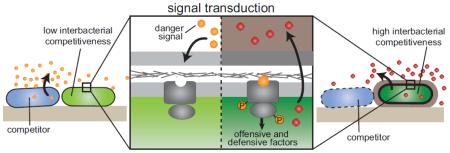
Abstract
Here we propose that bacteria detect and respond to threats posed by other bacteria via an innate immune-like process that we term danger sensing. We find support for this contention by reexamining existing literature from the perspective that intermicrobial antagonism, not opportunistic pathogenesis, is the major evolutionary force shaping the defensive behaviors of most bacteria. We conclude that many bacteria possess danger sensing pathways composed of a danger signal receptor and corresponding signal transduction mechanism that regulate pathways important for survival in the presence of the perceived competitor.
Graphical abstract
Introduction
Viewed classically, an immune system relies on specialized cell types to detect the presence of threats and orchestrate a response. The discovery of the CRISPR/CAS system in bacteria has challenged this long-held perspective and demonstrated that even adaptive immunity is feasible in a single-cell organism [1, 2]. Given that prokaryotes are among the most ancient and successful life forms, it would be surprising if they had not also evolved the ability to detect more generalized forms of danger and take protective measures, akin to the innate arm of immunity in eukaryotes.
Innate immune systems of multicellular organisms function by detecting molecules associated with invasion by pathogens. One such class of molecules consists of conserved cellular components of the pathogens themselves, termed pathogen-associated molecular patterns (PAMPs). Interaction between PAMPs and pattern recognition receptors (PRR) stimulates an inflammatory response during which effector cells are recruited to eliminate the danger and repair tissue. More recently, this paradigm has been extended to include a class of immunostimulatory molecules termed damage associated molecular patterns (DAMPs) [3]. DAMPs arise when endogenous molecules that make up a healthy cell are mislocalized to the extracellular milieu due to necrosis or cell damage. Many of the same PRRs that respond to PAMPs, including Toll- and Nod-like receptors, have the capacity to recognize these so-called danger signals [4, 5]. Eukaryotes thus detect exogenous and endogenous molecules that indicate danger and launch appropriate responses.
For a bacterium to sense and respond to danger, these actions must somehow occur within the context of an individual cell or a distributed population of genetically identical cells. This would also require the capacity to sense a set of cues that are associated with a threat and respond with measures that eliminate the source of danger, prevent an attack, or minimize damage resulting from an attack. Here we assemble an assortment of published studies and reevaluate their significance in light of the hypothesis that bacteria – in a process we term danger sensing – recognize exogenous self- and non-self-derived molecules indicative of the presence of a threat from a competitor and respond in a concerted fashion with protective measures, akin to the eukaryotic innate immune system.
Danger sensing in bacteria
Bacteria face constant threats to their survival, including environmental factors (e.g. extreme temperature, nutrient limitation, ultraviolent radiation) and antagonism by other organisms. For the purposes of this review, we will focus on biotic threats that derive from bacterial competitors. Interbacterial competition has been honed over eons as microbes battle for overlapping niches. Overt antagonism between bacteria is accomplished by diffusible small molecule antibiotics, antimicrobial peptides (AMPs), proteineaceous toxins, and contact-dependent toxin delivery pathways including the type VI secretion system (T6SS) and contact dependent inhibition (CDI), among other mechanisms [6-11]. Given that genes encoding antibacterial factors are virtually universal within bacterial genomes, it would be surprising if bacteria had not evolved generalized cellular programs, i.e., danger sensing pathways, for counteracting them.
Danger sensing is complementary to, but distinct from the concept of “competition sensing”, recently put forth by Cornforth and Foster. To explain the observation that toxin production is a component of many general stress responses, these authors propose that bacteria have evolved to recognize generalized cell damage as an indication of competitors [12]. In contrast, danger sensing is mediated through detection of exogenous molecules that coincide with the presence of a potential threat. As we describe further below, danger signals can derive either from damaged neighboring kin cells, or can be produced by the threatening species. However, in both of these cases, the danger signals themselves do not cause damage to the cells by which they are perceived.
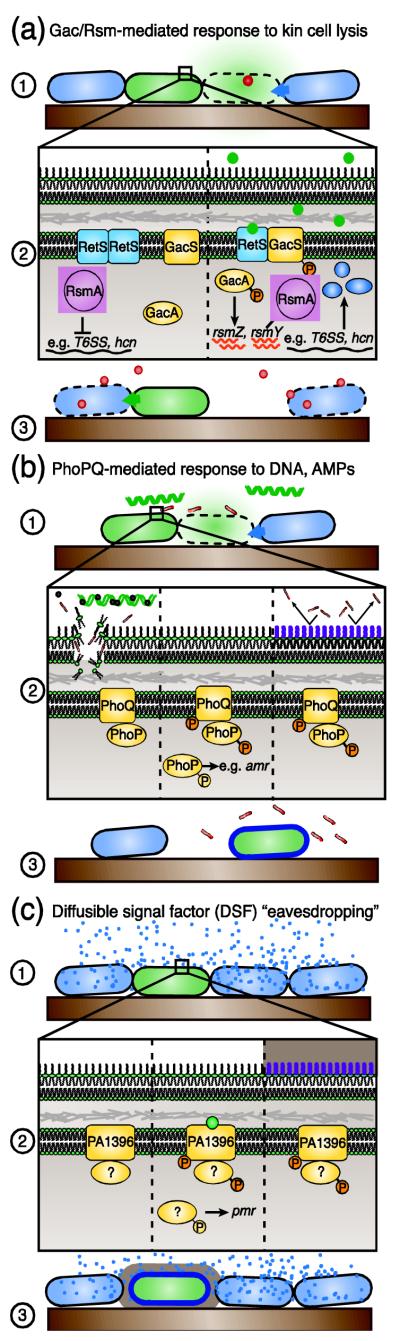
There are three components of the danger sensing process that we delineate. First are the molecular species that indicate a threat (Figure 1-1). Just as eukaryotes have the ability to sense PAMPs and mislocalized self-derived molecules (DAMPs), we find literature demonstrating that bacteria can also sense self- and non-self-derived cues and respond with measures to mitigate the danger. The second component is the receptor and signal transduction mechanism that detect and mediate a response to these danger signals (Figure 1-2). Where the signal transduction mechanism has been characterized, this is most often a two-component regulatory system (TCS), but other types of regulatory pathways are also employed. Last is the response, which is defined by the factor(s) modulated by the danger-responsive signal transduction pathway(s) (Figure 1-3). In the majority of examples we cite, responses consist of the elaboration of both offensive and defensive factors.
