Abstract
Cytosolic sulfotransferase 1C2 (SULT1C2) is expressed in the kidney, stomach, and liver of rats; however, the mechanisms regulating expression of this enzyme are not known. We evaluated transcriptional regulation of SULT1C2 by mevalonate (MVA)-derived intermediates in primary cultured rat hepatocytes using several cholesterol synthesis inhibitors. Blocking production of mevalonate with the 3-hydroxy-3-methylglutaryl-CoA reductase inhibitor pravastatin (30 μM), reduced SULT1C2 mRNA content by ∼40% whereas the squalene synthase inhibitor squalestatin (SQ1, 0.1 μM), which causes accumulation of nonsterol isoprenoids, increased mRNA content by 4-fold. Treatment with MVA (10 mM) strongly induced SULT1C2 mRNA by 12-fold, and this effect was blocked by inhibiting squalene epoxidase but not by more distal cholesterol inhibitors, indicating the effects of MVA are mediated by postsqualene metabolites. Using rapid amplification of cDNA ends (RACE), we characterized the 5′ end of SULT1C2 mRNA and used this information to generate constructs for promoter analysis. SQ1 and MVA increased reporter activity by ∼1.6- and 3-fold, respectively, from a construct beginning 49 base pairs (bp) upstream from the longest 5′-RACE product (-3140:-49). Sequence deletions from this construct revealed a hepatocyte nuclear factor 1 (HNF1) element (-2558), and mutation of this element reduced basal (75%) and MVA-induced (30%) reporter activity and attenuated promoter activation following overexpression of HNF1α or 1β. However, the effects of SQ1 were localized to a more proximal promoter region (-281:-49). Collectively, our findings demonstrate that cholesterol biosynthetic intermediates influence SULT1C2 expression in rat primary hepatocytes. Further, HNF1 appears to play an important role in mediating basal and MVA-induced SULT1C2 transcription.
Introduction
The cytosolic sulfotransferase (SULT) family of enzymes catalyzes the transfer of a sulfonate group from 3′-phosphoadenosine 5′-phosphosulfate to a hydroxyl or amino group on an accepting substrate (Strott, 2002). This conjugation reaction, termed sulfonation, is an important step in the biotransformation of many xenobiotic and endogenously produced compounds (Gamage et al., 2006). Sulfonation is generally associated with detoxification, leading to more hydrophilic compounds that are readily excreted in the urine or bile, and a number of pharmaceutical agents are inactivated in this manner (Gamage et al., 2006). In addition to a role in chemical defenses, SULTs are emerging as important mediators of lipid homeostasis (Cook et al., 2009; Bai et al., 2012) as well as endocrine responses, by modulating the biologic activity of various steroid and thyroid hormones (Strott, 2002; Lindsay et al., 2008). Six gene families (SULT1-6) have been identified in mammalian tissues that are divided into subfamilies based on amino acid sequence similarity (Blanchard et al., 2004). Among the major gene families, SULT1 members catalyze the conjugation of phenolic compounds, iodothyronines, catecholamines, and estrogens, whereas the SULT2 family sulfonates dehydroepiandrosterone, pregnenolone, bile acids, and cholesterol (Gamage et al., 2006; Runge-Morris et al., 2013).
Several genes belonging to the SULT1C family have been identified in humans and rodents; however, information regarding their function and regulation is limited (Runge-Morris and Kocarek, 2013). The SULT1C2 gene is localized to chromosome 2q11.2 in humans, 9q11 in rats, and 17 in mice, and orthologs have been cloned, expressed, and partially characterized (Freimuth et al., 2000; Xiangrong et al., 2000; Sugimura et al., 2002). SULT1C2 mRNA has been detected in a variety of human tissues, including liver, kidney, thyroid, stomach, and intestine (Her et al., 1997; Dooley et al., 2000; Stanley et al., 2005), whereas in rodents mRNA is predominately expressed in the kidneys, stomach, and liver (Xiangrong et al., 2000; Alnouti and Klaassen, 2006).
SULT1C2 catalyzes the sulfonation of the prototypical substrate p-nitrophenol as well as the procarcinogen N-hydroxy-2-acetylaminofluorene, suggesting a role in chemical carcinogenesis (Sakakibara et al., 1998; Xiangrong et al., 2000; Allali-Hassani et al., 2007). Additionally, sulfonation of iodothyronines, which reduces receptor binding affinity and accelerates deiodination, has been reported (Li et al., 2000; Strott, 2002; Allali-Hassani et al., 2007). A few studies have indicated that SULT1C2 expression is altered in various disease states. For example, hepatic SULT1C2 is increased as part of the stereotypical stress response after chemical exposure in human primary hepatocytes as well as in liver tissue from individuals with nonalcoholic steatohepatitis (NASH), cirrhosis, and hepatocellular cancer (Grinberg et al., 2014). In rats, SULT1C2 decreased during 2-amino-4,5-diphenylthiazole–induced polycystic kidney disease (Sugimura et al., 2002). Additionally, hepatic SULT1C2 expression is reduced in experimental models of diabetes and obesity-induced steatosis (Almon et al., 2009; Buque et al., 2010). These findings implicate altered expression of SULT1C2 in disease etiology; however transcriptional regulation has not been well characterized and is likely subject to tissue- and species-dependent mechanisms. Given the broad roles of SULTs in lipid, hormone, and xenobiotic metabolism, understanding the factors mediating transcriptional responses has important toxicologic as well as pharmaceutic implications.
In a microarray screen for novel genes differentially regulated by anticholesterol drugs in primary cultured rat hepatocytes, SULT1C2 mRNA was found to be significantly up-regulated by the squalene synthase inhibitor squalestatin 1 (SQ1) (Duniec-Dmuchowski et al., 2012). Squalene synthase catalyzes the first committed step in sterol biosynthesis whereby the branch-point isoprenoid farnesyl pyrophosphate (FPP), is converted to squalene (Fig. 1). Inhibition of squalene synthase causes accumulation of FPP (Jackson and Kocarek, 2008), which can be metabolized further to produce farnesol and several farnesol-derived dicarboxylic acids that are excreted in the urine (Bostedor et al., 1997). Isoprenoids and other cholesterol biosynthetic intermediates, such as oxysterols, function as biologically active signaling molecules that influence drug, lipid, and cholesterol metabolism through activation of one or more nuclear receptors (Kocarek et al., 1998; Edwards et al., 2002; Takahashi et al., 2002; Duniec-Dmuchowski et al., 2009). We previously reported that activators of the vitamin D receptor (VDR), liver X receptor (LXR), and pregnane X receptor (PXR) induce SULT1C2 mRNA and immunoreactive protein in a human colon cancer cell line (Rondini et al., 2014). However, whether similar signaling pathways regulate rat SULT1C2 has not been previously determined.
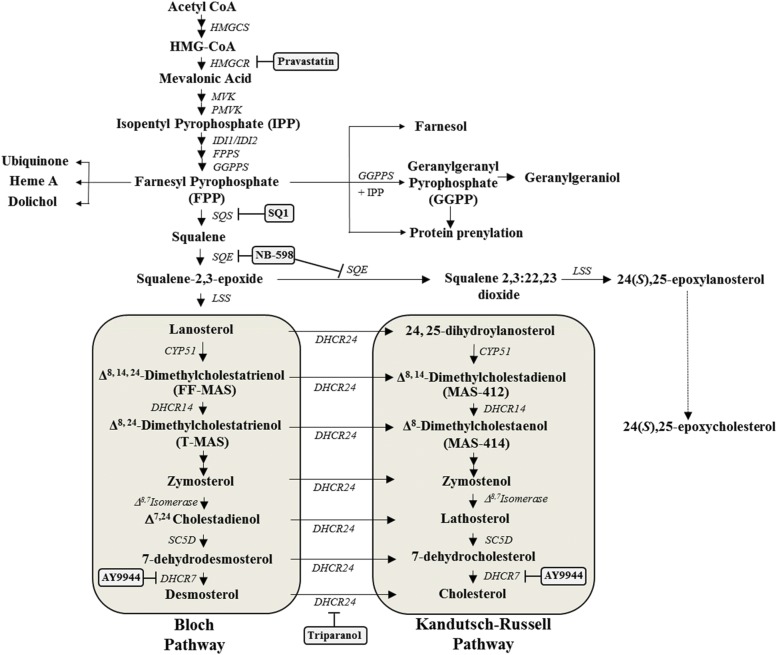
Fig. 1.
Overview of the cholesterol biosynthetic pathway and cholesterol synthesis inhibitors used in the current study. Cholesterol synthesis inhibitors used in this study and the enzymatic targets are indicated in gray-shaded boxes. Enzyme names are italicized. Abbreviations: CYP51, lanosterol 14α-demethylase; DHCR24, 24-dehydrocholesterol reductase; DHCR14, sterol delta(14)-reductase; SC5D, sterol-C5-desaturase; DHCR7, 7-dehydrocholesterol reductase; FPPS, farnesyl pyrophosphate synthase; FF-MAS follicular fluid meiosis activating sterol (Δ8,14,24-dimethylcholestatrienol); GGPP, geranylgeranyl pyrophosphate; GGPPS, geranylgeranyl pyrophosphate synthase; HMGCS, 3-hydroxy-3-methylglutaryl coenzyme A (CoA) synthase; IDI1, isopentenyl-diphosphate delta isomerase 1; IDI2, isopentenyl-diphosphate delta isomerase 2; LSS, lanosterol synthase; MAS-412 meiosis activating sterol-412 (Δ8,14-dimethylcholestanol); MAS-414 meiosis activating sterol-414 (Δ8-dimethylcholestadionel); MVK, mevalonate kinase; PMVK, phosphomevalonate kinase; SQS, squalene synthase; SQE, squalene epoxide; T-MAS testicular meiosis activating sterol (Δ8,24-dimethylcholestadionel).
In the current investigation, we further evaluated regulation of SULT1C2 by intermediates of the cholesterol biosynthetic pathway in primary cultured rat hepatocytes using mevalonate (MVA) and a variety of cholesterol synthesis inhibitors. Additional studies were conducted to characterize SULT1C2 mRNA and identify promoter regions mediating treatment-induced transcriptional responses.
Materials and Methods
Materials.
