HrpA two-partner secretion system is essential for intracellular survival of Neisseria meningitidis. Cell Microbiol. 2008;10:2461–2482. doi: 10.1111/j.1462-5822.2008.01222.x. [DOI] [PubMed] [Google Scholar]
- 72.Gottig N, Garavaglia BS, Garofalo CG, Orellano EG, Ottado J. A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS One. 2009;4:e4358. doi: 10.1371/journal.pone.0004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guilhabert MR, Kirkpatrick BC. Identification of Xylella fastidiosa antivirulence genes: hemagglutinin adhesins contribute X. fastidiosa biofilm maturation and colonization and attenuate virulence. Mol Plant Microbe Interact. 2005;18:856–868. doi: 10.1094/MPMI-18-0856. [DOI] [PubMed] [Google Scholar]
- 74.Voegel TM, Warren JG, Matsumoto A, Igo MM, Kirkpatrick BC. Localization and characterization of Xylella fastidiosa haemagglutinin adhesins. Microbiology. 2010;156:2172–2179. doi: 10.1099/mic.0.037564-0. [DOI] [PubMed] [Google Scholar]
- 75.Anderson MS, Garcia EC, Cotter PA. Kind discrimination and competitive exclusion mediated by contact-dependent growth inhibition systems shape biofilm community structure. PLoS Pathog. 2014;10:e1004076. doi: 10.1371/journal.ppat.1004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garcia EC, Anderson MS, Hagar JA, Cotter PA. Burkholderia BcpA mediates biofilm formation independently of interbacterial contact-dependent growth inhibition. Mol Microbiol. 2013;89:1213–1225. doi: 10.1111/mmi.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]