Figure 1. Examples of danger sensing pathways.
Danger sensing can be broken into three components: (1) one or more danger cues; (2) a danger sensing signal transduction mechanism; and (3) a danger response regulon.
(a) Gac/Rsm-mediated response to cell lysis. (1) A competitor (blue) lyses a subset of P. aeruginosa cells (green), e.g. through production of a toxin (red circle). (2) Left: The Gac/Rsm danger sensing signal transduction pathway in the inactive state. Right: A mislocalized self-derived molecule from lysed P. aeruginosa (green circles) stimulates this pathway to relieve negative regulation of danger response factors. (3) The outcome of danger sensing is the increased production of factors (brown circles) that target competitor cells and defend against antagonism.
(b) PhoPQ-mediated response to DNA and AMPs. (1) Competitor cells (blue) produce AMPs that damage cell membranes of target cells (green), which may lead to cell lysis and DNA release. (2) Left panel: eDNA from lysed cells chelates cations, reducing the local Mg2+ (black circles) concentration; additionally AMPs themselves can be sensed. Middle: Low Mg2+ concentrations and/or AMPs stimulate PhoPQ. Right panel: The PhoPQ regulon mediates outer membrane modifications that reduce its negative charge. (3) Resulting membrane alterations protect cells from assault by AMPs.
(c) Diffusible signal factor (DSF) “eavesdropping”. (1) The quorum sensing signal, DSF, is produced by S. maltophilia (blue) and sensed by P. aeruginosa (green). (2) Left: A putative sensor kinase (PA1396) in its inactive state. Middle: PA1396 detects DSF and stimulates its regulon. Right: Gene expression leads to outer membrane modifications and increased production of exopolysaccharides. (3) This cellular program may promote survival of P. aeruginosa in the presence of S. maltophilia.
Sensing danger as self (DAMP)
Gac/Rsm-mediated detection of kin cell lysis
One mechanism by which eukaryotic innate immune systems sense danger is through detection of DAMPs. Recently, an analogous system for detecting cellular damage and mounting a counterattack was discovered in the Gram-negative opportunistic pathogen, Pseudomonas aeruginosa (Figure 1A) [13]. During growth in the presence of an antagonistic competitor, a subset of the P. aeruginosa population lyse, releasing cell-associated molecules into the extracellular milieu. One or more of these molecules then stimulates a global regulatory pathway in surrounding cells that promotes the fitness of P. aeruginosa during interbacterial competition. When considered a danger sensing pathway, the molecules released by lysed cells are the danger signals (analogous to eukaryotic DAMPs) that launch a global response when sensed by the intact cells.
The pathway that responds to lysate from kin cells, and that we propose coordinates the danger response, is the Gac/Rsm pathway. At its foundation, Gac/Rsm consists of a sensor kinase and response regulator pair, GacS and GacA, respectively [14]. When GacS is stimulated, GacA positively regulates transcription of the small RNAs (sRNA) rsmY and rsmZ [15]. These sRNAs modulate activity of RsmA and RsmF, RNA binding proteins that interact with recognition sequences in a subset of messenger RNAs (mRNA) and prevent their translation [15, 16]. At sufficient abundance, rsmY and rsmZ sequester RsmA and RsmF from these mRNA targets, allowing translation to proceed. GacS is additionally modulated by upstream orphan sensor kinases [17-20], one of which, RetS, is required for the ability of P. aeruginosa to detect kin cell lysis [13, 17-20]. A point mutation in a conserved residue in the predicted periplasmic signal binding domain of RetS abrogates the ability of P. aeruginosa to sense kin cell lysate, suggesting that this sensor kinase may directly perceive the danger signal(s).
Viewing the Gac/Rsm pathway as a danger response, its regulon encompasses offensive factors (e.g. the T6SS and hydrogen cyanide) that have been demonstrated to directly antagonize the source of the threat and putative defensive factors (e.g. exopolysaccharides) that may provide resistance to antagonism [14, 18, 21]. Additionally, many uncharacterized proteins under Gac/Rsm control are secreted or predicted to localize to the cell surface, consistent with their participation in intercellular interactions [21, 22]. P. aeruginosa cells lacking a functional Gac/Rsm system undergo dramatic cell death in the presence of a bacterial competitor that cannot be attributed to any one component of the pathway, indicating that Gac-mediated danger sensing is a multi-faceted response [13]. Though we now appreciate that many factors under Gac/Rsm control mediate bacterial interactions, the pathway has received most attention as a key mediator of the switch between the chronic and acute infectious states of P. aeruginosa [19, 23]. As its populations are predominantly free-living in the environment, the selective pressure for the duality of factors under Gac/Rsm likely derived from intermicrobial antagonism.