NB-598 [(E)-N-ethyl-6,6-dimethyl-N-[[3-[(4-thiophen-3-ylthiophen-2-yl)methoxy]phenyl]methyl]hept-2-en-4-yn-1-amine] was a gift from Banyu Pharmaceutical (Tokyo, Japan). SQ1 and pravastatin (Prav) were gifts from GlaxoSmithKline (Research Triangle Park, NC) and Bristol-Myers Squibb Co. (Stamford, CT), respectively, and ciprofibrate (Cipro) was a gift from Sterling Winthrop Pharmaceuticals Research Division (Rensselaer, NY). AY-9944 (N-[(2-chlorophenyl)methyl]-1-[4-[[(2-chlorophenyl)methylamino]methyl]cyclohexyl]methanamine;hydrochloride) was purchased from Tocris Biosciences (Minneapolis, MN). Dimethylsulfoxide (DMSO), phenobarbital (PB), pregnenolone-16α-carbonitrile (PCN), R-mevalonolactone, GW3965 (2-[3-[3-[[2-chloro-3-(trifluoromethyl)phenyl]methyl-(2,2-diphenylethyl)amino]propoxy]phenyl]acetic acid), GW4064 (3-[(E)-2-[2-chloro-4-[[3-(2,6-dichlorophenyl)-5-propan-2-yl-1,2-oxazol-4-yl]methoxy]phenyl]ethenyl]benzoic acid), triparanol, and chenodeoxycholic acid (CDCA) were purchased from Sigma-Aldrich (St. Louis, MO). Matrigel was purchased from Corning (Bedford, MA), PureCol from Advanced BioMatrix (San Diego, CA), and recombinant human insulin (Novolin R) from Novo Nordisk Pharmaceuticals (Princeton, NJ). Culture medium and Lipofectamine 2000 reagent were purchased from Invitrogen (Carlsbad, CA). Additional sources of reagents are provided below.
Primary Cultures of Rat Hepatocytes.
All procedures were conducted in conformity with the regulatory guidelines of the Wayne State University Division of Laboratory Animal Resources (Detroit, MI). Adult male Sprague-Dawley rats (175–200 g) were purchased from Harlan Sprague-Dawley (Indianapolis, IN). Upon receipt, animals were housed in an Association for Assessment and Accreditation of Laboratory Animal Care International–approved animal facility and allowed to acclimatize for 1 week before use. Hepatocytes were isolated using a two-step collagen-perfusion technique, as described previously (Kocarek and Reddy, 1996). After isolation, hepatocytes were plated onto collagen I–coated 6-well plates (for RNA isolation experiments) or 12-well plates (for transfection experiments) at a cell density of 150,000 hepatocytes/cm2 and were cultured in Williams’ E medium containing 0.25 U/ml insulin, 0.1 µM triamcinolone acetonide, 100 U/ml penicillin, and 100 μg/ml streptomycin. Drugs were added to the culture medium as concentrated stock solutions (1000X) dissolved either in water (MVA, PB, Prav, SQ1, and AY-9944) or DMSO (Cipro, NB-598, PCN, GW3965, GW4064, CDCA, and triparanol).
Quantitative Reverse-Transcription Polymerase Chain Reaction for SULT1C2 in Primary Cultured Rat Hepatocytes.
Hepatocytes were plated onto 6-well plates as described previously, and 24 hours after plating cells were overlayed with Matrigel (1:50 dilution) diluted in Williams’ E medium. The next day, hepatocytes were treated with cholesterol synthesis inhibitors or transcription factor activators at the concentrations indicated in the individual figure legends. The medium containing the treatments was replaced once after 24 hours. Forty-eight hours after the initial treatment, the cells were lysed, and the total RNA was extracted using RNeasy columns (Qiagen, Valencia, CA). We then synthesized cDNA from total RNA using the High Capacity Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions.
Primers to detect SULT1C2 and TATA box-binding protein (Assay ID Rn.PT.51.24118050) were purchased from Integrated DNA Technologies (IDT, Coralville, IA). The sequences of the primers used to detect SULT1C2 were 5′-TCGTTCTACAGTCCCCAAGT-3′ (forward) and 5′-TCCGCCCATCTTTTGCTTA-3′ (reverse). Treatment-induced changes in the mRNA levels of SULT1C2 were determined using the StepOne Plus Real Time PCR system (Applied Biosystems). The reaction mixture (20 μL) contained 50 ng RNA equivalents, 10 μL SYBR green Master Mix (Applied Biosystems), and 150 nM of diluted primers. The real-time polymerase chain reaction (PCR) cycling conditions were as follows: initial activation step at 95°C (15 minutes) and 40 cycles of melting (95°C, 15 seconds) and annealing/extension (60°C, 1 minute). Assays were performed in duplicate, and the relative fold-changes were quantified using the comparative cycle threshold CtΔΔCt method. Results were normalized to untreated controls and are expressed as mean ± S.E.M.
Rapid Amplification of cDNA Ends.
Determination of the 5′ mRNA sequence for rat SULT1C2 was conducted using the SMARTer Rapid Amplification of cDNA Ends (RACE) cDNA Amplification Kit (Clontech Laboratories, Mountain View, CA) according to the manufacturer’s recommendations. Briefly, total RNA was isolated from primary cultured rat hepatocytes as described earlier and assessed for quality using the Bioanalyzer 2100 (Agilent, Santa Clara, CA). RACE-ready cDNA was prepared from 750 ng of high quality RNA, and the 5′-end of SULT1C2 was then amplified using the Advantage 2 PCR kit (Clontech Laboratories) and gene-specific reverse primers. Due to a high sequence homology with SULT1C2A in the coding region (Xiangrong et al., 2000), two different SULT1C2-specific reverse primers were designed targeting noncoding exon 3 of the reference sequence (NM_133547.4) and are as follows: primer 1, 5′-CCCTGCACAACAGAACAGAAGGCTG-3′; and primer 2, 5′-GGAGGGATGCTTTCCCCTAGCCTCT-3′. After amplification, the PCR products were resolved on agarose gels containing ethidium bromide, and individual bands were extracted and purified using the QiaQuick Gel Purification kit (Qiagen). Purified PCR products were then ligated into the pGEM-T Easy plasmid (Promega, Madison, WI), and individual clones were sequenced at the Wayne State University Applied Genomics Technology Center (Detroit, MI).
PCR validation of the SULT1C2 mRNA structure was conducted using Quickload Taq Master Mix (New England Biolabs, Ipswich, MA), 50-ng RNA equivalents of cDNA, and diluted primers (250 nM each). The sequence of the forward primer was 5′-CAGAGGGATACTTGCAATTTT-3′, corresponding to nucleotides 103–123 of the longest RACE product. The reverse primers were located either on exon 5 (5′-ACATCCCCATTCTGCTCAATCATGTC-3′) or on exons 8–9 (5′-TTTCACGCTTTGGGTCCCTCTTCACA-3′) of the reference mRNA. The PCR cycling conditions included an initial melting step at 94°C (3 minutes), followed by five cycles at 94°C for 20 seconds, 58°C for 20 seconds, and 68°C for 1 minute, and by 30 cycles at 94°C for 20 seconds, 50°C for 20 seconds, and 68°C for 1 minute with a final extension at 68°C for 7 minutes. The PCR bands were resolved on an agarose gel, and the individual bands were extracted and purified described earlier. Following purification, PCR products were ligated into the pGEM-T Easy plasmid, and individual clones were sequenced. Sequences of clones were then aligned using the multiple sequence alignment tool with the Clustal Omega program (Goujon et al., 2010).
Nuclear Receptor-Responsive Reporter Plasmids.
Generation of luciferase reporter plasmids responsive to the constitutive androstane receptor (CAR), PXR, peroxisome proliferator-activated receptor (PPAR), farnesoid X receptor (FXR), and LXR has been previously described (Kocarek et al., 1998; Kocarek and Mercer-Haines, 2002). Briefly, the CAR-responsive reporter plasmid contains a PB responsive element within a 2451-base pair (bp) fragment of the rat CYP2B1 gene (Kocarek et al., 1998); the PXR reporter contains proximal and distal xenobiotic-responsive elements from the human CYP3A4 5′-flanking region and was a gift from Dr. Bryan Goodwin (GlaxoSmithKline Research and Development; Research Triangle Park, NC) (Goodwin et al., 1999); and the PPAR reporter plasmid contains the peroxisome-proliferator responsive element from the rat CYP4A1 promoter (Aldridge et al., 1995) ligated into the pGL3-Promoter plasmid (Promega). The plasmid for FXR contains six copies of a consensus FXR-responsive inverted repeat 1 motif (Laffitte et al., 2000), and for LXR contains four copies of an LXR-responsive direct repeat 4 motif (Lehmann et al., 1997), both ligated upstream of a minimal thymidine kinase promoter in the pGL3-Basic luciferase reporter plasmid (Promega).
Generation of Rat SULT1C2 Luciferase Reporter Constructs.
A reporter construct containing ∼3.1 kb of the region upstream of the longest 5′-RACE product was prepared by PCR using rat genomic DNA as template, Herculase II Fusion DNA Polymerase (Agilent), and primers designed to amplify nt 4676640-4679731 of rat chromosome 9 reference sequence NC_005108.4 (the SULT1C2 gene is located at nt 4654278-4685416 in the reverse orientation). The 5′-terminus of the longest RACE product aligns to nt 8826 of the reported SULT1C2 gene sequence and is flanked by a hexameric repeat region. Therefore, the reverse primer was designed to begin just proximal to this repeat region (i.e., at nt -49 relative to the 5′-end of the RACE product). The PCR product was ligated into the promoterless pGL4.10 firefly luciferase reporter plasmid (Promega). This plasmid (termed r1C2-1) was then used as a template to prepare additional reporter constructs by PCR. A complete list of the primer pairs used to generate the constructs along with sequence alignments is available in Supplemental Table 1. After construction, inserts were verified by sequencing as described earlier.
Site-Directed Mutagenesis of a Hepatocyte Nuclear Factor 1 Element in the SULT1C2 Promoter.
Transcription factor binding sites within the responsive regions of the SULT1C2 promoter were predicted using the JASPAR database (Mathelier et al., 2014). A hepatocyte nuclear factor 1 (HNF1) binding site (5′-GGTTAATAATTTTT-3′) was identified within the MVA-responsive region located at nt -2545 to -2558 relative to the longest RACE product. Base pair mutations in the core region of this HNF1 site were introduced into the r1C2-2 reporter plasmid using the Quickchange II XL Site-Directed Mutagenesis Kit (Agilent) according to the manufacturer’s instructions. Mutagenic primers were purchased from IDT, and the sequence of the reverse primer is 5′-GGAGATTGGGAATTAGTTTACTTAGTGAATCCTGGTGtcccAcAAcgTTTTTAGTTTGGCACACCTTAGCATCACGAGTCTTAA-3′, with nucleotide changes indicated in lowercase letters. After cloning, the sequence of the mutated SULT1C2 plasmid was verified as indicated earlier.
Generation of Rat HNF1α and HNF1β Expression Plasmids.
Expression plasmids were generated by PCR using rat hepatocyte cDNA as the template, Pfu Fusion High Fidelity DNA Polymerase (Agilent), and primers designed to amplify the entire coding sequence of either rat HNF1α cDNA (nt 151-2130 of reference sequence NM_012669.1) or HNF1β cDNA (nt 152-1848 of reference sequence NM_001308148.1). The cDNA was prepared from high-quality RNA isolated from primary cultured rat hepatocytes as described earlier. Primer pairs were purchased from IDT, and the sequences are as follows: 5′-GCGAAGCTTGCCACCATGGTTTCTAAGTTGAGC-3′ (HNF1α forward), 5′-GCGGGATCCAGGCTCCAACACCCTCCA-3′ (HNF1α reverse), 5′-GCGAAGCTTGCCACCATGGTGTCCAAGCTCACG-3′ (HNF1β forward), and 5′-GCGGGATCCAGTGAGTGGTTATGTGGG-3′ (HNF1β reverse). The underscored nucleotides indicate restriction sites used for cloning into the pcDNA3.1 expression plasmid (Invitrogen), and the italicized nucleotides indicate an inserted Kozak consensus sequence. After construction, the inserted cDNA was verified by sequencing as described earlier.
Transient Transfection of Primary Cultured Rat Hepatocytes.
Primary cultures of rat hepatocytes were transiently transfected with reporter constructs as previously described (Kocarek and Mercer-Haines, 2002) with minor modifications. Hepatocytes were plated onto collagen I–coated 12-well plates (600,000 hepatocytes/well), and 24 hours after plating the culture medium was replaced with 1 ml Williams’ E medium and 0.2 ml of Opti-MEM containing a premixed complex of Lipofectamine 2000 reagent (3–4 μl), 1.6 µg of a firefly luciferase reporter plasmid, and the pRL-CMV reporter plasmid (0.75–1 ng) (Promega) to allow for normalization among wells. For HNF1 overexpression assays, 1.5 µg of the firefly luciferase reporter plasmid, 50 ng of an expression plasmid (pcDNA3.1, HNF1α, or HNF1β), and 1 ng of pRL-CMV were used in the transfection mixture. Five hours after transfection, culture medium was replaced with standard Williams’ E medium containing Matrigel (1:50 dilution), and the cells were further incubated overnight at 37°C. The next day, fresh medium either alone or containing drug treatments was added to each well as described in the individual figure legends. The medium containing the drugs was replaced once after 24 hours. Ninety-six hours after initial plating (48 hours after treatment), the hepatocytes were harvested, and luciferase activity (firefly and Renilla) was measured using the Dual Luciferase Reporter Assay System and a GloMax luminometer (Promega). Each treatment was performed in triplicate and repeated at least one independent time (1 rat hepatocyte preparation per independent experiment).
Transient Transfection of HepG2 Cells.
HepG2 human hepatoma cells were purchased from the American Type Culture Collection (Manassas, VA) and cultured in high glucose Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, nonessential amino acids, 100 U/ml penicillin, and 100 μg/ml streptomycin. For transfection experiments, cells were subcultured onto 12-well plates at a cell density of 150,000 cells per well. Forty-eight hours after plating (∼60% confluency), the cells were transiently transfected with a premixed complex containing Lipofectamine 2000 (3 μl), a SULT1C2 reporter or empty vector control (1.5 µg), an HNF1 expression plasmid or pcDNA3.1 control plasmid (50 ng), and 1 ng of pRL-CMV prepared in 0.2 mL of Opti-MEM medium. Cells were harvested for the measurement of luciferase activity (firefly and Renilla) 48 hours after transfection as described earlier. Treatments were performed in triplicate, and each experiment repeated at least one independent time (n = 3 wells/treatment/experiment).
Statistical Analyses.
Statistical analyses were conducted using SigmaStat Statistical Software (version 3.5; Point Richmond, CA). Data were analyzed using a one-way or two-way analysis of variance (ANOVA); when statistical differences were detected with the F statistic (P < 0.05), individual comparisons were made using the Student-Newman-Keuls test. All results are presented as mean ± S.E.M.
Results
SULT1C2 mRNA Levels Are Modulated by Intermediates of the Cholesterol Biosynthetic Pathway in Primary Cultured Rat Hepatocytes.
Preliminary results from our laboratory indicated that the anticholesterol drugs Prav and SQ1 differentially affected SULT1C2 mRNA levels in primary cultured rat hepatocytes. In the current investigation, we further evaluated transcriptional regulation of SULT1C2 by intermediates of the cholesterol biosynthetic pathway using a variety of cholesterol synthesis inhibitors (Fig. 1) with or without cotreatment with MVA, the immediate product of 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR). Primary cultured rat hepatocytes were treated for 48 hours, and relative changes in SULT1C2 mRNA levels were assessed by quantitative reverse-transcription PCR. The results are presented in Fig. 2.
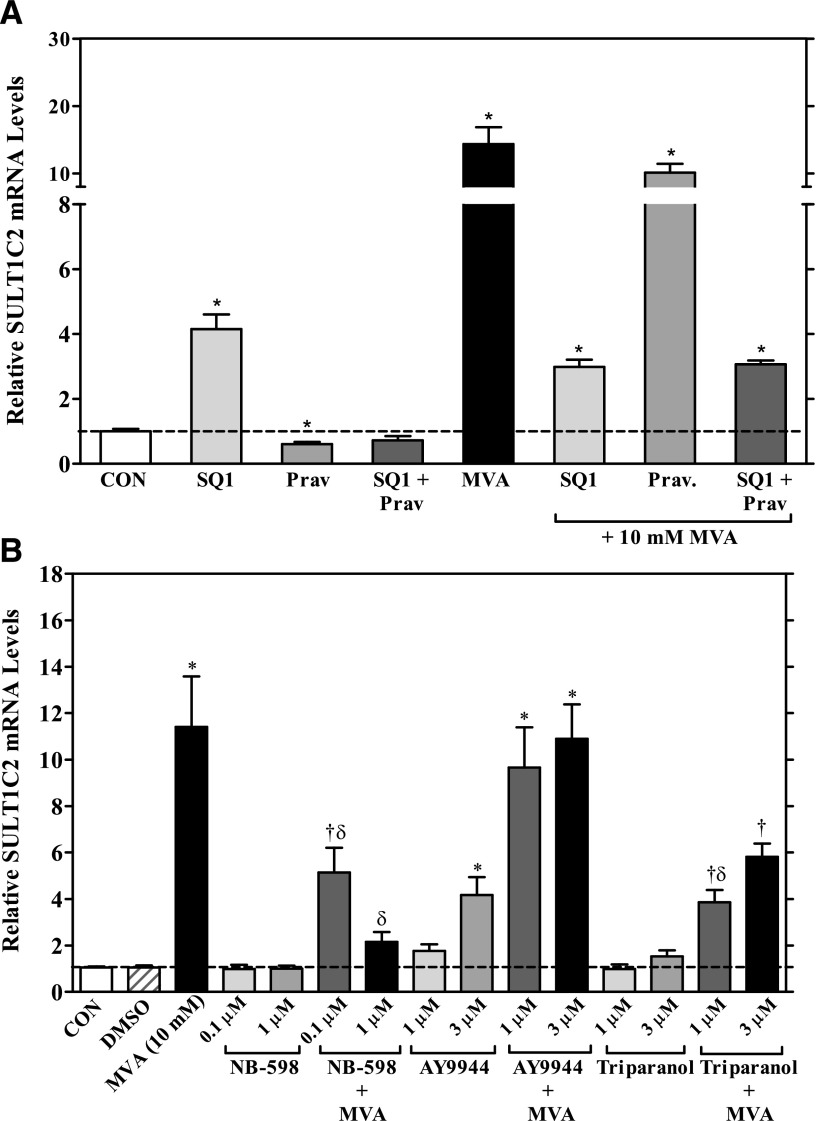
Fig. 2.
Effects of MVA and cholesterol synthesis inhibitors on SULT1C2 mRNA levels in primary cultured rat hepatocytes. Freshly isolated rat hepatocytes were plated onto six-well, collagen I–coated plates and maintained in Williams’ E medium. Twenty-four hours after plating, cells were overlayed with Matrigel. The next day, cells were treated with medium alone (CON: control) or containing one of the following: (A) SQ1 (0.1 µM), Prav (30 µM), SQ1 and Prav, or R-mevalonolactone (MVA, 10 mM) alone or in combination with SQ1, Prav, or SQ1 and Prav; (B) DMSO (0.1%), MVA (10 mM), or each of the following inhibitors: NB-598 (0.1 µM or 1 µM), AY-9944 (1 or 3 µM), or triparanol (1 or 3 µM) alone or in the presence of 10 mM MVA. Treatments were replaced once after 24 hours. Ninety-six hours after initial plating, cultures were processed for RNA isolation and cDNA synthesis, and SULT1C2 mRNA levels were quantified by quantitative reverse-transcription PCR, as described in the Materials and Methods section. Each bar represents the mean ± S.E.M. of normalized SULT1C2 values combined from three to six independent experiments (each independent experiment represents one rat hepatocyte preparation). For both panels: *statistically significantly different from untreated controls (CON); †statistically significantly different from DMSO controls; δsignificantly different from MVA-treated hepatocytes (significance shown for samples cotreated with MVA only) (P < 0.05).
We found that treatment of cells with the squalene synthase inhibitor SQ1 (0.1 μM), significantly increased SULT1C2 mRNA levels by ∼4-fold and treatment with 10 mM MVA by 12-fold, whereas the HMGCR inhibitor Prav decreased SULT1C2 mRNA levels by 40% (P < 0.05; Fig. 2A). In cotreatment experiments, Prav abolished the inducing effects of SQ1 on SULT1C2 gene expression, which could be restored by the addition of MVA. Supplementation with MVA also restored SULT1C2 mRNA in Prav-treated cells to levels comparable to that observed with MVA alone (Fig. 2A). However, the level of SULT1C2 induction (∼3-fold) that occurred when hepatocytes were cotreated with MVA and SQ1 was comparable to that seen after treatment with SQ1 alone. These findings indicate that the residual induction observed after cotreatment with MVA and SQ1 was attributable to SQ1-mediated isoprenoid accumulation, suggesting that the major portion of MVA’s effects was mediated by a metabolite(s) distal to FPP. Therefore, additional studies were conducted with inhibitors of downstream steps in the cholesterol biosynthetic pathway.
NB-598 inhibits squalene epoxidase (also referred to as squalene monooxygenase), which catalyzes the conversion of squalene to squalene-2,3-epoxide (Fig. 1) (Horie et al., 1990). Treatment of hepatocytes with either 0.1 or 1 μM of NB-598 alone had no significant effect on SULT1C2 mRNA levels (P > 0.05; Fig. 2B). However, NB-598 dose-dependently reduced the inducing effect of MVA, suggesting that a more distal metabolite(s) rather than squalene accumulation was mediating the effects of MVA (P < 0.05; Fig. 2B).