Both the core components of the Gac/Rsm pathway and the hybrid sensor kinases that modulate GacS activity, including RetS, are conserved among pseudomonads, perhaps providing a common mechanism for danger sensing [24, 25]. Supporting this hypothesis, Gac/Rsm modulates the ability of Pseudomonas protegens (previously Pseudomonas fluorescens) to inhibit plant pathogens, thus demonstrating that control of antibacterial factors by Gac/Rsm is not unique to P. aeruginosa [18, 26]. The core components of the Gac/Rsm pathway are more broadly conserved across γ-proteobacteria, but the potential for the pathway to serve in mediating a danger response outside of the pseudomonads is less clear [21]. In Escherichia coli for example, acetate and formate stimulate activity of the GacS homolog, BarA, suggesting the pathway in this organism serves to monitor metabolic status of the population [27]. However, Lapouge and colleagues noted that across the γ-proteobacteria, many Gac regulated factors have relevance to interbacterial interactions: production of antibiotics, extracellular proteases, quorum sensing factors, and exopolysaccharide production [21]. Furthermore, the RsmA homolog, CsrA, negatively regulates T6S in Yersinia pseudotuberculosis [28]. These observations may indicate a more general role for this conserved signaling pathway in mediating danger sensing.
Sensing extracellular DNA (eDNA) via PhoPQ
The PhoPQ pathway has been studied most extensively in the pathogen Salmonella enterica [29, 30]. The sensor kinase PhoQ recognizes cues associated with the intracellular niche of the bacterium (AMPs, low pH, and low Mg2+ concentration), and acting through its cognate response regulator PhoP, induces a multifaceted cellular program that enhances survival in vivo [31-33] (Figure 1B). However, the PhoPQ system is found in many bacteria that are not adapted to an intracellular, or even host-associated, lifestyle. We propose that in these bacteria, PhoPQ could serve as the signal transduction mechanism within a conserved danger sensing pathway. Under certain circumstances, each of known stimuli of PhoPQ listed above could indicate a proximal bacterial threat. One consequence of PhoPQ sensitivity to Mg2+ is that cells respond to the presence of extracellular DNA (eDNA), which can sequester large quantities of the cation [34, 35]. Since eDNA is generated when cells lyse, PhoPQ is in effect responsive to an endogenous signal that is released into the extracellular milieu during intercellular antagonism.
Consistent with a role for this pathway in mediating a response to danger, the PhoPQ regulon induces outer membrane modifications that provide protection from antimicrobials, most notably the AMPs [36]. For example, PhoPQ activates the PmrAB TCS, which induces the covalent modification of lipid A with aminoarabinose and ethanolamine. Collectively, these changes prevent the integration of AMPs into the membrane by an overall reduction of the negative charge of lipopolysaccharide [37]. Other defensive measures are also controlled by PhoPQ homologues in a variety of species, including formation of cellular aggregates by Yersinia pestis, and outer membrane localization of a protease of S. enterica that inactivates a subset of AMPs [38, 39].
Responses to exogenous cues that alert cells to danger
Sensing and responding to antibacterial molecules
In many tissues of eukaryotes, any sign of the presence of another organism indicates that a barrier has been breached and an immune response is warranted. Eukaryotes have thus evolved the capacity to sense conserved yet intrinsically harmless bacterial structures such as flagellin and lipopolysaccharide. In contrast, free-living bacteria rarely exist in isolation from other microbes and therefore responding to such molecules would lead to a constant state of “immune” activation. We reason that this is an explanation for the prevalence of pathways in bacteria that instead sense the effectors of interbacterial antagonism. Such cues signify the proximity of rival cells while simultaneously allowing bacteria to differentiate between friend and foe. Decades of literature regarding bacterial responses to antibiotics support the supposition that toxins can be detected. However, to assess whether such a detection mechanism constitutes a true danger response, it is important to distinguish between the responses that occur due to antibiotic-induced cellular damage (e.g. general stress responses) and those that occur at concentrations in which the antibiotic does not activate a general stress response and instead operates as a signaling molecule.
One type of response to antimicrobials consists of an antibiotic stimulating expression of a single operon encoding a corresponding resistance determinant, a process termed adaptive resistance. The cassettes that encode tetracycline and vancomycin resistance are well characterized examples of this process [40]. In both cases, a negative regulator directly interacts with the antibiotic to relieve repression of resistance genes encoded in the same operon. Specifically, when tetracycline binds to the repressor TetR, expression of the tetracycline efflux pump TetA, is induced [41]. Vancomycin is sensed by the VanRS TCS, which induces a modification of the antibiotic target, a precursor for peptidoglycan synthesis [42]. Accordingly, these gene clusters can be considered to function as highly specific danger sensing pathways wherein a given antibiotic is sensed by a signal transduction mechanism to mediate resistance.
Antimicrobials have also been reported to stimulate more complex regulons that encompass diverse protective mechanisms, including antibiotic production, spore formation, exopolysaccharide synthesis, or a combination of these functions [40, 43, 44]. In one of the first studies of the effects of sub-inhibitory antibiotics on bacterial physiology, it was demonstrated that tobramycin, a natural product synthesized by the bacterium Streptomyces tenebrarius [45], induces production of exopolysaccharides in P. aeruginosa [46]. Increased exopolysaccharide production by P. aeruginosa in turn confers enhanced resistance to tobramycin and other aminoglycoside antibiotics [47]. One interpretation of these findings is that production of exopolysaccharides by P. aeruginosa in response to tobramycin is a danger sensing behavior. However, without a specific sensor identified, it cannot be ruled-out that sub-inhibitory concentrations of tobramycin induce stress response pathways, yielding similar cellular aggregation outcomes [48].