To evaluate this further, we examined the effects of treatment with the distal cholesterol synthesis inhibitors AY-9944 and triparanol on SULT1C2 expression. AY-9944 inhibits DHCR7 (3β-hydroxysterol Δ7-reductase), primarily reducing the conversion of 7-dehydrocholesterol (7DHC) to cholesterol via the Kandutsch-Russell pathway and also 7-dehydrodesmosterol to desmosterol by the Bloch pathway (Horlick, 1966; Sanchez-Wandelmer et al., 2009; Meljon et al., 2013) (Fig. 1). Triparanol inhibits DHCR24 (sterol Δ24 reductase), resulting in reduced cholesterol synthesis and accumulation of desmosterol and zymosterol, depending on the dose (Fig. 1) (Gibbons and Pullinger, 1977; Popjak et al., 1989; Chevy et al., 2002). We found that treatment with AY-9944 (1 or 3 μM) dose dependently increased SULT1C2 mRNA levels, with a ∼4-fold induction observed at the highest dose tested (3 μM; P < 0.05). Cotreatment with MVA further increased SULT1C2 mRNA levels to that observed with MVA alone (Fig. 2B). By contrast, triparanol treatment alone (1 or 3 μM) had no significant effect on SULT1C2 levels (P > 0.05). Supplementation of triparanol-treated cells with MVA significantly increased SULT1C2 mRNA levels, but to a lesser extent than that observed in cells treated with AY-9944 (Fig. 2B). These findings indicate that SULT1C2 is transcriptionally regulated by at least two steps in the cholesterol biosynthesis pathway, through isoprenoid accumulation and a postsqualene step.
Several MVA and cholesterol pathway intermediates including nonsterol isoprenoids (e.g., FPP, farnesol, and geranylgeraniol), squalene metabolites (e.g., 2,3;22,23-diepoxysqualene), precholesterol sterols (e.g., desmosterol), and oxysterols (e.g., 24(S),25-epoxycholesterol) are known to modulate the activity of various nuclear receptors (Lehmann et al., 1997; Kocarek and Mercer-Haines, 2002; Takahashi et al., 2002; Yang et al., 2006; Wong et al., 2008; Duniec-Dmuchowski et al., 2009; Goto et al., 2011). Additionally, we recently showed that several nuclear receptors including VDR, PXR, and LXR are involved in the transcriptional regulation of human SULT1C2 in a colon cancer cell line (Rondini et al., 2014). Therefore, to evaluate whether the effects of SQ1 and MVA on SULT1C2 expression were mediated through similar signaling pathways, we first tested the effects of these treatments on several nuclear receptor-responsive reporter constructs in primary cultured rat hepatocytes.
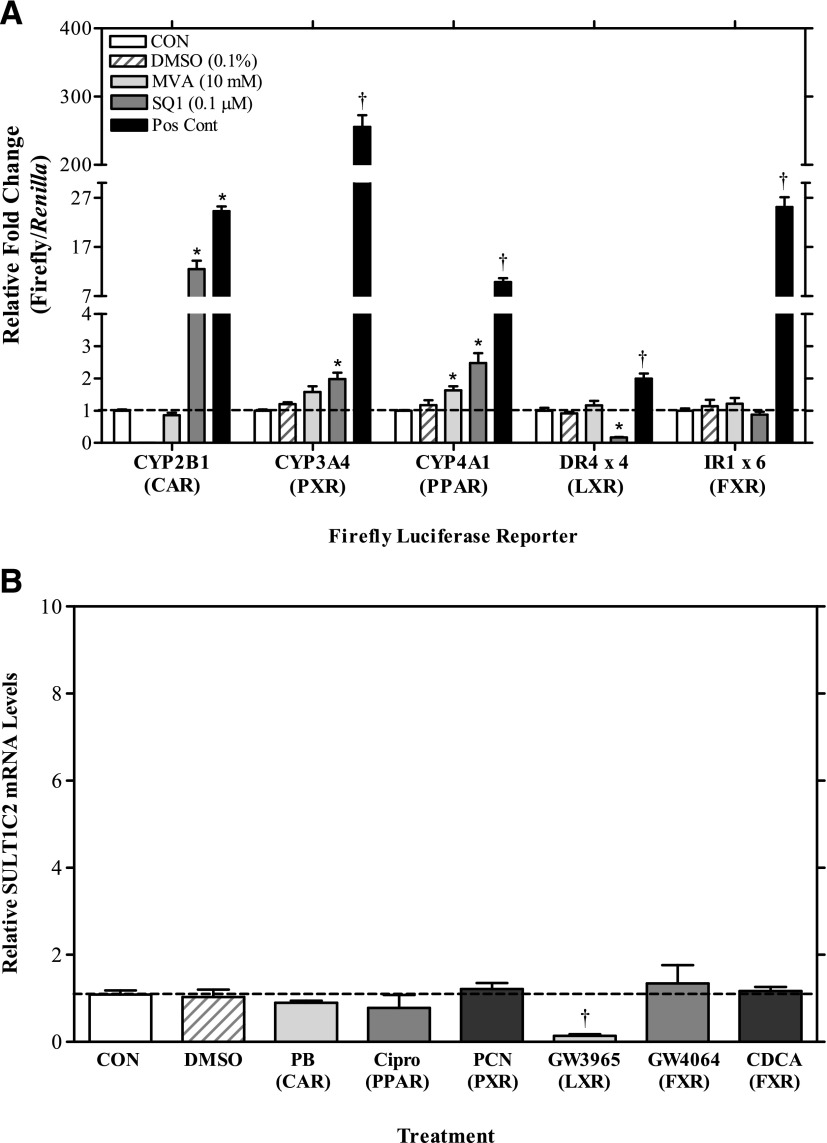
As shown in Fig. 3A, SQ1 robustly induced the activity of the CAR-responsive reporter, CYP2B1 (∼10-fold) and produced smaller increases from the PPAR- (3-fold) and PXR- (2-fold) responsive reporters (P < 0.05). SQ1 treatment also strongly inhibited LXR-mediated reporter activity by ∼80% (P < 0.05). In comparison, MVA treatment significantly induced activity only on the PPAR-responsive reporter (∼2-fold) (Fig. 3A; P < 0.05). To determine whether any of these nuclear receptors are directly involved in regulation of SULT1C2, we tested the effects of prototypical activators for CAR, PPAR, PXR, LXR, and FXR on SULT1C2 expression. As shown in Fig. 3B, activators for CAR (PB, 100 μM), PPAR (Cipro, 100 μM), PXR (PCN, 10 μM), and FXR (GW4064, 10 μM, and CDCA, 100 µM) had no significant effect on SULT1C2 mRNA levels, whereas the LXR activator GW3965 (10 μM) decreased SULT1C2 mRNA content by ∼85% (P < 0.05; Fig. 3B). These findings indicate that although SQ1, and to a lesser extent, MVA, activate some nuclear receptor signaling pathways, these effects do not appear to mediate the ability of these compounds to increase SULT1C2 expression.
Fig. 3.
Effects of nuclear receptor signaling pathways on SULT1C2 expression in primary cultured rat hepatocytes. (A) Twenty-four hours after plating, rat hepatocyte cultures were transfected with a nuclear receptor-responsive firefly reporter plasmid as described in Materials and Methods. Twenty-four hours after transfection, cultures were incubated in William’s E medium either alone (CON: control) or containing 0.1% DMSO, 10 mM MVA, 0.1 µM SQ1, or one of the following positive control nuclear receptor activators: 100 µM PB (CAR), 10 µM PCN (PXR), 100 µM Cipro (PPAR), 1 µM GW3965 (LXR), or 10 µM GW4064 (FXR). Forty-eight hours after treatment, hepatocytes were harvested for the measurement of luciferase activities as described in Materials and Methods. Each bar represents the mean ± S.E.M. of normalized luciferase values (firefly/Renilla) from two independent experiments (n = 3/treatment/experiment). (B) Forty eight hours after plating, cells were treated with medium alone (CON: control) or containing one of the following drug treatments: DMSO (0.1%), PB (100 µM), Cipro (100 µM), PCN (10 µM), GW3965 (10 µM), GW4064 (10 µM), or CDCA (100 µM). Treatments were replaced once after 24 hours. Ninety-six hours after initial plating, cultures were processed for RNA isolation and cDNA synthesis, and SULT1C2 mRNA levels were quantified by quantitative reverse-transcription PCR, as described in Materials and Methods. Each bar represents the mean ± S.E.M. of normalized SULT1C2 values combined from two independent experiments (each independent experiment represents one rat hepatocyte preparation). For all panels: *statistically significantly different from untreated controls (CON); †statistically significantly different from DMSO controls (P < 0.05).
Determination of the 5′ Untranslated Region of Rat SULT1C2 mRNA Using RACE.
Alignment of the reference sequence for SULT1C2 mRNA (NM_133547.4) to the rat chromosome 9 sequence suggested the presence of three noncoding exons in its 5′ structure, which would be unique compared with other SULT family members (Supplemental Fig. 1A; Fig. 4B). During the course of this study, 15 additional splice variants have been computationally predicted and indexed in the National Center for Biotechnology Information (NCBI) database. Among these, 12 of the 15 variants lack noncoding exon 3. Although exon 2 is highly conserved, exon 1 is predicted to align to seven different regions on the SULT1C2 gene, suggesting multiple transcription start sites.
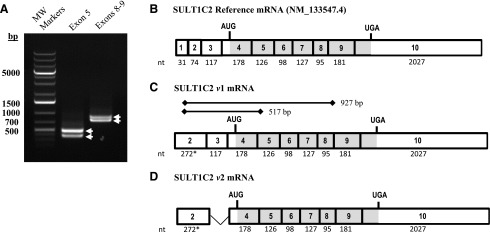
Fig. 4.
PCR validation of the rat hepatic SULT1C2 mRNA structure as determined by 5′-RACE. The 5′ mRNA sequence for rat SULT1C2 was determined by RACE using gene-specific reverse primers targeting exon 3 of the reference sequence mRNA (NM_133547.4), as described in Materials and Methods. (A) Agarose gel electrophoresis of PCR products obtained from amplification of the rat SULT1C2 cDNA using a forward primer targeting nt 103-123 on exon 2 of the longest 5′-RACE sequence and reverse primers targeting exon 5 and exons 8–9 as described in Materials and Methods and indicated in C. (B) Structure of the rat SULT1C2 reference sequence mRNA (NM_133547.4) and (C, D) structures of the two rat SULT1C2 mRNA species (variant 1 and 2) detected in primary rat hepatocytes. A splice variation was detected in noncoding exon 3. White boxes denote noncoding regions, and gray-shaded boxes denote coding regions. *The size of exon 2 (272 nt) is estimated based on the longest 5′-RACE product. The size of exon 10 is based on the reference sequence and includes nucleotides up to the polyadenylation site.
To validate the 5′ mRNA structure in preparation for promoter analysis, we conducted 5′-RACE on mRNA isolated from primary rat hepatocytes using two different gene-specific primers. The aligned sequences of the isolated clones (n = 4/primer) are presented in Supplemental Fig. 1B. In the eight clones that were evaluated, we did not find evidence for the previously predicted exon 1. Based on the longest 5′ sequence, exon 2 was determined to be 198-bp longer than the reference sequence’s exon 2, and its 5′-end was flanked by a hexameric nucleotide repeat region that varied slightly in length among clones (Supplemental Fig. 1B; Fig. 4, B and C).