Responding to subinhibitory concentrations of antibiotics by increasing exopolysaccharide production is a response to danger also exhibited by Gram-positive organisms. A recent study demonstrated that the thiocillin antibiotics – a class of peptide antibiotics produced by members of the Streptomyces and Bacilli – induce exopolysaccharide production in Bacillus subtilis [49]. The response occurs by an unknown mechanism that does not require KinD, an established regulator of exopolysaccharide production in B. subtilis. The authors of this study also elegantly established that the signaling function of thiocillin is independent of its toxicity; non-toxic versions of the peptides generated by genetically modifying the biosynthesis locus retained their exopolysaccharide-stimulatory activity.
In other Gram-positive organisms, sub-inhibitory antibiotics can induce a complex developmental program that consists of production of multicellular aggregates, spore formation, and antibiotic synthesis. For example, in Streptomyces coelicolor, the angucycline class of antibiotics induces production of the antibiotic undecylprodigiosin along with aerial hyphae that help distribute spores [50]. Angucyclines are synthesized by other Streptomyces species and are detected in S. coelicolor by a gamma-butyrolactone-like receptor, ScBR2, which in turn directly regulates the response [50]. A related phenotypic response is induced in S. coelicolor in the presence of the predatory bacterium Myxococcus xanthus, but the signal and receptor mediating this effect have not been identified [51]. M. xanthus also induces a range of phenotypic responses in environmental B. subtilis strains. This can include production of the antibiotic bacillaene or the formation of spore-filled aggregates, both of which confer protection from predation [51-54].
Many of the above pathways that we have recast as danger sensing pathways were originally studied in the context of virulence. Bacterial pathogenesis has also been the lens through which most studies have viewed the response of bacteria to the diverse group of antimicrobials known as AMPs, no doubt due to the important role these molecules play in eukaryotic innate immunity. However, AMPs are also synthesized by many species of bacteria. For example, lantibiotics such as nisin are ribosomally encoded, post-translationally modified peptide antibiotics that mediate antagonism between Gram-positive organisms [55]. Polymyxin B and colistin are cationic lipopeptides produced by Bacillus polymyxa that have been widely adopted for clinical uses [56].
Although diverse in structure, AMPs are commonly cationic, and exert toxicity by interacting with the negatively charged bacterial cell surface [57]. As a result of this common mechanism of action, responses to AMPs across bacterial species also share a number of common features. In contrast to the pathways that detect specific antibiotics discussed earlier in this section, the sensors that perceive AMPs are generally more promiscuous [11, 58]. For example, the aps three-component regulator/sensor system in Staphylococcus spp. is stimulated by a variety of cationic AMPs, including molecules produced by eukaryotes and others that are bacterially synthesized [59]. AMP recognition in this system is mediated by an interaction between the positively charged peptides and an extracellular acidic loop present on the membrane-associated sensor kinase ApsS [60]. In Gram-negative bacteria, a major player in the response to AMPs is the PhoPQ TCS introduced above. As in the case of ApsS activation, the positive charge of AMPs is crucial for stimulating activation through PhoPQ [61]. Bacteria can also have multiple, partially redundant pathways for detection of AMPs. In P. aeruginosa, for example, the TCSs PmrAB [62], ParRS [63], and CprRS [64] act along with PhoPQ [30, 65] to sense and respond to a wide range of AMPs.
AMP-sensing pathways in both Gram-negative and Gram-positive species respond by inducing modifications to the cell surface that confer enhanced AMP resistance [11]. The PhoPQ system, for instance, induces the synthesis and incorporation of the positively charged molecule aminoarabinose into lipid A, as described in the previous section [37]. The resulting loss of negative charge on the cell surface can then reduce interaction with AMPs. The Aps system in Staphyloccocus spp. also regulates functions that lead to decreased negative charge on the cell surface – in this case D-alanyl group addition to cell wall teichoic acids and substitution of negatively charged lipid phosphatidylglycerol in the cell membrane with the neutral species lysyl-phosphatidylglycerol [66]. Induction of export systems can also contribute to AMP resistance, as in the case of VraFG upregulation by the Aps system [59].
Owing to the fact that sub-inhibitory concentrations of AMPs induce expression of factors that protect cells from their toxicity, AMP-sensing pathways fall within the danger sensing framework. Indeed, they have previously been likened to a prokaryotic innate immune system [11]. Given the prevalence of AMP production by bacteria, we hypothesize that adaptive resistance pathways evolved primarily in response to bacterial AMPs and later contributed to the ability of environmental organisms to become opportunistic pathogens. Future experiments are needed to assess the extent to which AMP-sensing pathways affect fitness during interbacterial competition.
Responding to signaling molecules produced by other species (‘eavesdropping’)
As described above, direct detection of the effectors of bacterial antagonism is one means by which bacteria could distinguish friend from foe. Quorum sensing signals are a second class of molecules with characteristics that make them attractive candidates to serve a PAMP-like function for bacteria. We use the term “quorum sensing signal” here to refer specifically to molecules perceived by a receptor that are dedicated to intercellular signaling and lead to initiation of a regulatory cascade. Most quorum sensing signals are species or even strain-specific, which makes detection of foreign signals a good proxy for the presence of another species. Additionally, quorum sensing often regulates the production of antagonistic factors; foreign signal detection could thus be a means of detecting the presence of a potentially antagonistic neighbor [67].
A molecule termed diffusible signaling factor (DSF) (cis-2-dodecanoic acid) is a recently characterized quorum sensing signal produced by Xanthomonas spp., Stenotrophomonas maltophilia and Burkholderia cenocepacia [68, 69]. Ryan and colleagues demonstrated that P. aeruginosa detects DSF produced by Stenotrophomonas maltophilia (Figure 1C) using a putative sensor kinase that contains a signal binding domain with homology to known DSF receptors [70]. Upon detection of DSF, P. aeruginosa synthesizes increased quantities of exopolysaccharides and induces expression of the aminoarabinose membrane modification pathway that protects against AMPs. Recasting this pathway from the perspective of danger sensing, DSF is a signal that coincides with the presence of a competitor to stimulate behaviors that can be considered protective, or defensive.