To validate the mRNA structure further, we conducted PCR using a forward primer immediately adjacent to the repeat region and reverse primers located on exon 5 and exons 8–9 (Fig. 4C). For both primer pairs, two PCR bands were detected that differed in size by ∼100 bp, which we subsequently cloned and sequenced (Fig. 4A). As shown in Fig. 4C, the PCR results confirmed the structure of exon 2 that was obtained by 5′-RACE. An mRNA splice variant lacking the noncoding exon 3 was additionally detected in the rat hepatocytes (Fig. 4D). The fully aligned sequences of the clones obtained from PCR are available in Supplemental Fig. 2.
Rat SULT1C2 Promoter Activity Is Regulated by MVA and SQ1.
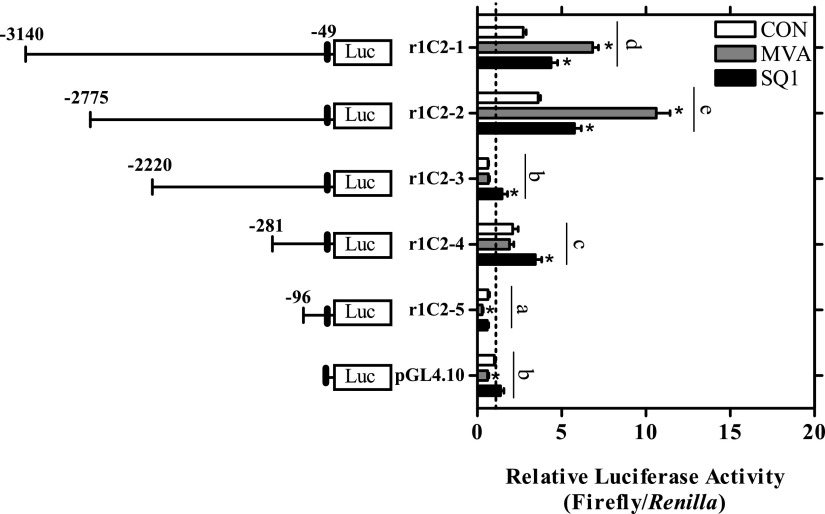
Reporter constructs were generated containing progressive deletions of the 5′-flanking region of the SULT1C2 gene identified through RACE to identify elements potentially mediating basal and treatment-induced transcriptional responses. The largest construct, r1C2-1 (-3140:-49) was 3.1 kb in length and began 49 bp upstream from the longest 5′-RACE product (-49 bp; see Supplemental Table 1), at the start of the hexameric repeat region. Primary cultured rat hepatocytes were transiently transfected with each reporter plasmid, and luciferase activity was measured 48 hours after treatment. All results were normalized to the expression values of untreated empty vector controls.
As shown in Fig. 5, basal luciferase activity differed to some extent among the constructs evaluated. Transfection with either the r1C2-1 (-3140:-49), r1C2-2 (-2775:-49), or r1C2-4 (-281:-49) plasmid increased reporter activity by ∼2- to 4-fold compared with empty vector controls (P < 0.05; Fig. 5). The r1C2-3 (-2220:-49) construct had substantially lower activity than that of the r1C2-2 and r1C2-4 plasmids, suggesting loss of an enhancer and/or presence of a transcriptional repressor element within this region (Fig. 5). The absolute and relative activity of the shortest construct tested, r1C-5 (-96:-49), was also significantly lower than that of the empty vector control. Evaluation of this region using JASPAR predicted a putative TATA box binding protein site beginning 11-bp upstream of the start of the r1C2-5 construct on the SULT1C2 gene, indicating that this region may contain the core promoter for SULT1C2 (Fig. 7B). Treatment-induced changes in reporter activity were also evaluated for each construct (Fig. 5). MVA (5 mM) significantly increased reporter activity by ∼3-fold on the r1C2-1 (-3140:-49) and r1C2-2 (-2775:-49) constructs only (P < 0.05; Fig. 5). In comparison, all the constructs from r1C2-1 through r1C2-4 displayed some degree of responsiveness to SQ1 although the fold-changes (≤2) were generally less than that observed for MVA (Fig. 5). However, neither MVA nor SQ1 influenced reporter activity on the r1C2-5 (-96:-49) construct (P>0.05).
Fig. 5.
Effects of MVA and SQ1 treatment on rat SULT1C2 promoter activity. A firefly luciferase reporter construct containing ∼3.1 kb of the SULT1C2 5′-flanking region (r1C2-1) and 4 deletion reporter constructs (r1C2-2 to r1C2-5) were prepared as described in Materials and Methods. Twenty-four hours after plating, primary cultured rat hepatocytes were transfected with a SULT1C2 reporter plasmid or with the empty vector as a control (pGL4.10). The next day, the hepatocytes were treated with Williams’ E medium alone (CON: control) or containing SQ1 (0.1 µM) or MVA (5 mM). The medium containing the treatments was replaced after 24 hours. Ninety-six hours after plating, cells were harvested for the measurement of luciferase activity. Bars represent mean ± S.E.M. of luciferase measurements (firefly/Renilla) normalized to untreated, empty vector controls. The results are combined from three independent experiments, with each independent experiment representing one rat hepatocyte preparation (n = 3 wells/treatment/experiment). A statistically significant treatment X plasmid interaction (P < 0.05) was detected. For clarity, only statistically significant treatment effects within individual plasmids and overall significant differences between plasmids (constructs) are shown. *Statistically significant effect of treatment (MVA or SQ1) within each reporter construct compared with the untreated, construct-matched controls (CON) (P < 0.05). Different letters denote significant differences between constructs on luciferase reporter activity (P < 0.05).
Fig. 7.
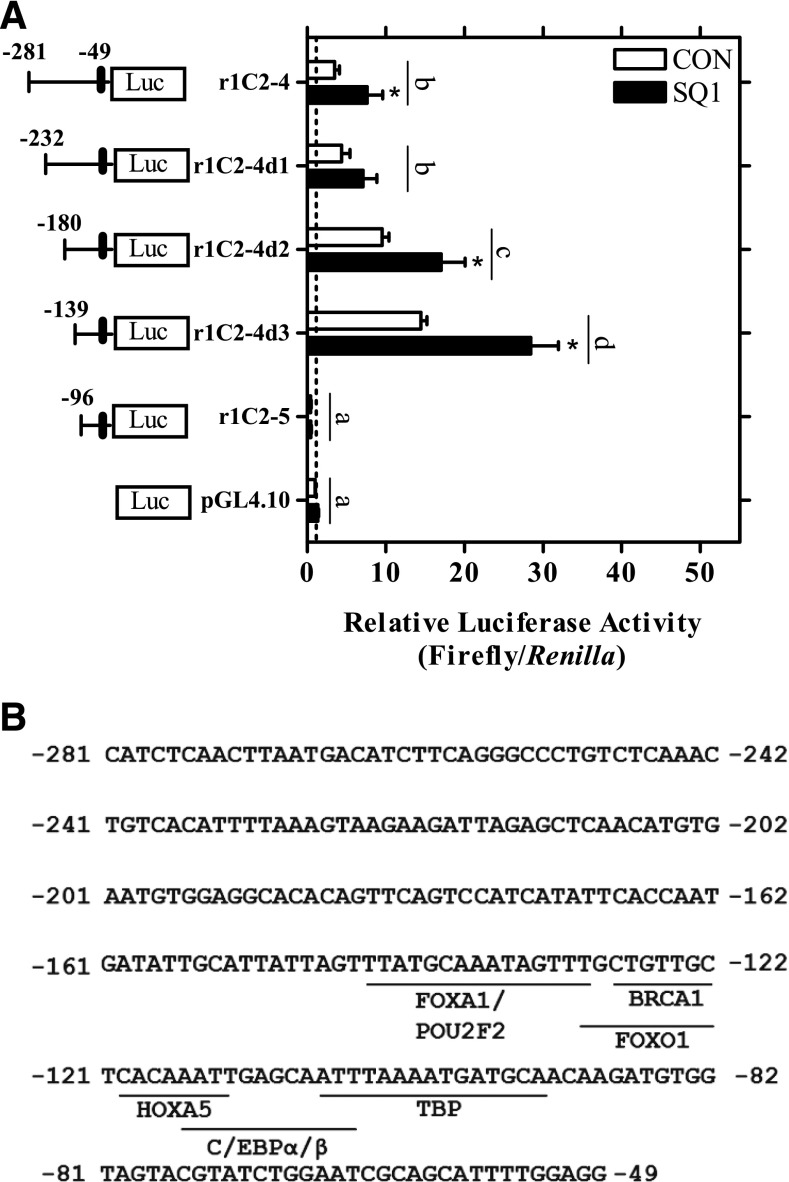
Evaluation of the SQ1-responsive region of the SULT1C2 promoter. (A) Three firefly luciferase deletion constructs based on the r1C2-4 sequence (-281:-49) were prepared. Twenty-four hours after plating, primary cultured rat hepatocytes were transfected with a SULT1C2 reporter plasmid (r1C2-4, deletion constructs r1C2-4d1-3, or r1C2-5) or with the empty vector as a control (pGL4.10). The next day, the hepatocytes were treated with Williams’ E medium alone (CON, control) or containing 0.1 µM SQ1. The medium containing the treatments was replaced once after 24 hours. Ninety-six hours after plating, cells were harvested for the measurement of luciferase activity. Bars represent mean ± S.E.M. of luciferase measurements (firefly/Renilla) normalized to untreated, empty vector controls. Results are combined from three independent experiments, with each independent experiment representing one rat hepatocyte preparation (n = 3 wells/treatment/experiment). *Statistically significant effect of SQ1 treatment within each reporter construct compared with the untreated, construct-matched control (CON) (P < 0.05). Different letters denote significant differences between the individual constructs on luciferase reporter activity (P < 0.05). (B) Transcription factor binding sites identified in the (-145:-49) promoter region predicted using the JASPAR database.
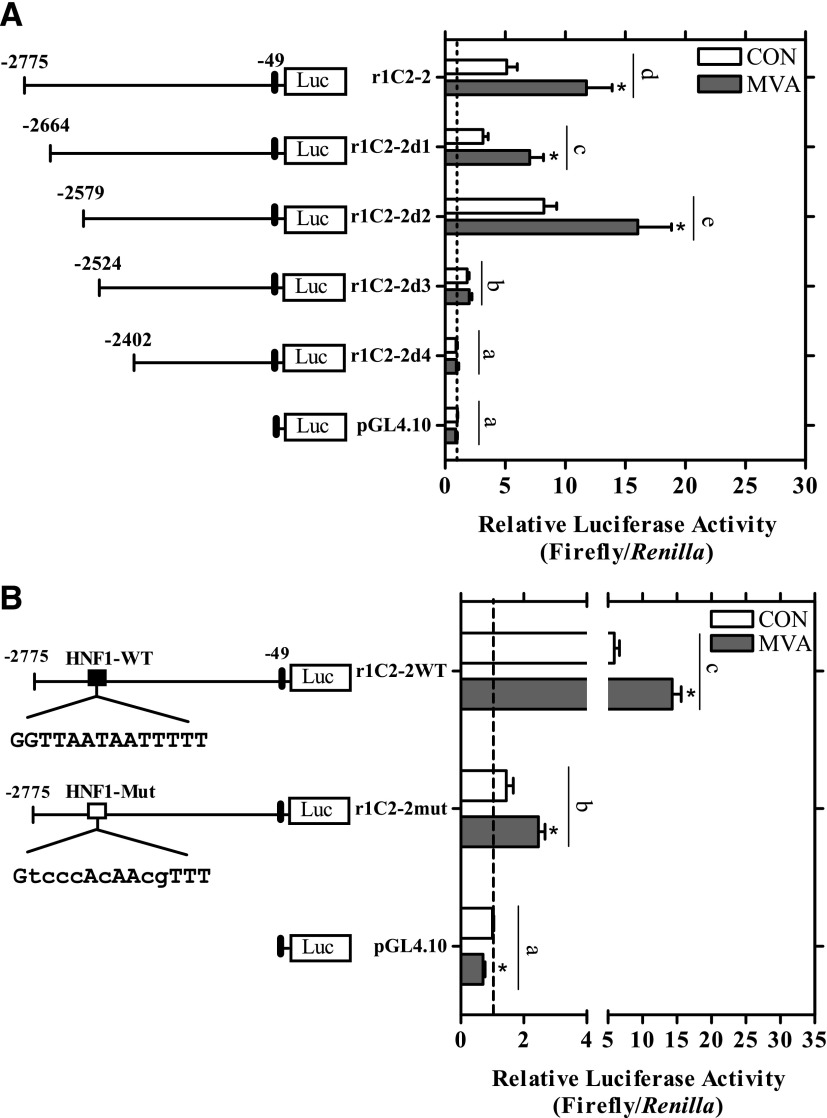
Further deletion of constructs r1C2-2 and r1C2-4 was conducted to identify minimal responsive regions mediating the MVA- and SQ1-induced effects, respectively (Figs. 6 and 7). As shown in Fig. 6A, deletion of the first 200 bp of the r1C2-2 plasmid (-2775:-49) did not significantly alter MVA responsiveness (P>0.05). However, deletion of an additional 55 bp (r1C2-2d3) strongly and significantly reduced both basal and MVA-induced reporter activity (P < 0.05, Fig. 6A). Using JASPAR, an HNF1 element (5′-GGTTAATAATTTTT-3′) was strongly predicted within this 55-bp region (nt -2558:-2545) and the conserved sequences for this site were then mutated to determine the effects on luciferase activity. As shown in Fig. 6B, basal reporter activity was significantly reduced by ∼75% and MVA-induced activity by 30% compared with the r1C2-2 construct, suggesting the HNF1 site is an important determinant of basal and MVA-induced transcription (P < 0.05; Fig. 6B).
Fig. 6.
Localization of the MVA-responsive region of the SULT1C2 promoter and impact of an HNF1-binding site. (A) Four firefly luciferase deletion constructs based on the r1C2-2 sequence (-2775:-49) were prepared as described in Materials and Methods. (B) Conserved elements of a computationally predicted HNF1 site (nt -2558:-2545) were mutated in the r1C2-2 plasmid (-2775:-49) using site-directed mutagenesis. Twenty-four hours after plating, primary cultured rat hepatocytes were transfected with a SULT1C2 reporter plasmid (r1C2-2 or deletion constructs r1C2d1-4), an HNF1 site-mutated plasmid (r1C2-2mut), or with an empty vector control (pGL4.10). The next day, cells were treated with Williams’ E medium alone (CON, control) or containing MVA (5 mM). The medium containing the treatments was replaced once after 24 hours. Ninety-six hours after plating, cells were harvested for the measurement of luciferase activity. Bars represent the mean ± S.E.M. of luciferase measurements (firefly/Renilla) normalized to untreated, empty vector controls. All results are combined from two to four independent experiments, with each independent experiment representing one rat hepatocyte preparation (n = 3 wells/treatment/experiment). For clarity, only significant treatment effects within individual plasmids and significant differences between plasmids (constructs) are shown. *Statistically significant effect of MVA treatment within each reporter construct compared with the untreated, construct-matched control (CON) (P < 0.05). Different letters denote significant differences between the individual constructs on luciferase reporter activity (P < 0.05).
To identify minimal SQ1-responsive regions, three additional deletions of the r1C2-4 (-281:-49) plasmid were generated (Fig. 7A). All the deletion constructs generated from the r1C2-4 (-281:-49) plasmid retained some responsiveness to SQ1 treatment (P < 0.05; Fig. 7A). Mutation of a predicted FOXA1-binding site did not affect SQ1-inducible luciferase activity (data not shown), indicating that SQ1 regulates transcription from a different site that we were unable to identify in this study. The SQ1 responsive region along with predicted transcription binding sites is presented in Fig. 7B.
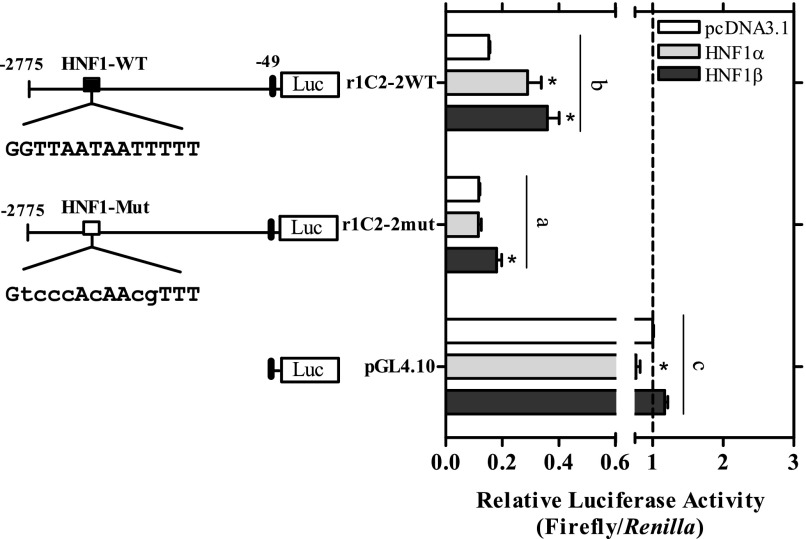
Two homologs of HNF1, HNF1α and HNF1β, are expressed in human and rodent tissues (Bach et al., 1991; Harries et al., 2009) that can bind to the same DNA element to influence transcriptional responses (Baumhueter et al., 1988; Mendel et al., 1991). To assess the functionality of the HNF site on the r1C2-2 construct further, we initially overexpressed either HNF1α or HNF1β in primary cultured rat hepatocytes and determined the effect on reporter activity. As shown in Supplemental Fig. 3, transient transfection with 50 ng of either of the HNF1 expression plasmids had no inducing effect on reporter activity from the r1C2-2 construct, but rather it decreased basal activity of both the r1C-2 and r1C2-2mut reporters (P < 0.05; Supplemental Fig. 3). Different amounts of expression plasmid (25 and 75 ng) produced a similar trend (data not shown), suggesting that overexpression of HNF1 in primary rat hepatocytes likely squelched limiting amounts of transcriptional coactivators, as has been previously reported in this cell type (Metz et al., 2000).
Additional studies were therefore conducted in HepG2 human hepatoma cells. HepG2 cells highly express HNF1α; however, basal transcription of HNF1-dependent reporters is low and inducible when cotransfected with HNF1 expression plasmids (Rollier et al., 1993; Metz et al., 2000). As shown in Fig. 8, overexpression of either HNF1α or 1β in HepG2 cells significantly induced reporter activity from the r1C2-2 construct by ∼≤2-fold (P < 0.05), whereas this effect was significantly attenuated when the HNF1 element was mutated (P < 0.05; Fig. 8). The HNF1β plasmid had a significant residual effect on the r1C2-2mut promoter, but this residual activity was still significantly less than that observed in the r1C2-2 promoter construct (P < 0.05). These findings demonstrate that the HNF1 site is functional and implies some redundancy of HNF1 proteins on SULT1C2 promoter activity.
Fig. 8.
Overexpression of HNF1α and HNF1β activates the rat SULT1C2 promoter in HepG2 cells. HepG2 cells were transiently transfected with 50 ng of an HNF1α or HNF1β expression plasmid or pcDNA3.1 control plasmid together with the r1C2-2 reporter, the r1C2-2 reporter containing a mutated HNF1 site (r1C2-2mut), or with an empty vector control (pGL4.10), as described in the Materials and Methods. Forty-eight hours after transfection, cells were harvested for the measurement of luciferase activity. Bars represent the mean ± S.E.M. of luciferase measurements (firefly/Renilla) normalized to the empty vector controls. All results are combined from three independent experiments (n = 3 wells/treatment/experiment). For clarity, only significant treatment effects within individual plasmids and significant differences among plasmids (constructs) are shown. *Statistically significant effect of the HNF1 expression plasmid within each reporter construct compared with the construct-matched pcDNA3.1 control (CON) (P < 0.05). Different letters denote significant differences among the individual constructs on luciferase reporter activity (P < 0.05).
Discussion
The cholesterol biosynthetic pathway is composed of at least 20 enzymatic reactions whereby cellular acetate is converted to cholesterol (Bloch, 1965). Aside from a role in cholesterol production, a number of biosynthetic intermediates are increasingly recognized to function as signaling molecules that influence diverse cellular processes (Edwards and Ericsson, 1999). For example, the branch point intermediate FPP and its isoprenoid metabolites are precursors for components of cellular respiration (ubiquinone and heme), glycoprotein synthesis (dolichol), and prenylation (FPP or geranylgeranyl pyrophosphate) of small GTP-binding proteins involved in cell signaling (Grunler et al., 1994). Isoprenoids can also activate the nuclear receptors CAR (farnesol), PPAR (FPP, geranylgeraniol, farnesol), and FXR (farnesol) to influence enzymes that metabolize drugs, lipids, and bile acids (Kocarek and Mercer-Haines, 2002; Takahashi et al., 2002; Goto et al., 2011). Among postsqualene metabolites, activation of LXR by endogenous oxysterols regulates various aspects of cholesterol homeostasis (Janowski et al., 1996; Janowski et al., 1999; Wong et al., 2008; Zhao and Dahlman-Wright, 2010) whereas methyl sterols, produced from lanosterol have meiosis-stimulating activity (Byskov et al., 1995). Finally, desmosterol and other postsqualene metabolites have been linked to TH17 lymphocyte differentiation through activation of retinoic acid–related orphan receptor γ (Hu et al., 2015; Santori et al., 2015).