A second example of interspecies signal detection that we propose can be interpreted as danger sensing has been described for the bacterium Chromobacterium violaceum [71]. The acyl-homoserine lactone (acyl-HSL) receptor (LuxR homolog CviR) of this bacterium senses not only its cognate acyl-HSL, but also related signals produced by other species, including Burkholderia thailandensis. Both C. violaceum and B. thailandensis regulate production of antibiotics by quorum sensing. In co-cultures, early activation of the CviR regulon by B. thailandensis provides increased fitness for C. violaceum through its production of antibiotics. This ability to mount an antagonistic response to the detection of a signaling molecule that is used by another species to regulate antibiotic production conforms to our definition of bacterial danger sensing.
Finally, a number of so-called “orphan” or “solo” acyl-HSL receptors (LuxR homologs) have been identified that lack a linked quorum sensing signal synthase [72]. While the function of these proteins remains largely unclear, it has been previously proposed that a subset of orphan LuxR homologues could function in interspecies signaling [72, 73]. We suggest orphan LuxR proteins could function in danger sensing pathways by providing a means of recognizing foreign signals and inducing protective functions, a possibility that remains to be experimentally tested.
Competence as a response to danger
Competence and competition among closely related organisms
Bacteria possess a behavior that is altogether different from those described above, but that we argue also constitutes a response to danger – the uptake of DNA through the process of competence. One cannot a priori categorize competence as either offensive or defensive, as this designation would depend on the precise function conferred by the incoming DNA. If this genetic material promotes DNA repair by serving as a template, competence would be playing a defensive role. Foreign DNA can also become stably incorporated into the genome of the recipient organism, resulting in horizontal gene transfer (HGT). If such horizontally exchanged DNA encodes an immunity determinant to a toxin from a competitor, this would again fulfill a defensive function and promote survival in the presence of competition. In contrast, competence could be thought of as offensive if it promotes the acquisition of factors such as toxins that directly counter a threat. In certain cases, including T6SS- and CDI-associated toxins, horizontal exchange among divergent bacterial species is almost certainly facilitated by the placement of modular toxin domains within repetitive flanking sequences [9, 74, 75]. Such regions frequently include multiple immunity proteins, suggesting that HGT may concomitantly promote expansion of the immunity repertoire of an organism. Experimental evidence suggests that immunity genes acquired within these clusters can be expressed and provide protection against cognate incoming toxins [76]. Below we present a recent example from the literature in which we view the induction of competence as part of a danger sensing pathway.
Vibrio cholerae and competence
It was recently discovered that V. cholerae coordinately regulates the production of antagonistic factors with competence in response to chitin. Here we propose that chitin could be a danger signal that is unlike the cues discussed previously in that it does not derive from bacterial competitors, but nonetheless coincides with their proximity. In the marine environment, Vibrio species preferentially associate with nutritionally rich microenvironments such as the chitinous surfaces of invertebrates or their debris [85, 86]. The resulting elevated bacterial density would necessarily lead to an increase in competition for nutrients and substrate; therefore the presence of chitin may be a cue that indicates a need for increased antagonism and protective measures. Growth on chitin has long been known to induce competence in Vibrio cholerae via the tfoX regulator [87]. Recently, characterization of the full TfoX regulon revealed that additionally, expression of the T6SS is induced [88]. This led Borgeaud and colleagues to hypothesize that similar to the effect of bacteriocin production by S. pneumoniae, T6S-dependent lysis of competitors releases DNA that can be taken up by competence machinery. They went on to demonstrate that expression of T6S genes enhanced the frequency of natural transformation during co-culture with a susceptible competitor.
The coordination of antagonism with competence could facilitate the exchange of genes encoding either toxins that mediate competition or immunity determinants that protect from antagonism. Consistent with this hypothesis, T6S-associated effector gene clusters vary among Vibrio cholerae strains, whereas the T6SS apparatus is conserved [75]. The variability of T6S-associated gene clusters is observed in a wide range of organisms [89]. It will therefore be interesting to investigate whether toxin or immunity acquisition occurs more rapidly in organisms that are naturally competent and coordinate this property with a lysis pathway.
Subjects for future study
Here we have reexamined existing literature from the perspective of a danger sensing hypothesis. We find evidence of pathways that function to sense microbial threats and coordinate responses in a range of bacteria, from professional and opportunistic pathogens (e.g. V. cholerae, S. enterica, P. aeruginosa) to purely environmental organisms (e.g. S. coelicolor, B. subtilis). Based on the apparent prevalence of danger sensing pathways, we predict that targeted investigations will reveal that the capacity for danger sensing is a widespread and integral part of bacterial physiology.
Many of the danger response regulons described in this review are replete with hypothetical genes and genes of unknown function. This may not be happenstance, but rather a result of the tendency to study bacteria in monoculture or through the lens of pathogenesis. The components of danger response regulons are necessarily factors that function in genetically heterogenous populations and thus may have been systematically overlooked or their evolutionary underpinnings obscured in both of these settings. For example, the Gac/Rsm pathway was interpreted as a quorum-like system owing to the observation that in monoculture this pathway responds to self-derived signals. However, the recent investigation of the pathway in the presence of competing bacteria offers an alternative explanation for its sensitivity to self-derived cues and provides a physiological context in which to define the function of factors under RsmA control. We suggest that interrogating the components of danger response regulons within mixed communities that more accurately recapitulate the natural setting in which they operate could be a productive approach to reducing the number of currently uncharacterized proteins.
Highlights.
Bacteria can sense and respond to exogenous molecules associated with danger.
Danger signals can be damage-associated molecules derived from kin cells.
Various pathways transduce danger signals to launch protective cellular responses.
The response to danger may consist of offensive and/or defensive factors.
Acknowledgements
The authors thank members of the Mougous laboratory for insights and discussion. This work was supported by grants from the US National Institutes of Health (AI080609, AI105268, and AI114923) and the Defense Threat Reduction Agency (HDTRA-1-13-014). M.L. was supported by an NIH Cellular and Molecular Training Grant (T32GM007270), J.D.M. holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund, and is a Howard Hughes Medical Institute Investigator.
Abbreviations
- AMP
antimicrobial peptide
- CDI
contact dependent inhibition
- DAMP
damage associated molecular pattern
- DSF
diffusible signal factor
- eDNA
extracellular DNA
- HGT
horizontal gene transfer
- LPS
lipopolysaccharide
- PAMP
pathogen associated molecular pattern
- PRR
pattern recognition receptor
- sRNA
small RNA
- T6SS
type VI secretion system
- TCS
Two component system
Glossary
- Contact dependent inhibition (CDI)
A two-component toxin delivery pathway that mediates competition between closely related bacteria.
- Chitin
An N-acetylglucosamine polymer that makes up the exoskeleton of many invertebrates, including choanoflagellates, insects, and the cell walls of fungi.
- Competence
The ability of a cell to take up extracellular DNA.
- CRISPR/CAS (Clustered regularly interspaced short palindromic repeats/CRISPR associated protein)
A form of adaptive immunity that inactivates and remembers foreign DNA from viruses or plasmids.
- Quorum sensing
A signaling system by which bacteria monitor cell density to coordinate behavior.
- Horizontal gene transfer (HGT)
Incorporation of genes into the genome by a mechanism other than reproduction (vertical gene transfer).
- Natural transformation
The transfer of exogenous genetic material into a cell.
- Two component system
A class of signal transduction pathways consisting of a sensor kinase, that senses environmental cues, and a cognate response regulator, that coordinates a response.
- Type VI secretion system
A bacterial contact-dependent toxin delivery pathway that most commonly mediates interbacterial antagonism, but can also influence host cell interactions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–97. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- [2].Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- [3].Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008;8:279–89. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shi Y, Evans JE, Rock KL. Molecular identification of a danger signal that alerts the immune system to dying cells. Nature. 2003;425:516–21. doi: 10.1038/nature01991. [DOI] [PubMed] [Google Scholar]
- [5].Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–41. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- [6].Hayes CS, Aoki SK, Low DA. Bacterial contact-dependent delivery systems. Annu Rev Genet. 2010;44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- [7].Gabrielsen C, Brede DA, Nes IF, Diep DB. Circular bacteriocins: biosynthesis and mode of action. Appl Environ Microbiol. 2014;80:6854–62. doi: 10.1128/AEM.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jamet A, Nassif X. New Players in the Toxin Field: Polymorphic Toxin Systems in Bacteria. MBio. 2015;6 doi: 10.1128/mBio.00285-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ruhe ZC, Low DA, Hayes CS. Bacterial contact-dependent growth inhibition. Trends Microbiol. 2013;21:230–7. doi: 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Coulthurst SJ, Barnard AM, Salmond GP. Regulation and biosynthesis of carbapenem antibiotics in bacteria. Nat Rev Microbiol. 2005;3:295–306. doi: 10.1038/nrmicro1128. [DOI] [PubMed] [Google Scholar]
- [11].Draper LA, Cotter PD, Hill C, Ross RP. Lantibiotic resistance. Microbiol Mol Biol Rev. 2015;79:171–91. doi: 10.1128/MMBR.00051-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cornforth DM, Foster KR. Competition sensing: the social side of bacterial stress responses. Nature reviews Microbiology. 2013;11:285–93. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- [13].LeRoux M, Kirkpatrick RL, Montauti EI, Tran BQ, Peterson SB, Harding BN, et al. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. Elife. 2015;4 doi: 10.7554/eLife.05701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, et al. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol. 1997;24:309–19. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- [15].Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, et al. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol. 2009;73:434–45. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Marden JN, Diaz MR, Walton WG, Gode CJ, Betts L, Urbanowski ML, et al. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2013;110:15055–60. doi: 10.1073/pnas.1307217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kong W, Chen L, Zhao J, Shen T, Surette MG, Shen L, et al. Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol Microbiol. 2013;88:784–97. doi: 10.1111/mmi.12223. [DOI] [PubMed] [Google Scholar]
- [18].Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–54. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- [19].Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009;23:249–59. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, et al. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A. 2006;103:171–6. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67:241–53. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- [22].Robert-Genthon M, Casabona MG, Neves D, Coute Y, Ciceron F, Elsen S, et al. Unique features of a Pseudomonas aeruginosa alpha2-macroglobulin homolog. MBio. 2013;4 doi: 10.1128/mBio.00309-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ Microbiol. 2011;13:3128–38. doi: 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]
- [24].Records AR, Gross DC. Sensor kinases RetS and LadS regulate Pseudomonas syringae type VI secretion and virulence factors. J Bacteriol. 2010;192:3584–96. doi: 10.1128/JB.00114-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Workentine ML, Chang L, Ceri H, Turner RJ. The GacS-GacA two-component regulatory system of Pseudomonas fluorescens: a bacterial two-hybrid analysis. FEMS Microbiol Lett. 2009;292:50–6. doi: 10.1111/j.1574-6968.2008.01445.x. [DOI] [PubMed] [Google Scholar]