In our current investigation, we found that MVA-derived intermediates also regulate SULT1C2 expression in primary rat hepatocytes. Depletion of MVA production with the HMGCR inhibitor Prav significantly reduced basal SULT1C2 expression. To determine which metabolites influence SULT1C2 expression, hepatocytes were treated with downstream cholesterol synthesis inhibitors with or without cotreatment with MVA. We found that isoprenoid accumulation, mediated through squalene synthase inhibition (SQ1), moderately induced and MVA treatment strongly induced SULT1C2 expression. This latter effect was blocked by the squalene monooxygenase inhibitor NB-598, but not the distal cholesterol synthesis inhibitors AY-9944 or triparanol. Therefore, in addition to isoprenoids, SULT1C2 is regulated by one or more postsqualene metabolites, possibly desmosterol, 7DHC, and/or 7-dehydrodesmosterol.
As previously observed, SQ1 treatment strongly activated CAR (Kocarek and Mercer-Haines, 2002) and more moderately activated PPAR and PXR whereas MVA significantly activated PPAR only. However, prototypical agonists for CAR, PPAR, PXR, or FXR did not influence SULT1C2 mRNA levels. These findings are consistent with those of Alnouti and Klaassen (2008) who reported that activators of CAR, PPAR, and PXR had no significant effect on SULT1C2 expression in the livers of male mice, and further suggest that there are some species differences in the regulation of SULT1C2 by nuclear receptors (Rondini et al., 2014).
We also found that SQ1 significantly inhibited LXR reporter activity and that the LXR agonist GW3965 suppressed SULT1C2 expression. Mice deficient in hepatic squalene synthase were also found to have reduced LXR signaling in the liver (Nagashima et al., 2015), potentially due to decreased oxysterol synthesis as well as the antagonistic actions of the isoprenoid geranylgeranyl pyrophosphate on LXR activity (Forman et al., 1997). However, inhibition of basal LXR activity is unlikely to account for the SQ1-mediated SULT1C2 induction that was observed in this study because both Prav and NB-598 treatments would also reduce endogenous oxysterol levels but had differing effects on SULT1C2 expression. Also our experiments with MVA and triparanol implicated desmosterol, a known LXR agonist (Yang et al., 2006), as a potential activator of SULT1C2, indicating that endogenous sterols do not suppress SULT1C2 at the levels achieved in these experiments.
The 5′-untranslated region (UTR) of SULT1C2 mRNA present in hepatocytes differed to some extent from the reference and predicted mRNA sequences. Based on the longest RACE product, the 5′-UTR extended upstream from the reported 5′-end of exon 2 and was flanked by a hexameric nucleotide repeat that aligned to a region spanning 156 bp on the SULT1C2 gene; therefore, the actual transcription start site is likely proximal to this region. The findings from 5′-RACE were validated using PCR, and, consistent with several of the computationally derived sequences, we identified a splice variant in exon 3. By end-point PCR analysis, both transcripts appeared to be expressed at similar levels in hepatocytes; however, quantitative expression was not further determined. Alternative splicing in the 5′ UTR has been suggested to influence translation efficiency, mRNA stability, and tissue-specific expression levels (Hughes, 2006; Palaniswamy et al., 2010). Thus, future evaluation of these variants in hepatic as well as extrahepatic tissue is warranted.
A construct containing ∼3.1 kb of the SULT1C2 5′-flanking region was found to contain both basal and inducible-responsive transcriptional elements, and deletion analysis revealed two different regions mediating responsiveness to MVA and SQ1. MVA-inducible reporter activity was localized to a region that was -2525 to -2775 nt upstream from the start of the hexameric repeat, and mutation of a predicted HNF1-binding element within that region strongly reduced basal and partially that of MVA-induced reporter activity. The inducing effect of SQ1 on SULT1C2 promoter activity was localized to a more proximal region (nt -49 to -281), and the element(s) mediating SQ1-inducible responsiveness were not conclusively determined. Several binding sites for transcription factors including Forkhead box protein O1 (FOXO1), Homeobox A5 (HOXA5), and the CCAAT/enhancer binding proteins (C/EBP) α and β were predicted in this region. C/EBP-α and -β, in particular, are liver-enriched transcription factors involved in glucose homeostasis and numerous hepatocellular functions (Wang et al., 1995; Lekstrom-Himes and Xanthopoulos, 1998). C/EBP regulates SULT2A1 expression in rat hepatocytes (Fang et al., 2005); however, additional studies are needed to determine the functional roles of these sites in the regulation of SULT1C2.
HNF1α and HNF1β are homeodomain-containing transcription factors that are highly expressed in the liver, kidney, intestine, and pancreas and control the expression of a number of metabolic genes (Kuo et al., 1990; Blumenfeld et al., 1991; Pontoglio, 2000). Both proteins share strong similarities in their dimerization and DNA-binding domains and therefore can bind the same DNA sequence either as hetero or homodimers (Baumhueter et al., 1988; Mendel et al., 1991). Although there is some overlap in tissue expression, HNF1α comprises a majority of total HNF1 protein present in the adult liver (Pontoglio, 2000). Knockout studies in mice have suggested a role for HNF1 proteins in regulating bile acid, bilirubin, and various aspects of glucose, amino acid, and lipid homeostasis (Pontoglio et al., 1996; Shih et al., 2001; Coffinier et al., 2002; Kornfeld et al., 2013). In the current study, we identified an HNF1 element in the SULT1C2 promoter, and overexpressing either HNF1α or 1β significantly induced reporter activity in HepG2 cells, indicating both proteins are capable of activating SULT1C2 transcription. Consistent with these findings, Servitja et al. (2009) reported that mRNA expression of SULT1C2 is reduced by 8.3-fold in pancreatic islet cells isolated from Hnf1α−/− mice (Servitja et al., 2009). An HNF1-binding element was also identified 522 nt upstream of the transcription start site of murine SULT1C2 (Servitja et al., 2009), further implicating HNF1 as an important mediator of basal SULT1C2 expression in rodents. It is noteworthy that HNF1 is coexpressed in many of the same tissues as rodent and human SULT1C2 (Cereghini, 1996; Dooley et al., 2000; Xiangrong et al., 2000). Therefore, a more detailed analysis of HNF1 on transcriptional regulation of SULT1C2 in different tissues and during development would further strengthen our findings and lead to a better understanding of this enzyme in both physiologic and pathophysiologic conditions.
In conclusion, our findings demonstrate that SULT1C2 is regulated by cholesterol biosynthetic intermediates and that HNF1 is an important mediator of SULT1C2 transcription in primary rat hepatocytes. Although the biologic function of SULT1C2 has not been fully elucidated, our findings indicate that hepatic expression is closely linked to cholesterol homeostasis and provide a basis for future studies aimed at identifying additional regulatory elements for this enzyme. Regulation of human hepatic SULT1C2 has not been previously evaluated, although mRNA levels are reportedly elevated in the livers of individuals with NASH, cirrhosis, and hepatocellular carcinoma (Lake et al., 2011; Grinberg et al., 2014). Relevant to our findings, cholesterol biosynthetic rates are increased in NASH, and HMGCR levels positively correlate with disease severity (Min et al., 2012), suggesting that some of the observed up-regulation may be related to changes in cholesterol biosynthetic intermediates.
Several endogenous metabolites involved in regulating the expression of SULTs are also known substrates for sulfonation. For example, SULT2A1 has high sulfonating activity toward the secondary bile acid lithocholic acid (Huang et al., 2010). VDR and PXR are activated by lithocholate and regulate the transcription of SULT2A1 (Makishima et al., 2002; Echchgadda et al., 2004; Fang et al., 2007). Additionally, LXR regulates the expression of SULT2A1 and 2B1b (Jiang et al., 2005; Ou et al., 2014), and several oxysterols that are ligands for LXR are also sulfonated by these enzymes (Fuda et al., 2007; Cook et al., 2009). Although cholecalciferol, produced from 7DHC in the skin, was not an efficient substrate (Xiangrong et al., 2000), desmosterol, 7DHC, and other postsqualene metabolites can be sulfonated (Masse et al., 1982; Axelson, 1987; Hu et al., 2015). Based on our findings, it would be interesting to determine whether any of these compounds are also endogenous substrates for SULT1C2.
Supplementary Material
Acknowledgments
The authors thank Mary Gargano for her technical expertise in isolating the rat hepatocytes.
Abbreviations
- AY-9944
N-[(2-chlorophenyl)methyl]-1-[4-[[(2-chlorophenyl)methylamino]methyl]cyclohexyl]methanamine dihydrochloride
- C/EBP
CCAAT enhancer-binding protein
- CAR
constitutive androstane receptor
- CDCA
chenodeoxycholic acid
- Cipro
ciprofibrate
- 7DHC
7-dehydrocholesterol
- DMSO
dimethylsulfoxide
- FPP
farnesyl pyrophosphate
- FXR
farnesoid X receptor
- GW3965
2-[3-[3-[[2-chloro-3-(trifluoromethyl)phenyl]methyl-(2,2-diphenylethyl)amino]propoxy]phenyl]acetic acid
- GW4064
3-[(E)-2-[2-chloro-4-[[3-(2,6-dichlorophenyl)-5-propan-2-yl-1,2-oxazol-4-yl]methoxy]phenyl]ethenyl]benzoic acid
- HMGCR
3-hydroxy-3-methylglutaryl-CoA reductase
- HNF
hepatocyte nuclear factor
- LXR
liver X receptor
- MVA
mevalonate
- NASH
nonalcoholic steatohepatitis
- NB-598
(E)-N-ethyl-6,6-dimethyl-N-[[3-[(4-thiophen-3-ylthiophen-2-yl)methoxy]phenyl]methyl]hept-2-en-4-yn-1-amine
- PPAR
peroxisome proliferator-activated receptor
- PB
phenobarbital
- Prav
pravastatin
- PXR
pregnane X receptor
- PCN
pregnenolone-16α-carbonitrile
- PCR
polymerase chain reaction
- RACE
rapid amplification of cDNA ends
- RXR
retinoid X receptor
- SQ1
squalestatin 1
- SULT
cytosolic sulfotransferase
- UTR
untranslated region
- VDR
vitamin D receptor
Authorship Contributions
Participated in research design: Rondini, Kocarek.
Conducted experiments: Rondini, Pant.
Performed data analysis: Rondini.
Wrote or contributed to the writing of the manuscript: Rondini, Kocarek, Pant.
Footnotes
This work was supported in part through a postdoctoral fellowship awarded through the Office of Vice President of Research (Wayne State University, Detroit, MI) (to E.A.R); and supported by the National Institutes of Health National Heart, Lung, and Blood Institute [Grant R01 HL050710 (to T.A.K.)] and National Institute of Environmental Health Sciences [Grant R01 ES022606 and Center Grant P30 ES020957].