- [26].Kay E, Dubuis C, Haas D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc Natl Acad Sci U S A. 2005;102:17136–41. doi: 10.1073/pnas.0505673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chavez RG, Alvarez AF, Romeo T, Georgellis D. The physiological stimulus for the BarA sensor kinase. J Bacteriol. 2010;192:2009–12. doi: 10.1128/JB.01685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Avican K, Fahlgren A, Huss M, Heroven AK, Beckstette M, Dersch P, et al. Reprogramming of Yersinia from virulent to persistent mode revealed by complex in vivo RNA-seq analysis. PLoS Pathog. 2015;11:e1004600. doi: 10.1371/journal.ppat.1004600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kato A, Groisman EA. Howard Hughes Medical I. The PhoQ/PhoP regulatory network of Salmonella enterica. Adv Exp Med Biol. 2008;631:7–21. doi: 10.1007/978-0-387-78885-2_2. [DOI] [PubMed] [Google Scholar]
- [30].Macfarlane EL, Kwasnicka A, Ochs MM, Hancock RE. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol Microbiol. 1999;34:305–16. doi: 10.1046/j.1365-2958.1999.01600.x. [DOI] [PubMed] [Google Scholar]
- [31].Miller SI, Kukral AM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A. 1989;86:5054–8. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Garcia Vescovi E, Soncini FC, Groisman EA. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–74. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- [33].Groisman EA. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol. 2001;183:1835–42. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Johnson L, Horsman SR, Charron-Mazenod L, Turnbull AL, Mulcahy H, Surette MG, et al. Extracellular DNA-induced antimicrobial peptide resistance in Salmonella enterica serovar Typhimurium. BMC Microbiol. 2013;13:115. doi: 10.1186/1471-2180-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mulcahy H, Charron-Mazenod L, Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008;4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dalebroux ZD, Miller SI. Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr Opin Microbiol. 2014;17:106–13. doi: 10.1016/j.mib.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, et al. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–82. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- [38].Guina T, Yi EC, Wang H, Hackett M, Miller SI. A PhoP-regulated outer membrane protease of Salmonella enterica serovar typhimurium promotes resistance to alpha-helical antimicrobial peptides. J Bacteriol. 2000;182:4077–86. doi: 10.1128/jb.182.14.4077-4086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rebeil R, Jarrett CO, Driver JD, Ernst RK, Oyston PC, Hinnebusch BJ. Induction of the Yersinia pestis PhoP-PhoQ regulatory system in the flea and its role in producing a transmissible infection. J Bacteriol. 2013;195:1920–30. doi: 10.1128/JB.02000-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bernier SP, Surette MG. Concentration-dependent activity of antibiotics in natural environments. Front Microbiol. 2013;4:20. doi: 10.3389/fmicb.2013.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hillen W, Berens C. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu Rev Microbiol. 1994;48:345–69. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- [42].Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis. 2006;42(Suppl 1):S25–34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- [43].Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9:445–53. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- [44].Romero D, Traxler MF, Lopez D, Kolter R. Antibiotics as signal molecules. Chem Rev. 2011;111:5492–505. doi: 10.1021/cr2000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kharel MK, Basnet DB, Lee HC, Liou K, Woo JS, Kim BG, et al. Isolation and characterization of the tobramycin biosynthetic gene cluster from Streptomyces tenebrarius. FEMS Microbiol Lett. 2004;230:185–90. doi: 10.1016/S0378-1097(03)00881-4. [DOI] [PubMed] [Google Scholar]
- [46].Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–5. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- [47].Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GC, et al. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011;7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Oliveria NM, Martinez-Garcia E, Xavier J, Durham WM, Kolter R, Kim W, et al. Biofilm Formation As a Response to Ecological Competition. PLoS Biol. 2015;13:e1002191. doi: 10.1371/journal.pbio.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bleich R, Watrous JD, Dorrestein PC, Bowers AA, Shank EA. Thiopeptide antibiotics stimulate biofilm formation in Bacillus subtilis. Proc Natl Acad Sci U S A. 2015;112:3086–91. doi: 10.1073/pnas.1414272112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang W, Ji J, Li X, Wang J, Li S, Pan G, et al. Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc Natl Acad Sci U S A. 2014;111:5688–93. doi: 10.1073/pnas.1324253111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Perez J, Munoz-Dorado J, Brana AF, Shimkets LJ, Sevillano L, Santamaria RI. Myxococcus xanthus induces actinorhodin overproduction and aerial mycelium formation by Streptomyces coelicolor. Microb Biotechnol. 2011;4:175–83. doi: 10.1111/j.1751-7915.2010.00208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Muller S, Strack SN, Ryan SE, Kearns DB, Kirby JR. Predation by Myxococcus xanthus induces Bacillus subtilis to form spore-filled megastructures. Appl Environ Microbiol. 2015;81:203–10. doi: 10.1128/AEM.02448-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Muller S, Strack SN, Hoefler BC, Straight PD, Kearns DB, Kirby JR. Bacillaene and sporulation protect Bacillus subtilis from predation by Myxococcus xanthus. Appl Environ Microbiol. 2014;80:5603–10. doi: 10.1128/AEM.01621-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Straight PD, Willey JM, Kolter R. Interactions between Streptomyces coelicolor and Bacillus subtilis: Role of surfactants in raising aerial structures. J Bacteriol. 2006;188:4918–25. doi: 10.1128/JB.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].McAuliffe O, Ross RP, Hill C. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol Rev. 2001;25:285–308. doi: 10.1111/j.1574-6976.2001.tb00579.x. [DOI] [PubMed] [Google Scholar]
- [56].Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bahar AA, Ren D. Antimicrobial peptides. Pharmaceuticals (Basel) 2013;6:1543–75. doi: 10.3390/ph6121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol. 2006;4:529–36. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- [59].Li M, Cha DJ, Lai Y, Villaruz AE, Sturdevant DE, Otto M. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol. 2007;66:1136–47. doi: 10.1111/j.1365-2958.2007.05986.x. [DOI] [PubMed] [Google Scholar]
- [60].Li M, Lai Y, Villaruz AE, Cha DJ, Sturdevant DE, Otto M. Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci U S A. 2007;104:9469–74. doi: 10.1073/pnas.0702159104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Prost LR, Miller SI. The Salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cell Microbiol. 2008;10:576–82. doi: 10.1111/j.1462-5822.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- [62].McPhee JB, Lewenza S, Hancock RE. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol. 2003;50:205–17. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- [63].Fernandez L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock RE. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother. 2010;54:3372–82. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Fernandez L, Jenssen H, Bains M, Wiegand I, Gooderham WJ, Hancock RE. The two-component system CprRS senses cationic peptides and triggers adaptive resistance in Pseudomonas aeruginosa independently of ParRS. Antimicrob Agents Chemother. 2012;56:6212–22. doi: 10.1128/AAC.01530-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Macfarlane EL, Kwasnicka A, Hancock RE. Role of Pseudomonas aeruginosa PhoP-phoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology. 2000;146(Pt 10):2543–54. doi: 10.1099/00221287-146-10-2543. [DOI] [PubMed] [Google Scholar]
- [66].Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med. 2001;193:1067–76. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].He YW, Zhang LH. Quorum sensing and virulence regulation in Xanthomonas campestris. FEMS Microbiol Rev. 2008;32:842–57. doi: 10.1111/j.1574-6976.2008.00120.x. [DOI] [PubMed] [Google Scholar]
- [69].Twomey KB, O'Connell OJ, McCarthy Y, Dow JM, O'Toole GA, Plant BJ, et al. Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa. ISME J. 2012;6:939–50. doi: 10.1038/ismej.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ryan RP, Fouhy Y, Garcia BF, Watt SA, Niehaus K, Yang L, et al. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol Microbiol. 2008;68:75–86. doi: 10.1111/j.1365-2958.2008.06132.x. [DOI] [PubMed] [Google Scholar]
- [71].Chandler JR, Heilmann S, Mittler JE, Greenberg EP. Acyl-homoserine lactone-dependent eavesdropping promotes competition in a laboratory co-culture model. ISME J. 2012;6:2219–28. doi: 10.1038/ismej.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Subramoni S, Venturi V. LuxR-family 'solos': bachelor sensors/regulators of signalling molecules. Microbiology. 2009;155:1377–85. doi: 10.1099/mic.0.026849-0. [DOI] [PubMed] [Google Scholar]
- [73].Lee JH, Lequette Y, Greenberg EP. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol Microbiol. 2006;59:602–9. doi: 10.1111/j.1365-2958.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- [74].Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, et al. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe. 2012;11:538–49. doi: 10.1016/j.chom.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Unterweger D, Miyata ST, Bachmann V, Brooks TM, Mullins T, Kostiuk B, et al. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat Commun. 2014;5:3549. doi: 10.1038/ncomms4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Poole SJ, Diner EJ, Aoki SK, Braaten BA, t'Kint de Roodenbeke C, Low DA, et al. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet. 2011;7:e1002217. doi: 10.1371/journal.pgen.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Slager J, Kjos M, Attaiech L, Veening JW. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell. 2014;157:395–406. doi: 10.1016/j.cell.2014.01.068. [DOI] [PubMed] [Google Scholar]
- [78].Claverys JP, Martin B, Havarstein LS. Competence-induced fratricide in streptococci. Mol Microbiol. 2007;64:1423–33. doi: 10.1111/j.1365-2958.2007.05757.x. [DOI] [PubMed] [Google Scholar]
- [79].Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, et al. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol. 2004;51:1051–70. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- [80].Dagkessamanskaia A, Moscoso M, Henard V, Guiral S, Overweg K, Reuter M, et al. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol Microbiol. 2004;51:1071–86. doi: 10.1111/j.1365-2958.2003.03892.x. [DOI] [PubMed] [Google Scholar]
- [81].Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science. 2006;313:89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- [82].Whatmore AM, Barcus VA, Dowson CG. Genetic diversity of the streptococcal competence (com) gene locus. J Bacteriol. 1999;181:3144–54. doi: 10.1128/jb.181.10.3144-3154.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kreth J, Merritt J, Shi W, Qi F. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol Microbiol. 2005;57:392–404. doi: 10.1111/j.1365-2958.2005.04695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Claverys JP, Havarstein LS. Cannibalism and fratricide: mechanisms and raisons d'etre. Nat Rev Microbiol. 2007;5:219–29. doi: 10.1038/nrmicro1613. [DOI] [PubMed] [Google Scholar]
- [85].Takemura AF, Chien DM, Polz MF. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front Microbiol. 2014;5:38. doi: 10.3389/fmicb.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Martinez-Urtaza J, Blanco-Abad V, Rodriguez-Castro A, Ansede-Bermejo J, Miranda A, Rodriguez-Alvarez MX. Ecological determinants of the occurrence and dynamics of Vibrio parahaemolyticus in offshore areas. ISME J. 2012;6:994–1006. doi: 10.1038/ismej.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Yamamoto S, Morita M, Izumiya H, Watanabe H. Chitin disaccharide (GlcNAc)2 induces natural competence in Vibrio cholerae through transcriptional and translational activation of a positive regulatory gene tfoXVC. Gene. 2010;457:42–9. doi: 10.1016/j.gene.2010.03.003. [DOI] [PubMed] [Google Scholar]
- [88].Borgeaud S, Metzger LC, Scrignari T, Blokesch M. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science. 2015;347:63–7. doi: 10.1126/science.1260064. [DOI] [PubMed] [Google Scholar]
- [89].Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nature reviews Microbiology. 2014;12:137–48. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]