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Aldridge TC, Tugwood JD, Green S. (1995) Identification and characterization of DNA elements implicated in the regulation of CYP4A1 transcription. Biochem J 306:473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong A, Loppnau P, Martin F, Thornton J, Edwards AM, et al. (2007) Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol 5:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almon RR, DuBois DC, Lai W, Xue B, Nie J, Jusko WJ. (2009) Gene expression analysis of hepatic roles in cause and development of diabetes in Goto-Kakizaki rats. J Endocrinol 200:331–346. [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. (2006) Tissue distribution and ontogeny of sulfotransferase enzymes in mice. Toxicol Sci 93:242–255. [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. (2008) Regulation of sulfotransferase enzymes by prototypical microsomal enzyme inducers in mice. J Pharmacol Exp Ther 324:612–621. [DOI] [PubMed] [Google Scholar]
- Axelson M. (1987) The cholecalciferol sulphate system in mammals. J Steroid Biochem 26:369–373. [DOI] [PubMed] [Google Scholar]
- Bach I, Mattei MG, Cereghini S, Yaniv M. (1991) Two members of an HNF1 homeoprotein family are expressed in human liver. Nucleic Acids Res 19:3553–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Q, Zhang X, Xu L, Kakiyama G, Heuman D, Sanyal A, Pandak WM, Yin L, Xie W, Ren S. (2012) Oxysterol sulfation by cytosolic sulfotransferase suppresses liver X receptor/sterol regulatory element binding protein-1c signaling pathway and reduces serum and hepatic lipids in mouse models of nonalcoholic fatty liver disease. Metabolism 61:836–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumhueter S, Courtois G, Crabtree GR. (1988) A variant nuclear protein in dedifferentiated hepatoma cells binds to the same functional sequences in the beta fibrinogen gene promoter as HNF-1. EMBO J 7:2485–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MWH. (2004) A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics 14:199–211. [DOI] [PubMed] [Google Scholar]
- Bloch K. (1965) The biological synthesis of cholesterol. Science 150:19–28. [DOI] [PubMed] [Google Scholar]
- Blumenfeld M, Maury M, Chouard T, Yaniv M, Condamine H. (1991) Hepatic nuclear factor 1 (HNF1) shows a wider distribution than products of its known target genes in developing mouse. Development 113:589–599. [DOI] [PubMed] [Google Scholar]
- Bostedor RG, Karkas JD, Arison BH, Bansal VS, Vaidya S, Germershausen JI, Kurtz MM, Bergstrom JD. (1997) Farnesol-derived dicarboxylic acids in the urine of animals treated with zaragozic acid A or with farnesol. J Biol Chem 272:9197–9203. [DOI] [PubMed] [Google Scholar]
- Buqué X, Martínez MJ, Cano A, Miquilena-Colina ME, García-Monzón C, Aspichueta P, Ochoa B. (2010) A subset of dysregulated metabolic and survival genes is associated with severity of hepatic steatosis in obese Zucker rats. J Lipid Res 51:500–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byskov AG, Andersen CY, Nordholm L, Thøgersen H, Xia G, Wassmann O, Andersen JV, Guddal E, Roed T. (1995) Chemical structure of sterols that activate oocyte meiosis. Nature 374:559–562. [DOI] [PubMed] [Google Scholar]
- Cereghini S. (1996) Liver-enriched transcription factors and hepatocyte differentiation. FASEB J 10:267–282. [PubMed] [Google Scholar]
- Chevy F, Illien F, Wolf C, Roux C. (2002) Limb malformations of rat fetuses exposed to a distal inhibitor of cholesterol biosynthesis. J Lipid Res 43:1192–1200. [PubMed] [Google Scholar]
- Coffinier C, Gresh L, Fiette L, Tronche F, Schütz G, Babinet C, Pontoglio M, Yaniv M, Barra J. (2002) Bile system morphogenesis defects and liver dysfunction upon targeted deletion of HNF1beta. Development 129:1829–1838. [DOI] [PubMed] [Google Scholar]
- Cook IT, Duniec-Dmuchowski Z, Kocarek TA, Runge-Morris M, Falany CN. (2009) 24-hydroxycholesterol sulfation by human cytosolic sulfotransferases: formation of monosulfates and disulfates, molecular modeling, sulfatase sensitivity, and inhibition of liver x receptor activation. Drug Metab Dispos 37:2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley TP, Haldeman-Cahill R, Joiner J, Wilborn TW. (2000) Expression profiling of human sulfotransferase and sulfatase gene superfamilies in epithelial tissues and cultured cells. Biochem Biophys Res Commun 277:236–245. [DOI] [PubMed] [Google Scholar]
- Duniec-Dmuchowski Z, Dombkowski AA, Kocarek TA, Runge-Morris M. (2012) Regulation of SULT1C2 expression by endogenous isoprenoids in primary cultured rat hepatocytes (Abstract). FASEB J 26(Suppl):673.11. [Google Scholar]
- Duniec-Dmuchowski Z, Fang HL, Strom SC, Ellis E, Runge-Morris M, Kocarek TA. (2009) Human pregnane X receptor activation and CYP3A4/CYP2B6 induction by 2,3-oxidosqualene:lanosterol cyclase inhibition. Drug Metab Dispos 37:900–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echchgadda I, Song CS, Roy AK, Chatterjee B. (2004) Dehydroepiandrosterone sulfotransferase is a target for transcriptional induction by the vitamin D receptor. Mol Pharmacol 65:720–729. [DOI] [PubMed] [Google Scholar]
- Edwards PA, Ericsson J. (1999) Sterols and isoprenoids: signaling molecules derived from the cholesterol biosynthetic pathway. Annu Rev Biochem 68:157–185. [DOI] [PubMed] [Google Scholar]
- Edwards PA, Kennedy MA, Mak PA. (2002) LXRs; oxysterol-activated nuclear receptors that regulate genes controlling lipid homeostasis. Vascul Pharmacol 38:249–256. [DOI] [PubMed] [Google Scholar]
- Fang HL, Abdolalipour M, Duanmu Z, Smigelski JR, Weckle A, Kocarek TA, Runge-Morris M. (2005) Regulation of glucocorticoid-inducible hydroxysteroid sulfotransferase (SULT2A-40/41) gene transcription in primary cultured rat hepatocytes: role of CCAAT/enhancer-binding protein liver-enriched transcription factors. Drug Metab Dispos 33:147–156. [DOI] [PubMed] [Google Scholar]
- Fang HL, Strom SC, Ellis E, Duanmu Z, Fu J, Duniec-Dmuchowski Z, Falany CN, Falany JL, Kocarek TA, Runge-Morris M. (2007) Positive and negative regulation of human hepatic hydroxysteroid sulfotransferase (SULT2A1) gene transcription by rifampicin: roles of hepatocyte nuclear factor 4alpha and pregnane X receptor. J Pharmacol Exp Ther 323:586–598. [DOI] [PubMed] [Google Scholar]
- Forman BM, Ruan B, Chen J, Schroepfer GJ, Jr, Evans RM. (1997) The orphan nuclear receptor LXRalpha is positively and negatively regulated by distinct products of mevalonate metabolism. Proc Natl Acad Sci USA 94:10588–10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimuth RR, Raftogianis RB, Wood TC, Moon E, Kim UJ, Xu J, Siciliano MJ, Weinshilboum RM. (2000) Human sulfotransferases SULT1C1 and SULT1C2: cDNA characterization, gene cloning, and chromosomal localization. Genomics 65:157–165. [DOI] [PubMed] [Google Scholar]
- Fuda H, Javitt NB, Mitamura K, Ikegawa S, Strott CA. (2007) Oxysterols are substrates for cholesterol sulfotransferase. J Lipid Res 48:1343–1352. [DOI] [PubMed] [Google Scholar]
- Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, McManus ME. (2006) Human sulfotransferases and their role in chemical metabolism. Toxicol Sci 90:5–22. [DOI] [PubMed] [Google Scholar]
- Gibbons GF, Pullinger CR. (1977) Measurement of the absolute rates of cholesterol biosynthesis in isolated rat liver cells. Biochem J 162:321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin B, Hodgson E, Liddle C. (1999) The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol 56:1329–1339. [DOI] [PubMed] [Google Scholar]
- Goto T, Nagai H, Egawa K, Kim YI, Kato S, Taimatsu A, Sakamoto T, Ebisu S, Hohsaka T, Miyagawa H, et al. (2011) Farnesyl pyrophosphate regulates adipocyte functions as an endogenous PPARγ agonist. Biochem J 438:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38(Web server issue):W695-W699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg M, Stöber RM, Edlund K, Rempel E, Godoy P, Reif R, Widera A, Madjar K, Schmidt-Heck W, Marchan R, et al. (2014) Toxicogenomics directory of chemically exposed human hepatocytes. Arch Toxicol 88:2261–2287. [DOI] [PubMed] [Google Scholar]
- Grünler J, Ericsson J, Dallner G. (1994) Branch-point reactions in the biosynthesis of cholesterol, dolichol, ubiquinone and prenylated proteins. Biochim Biophys Acta 1212:259–277. [DOI] [PubMed] [Google Scholar]
- Harries LW, Brown JE, Gloyn AL. (2009) Species-specific differences in the expression of the HNF1A, HNF1B and HNF4A genes. PLoS One 4:e7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her C, Kaur GP, Athwal RS, Weinshilboum RM. (1997) Human sulfotransferase SULT1C1: cDNA cloning, tissue-specific expression, and chromosomal localization. Genomics 41:467–470. [DOI] [PubMed] [Google Scholar]
- Horie M, Tsuchiya Y, Hayashi M, Iida Y, Iwasawa Y, Nagata Y, Sawasaki Y, Fukuzumi H, Kitani K, Kamei T. (1990) NB-598: a potent competitive inhibitor of squalene epoxidase. J Biol Chem 265:18075–18078. [PubMed] [Google Scholar]
- Horlick L. (1966) Effect of a new inhibitor of cholesterol biosynthesis (AY 9944) on serum and tissue sterols in the rat. J Lipid Res 7:116–121. [PubMed] [Google Scholar]
- Hu X, Wang Y, Hao LY, Liu X, Lesch CA, Sanchez BM, Wendling JM, Morgan RW, Aicher TD, Carter LL, et al. (2015) Sterol metabolism controls T(H)17 differentiation by generating endogenous RORγ agonists. Nat Chem Biol 11:141–147. [DOI] [PubMed] [Google Scholar]
- Huang J, Bathena SP, Tong J, Roth M, Hagenbuch B, Alnouti Y. (2010) Kinetic analysis of bile acid sulfation by stably expressed human sulfotransferase 2A1 (SULT2A1). Xenobiotica 40:184–194. [DOI] [PubMed] [Google Scholar]
- Hughes TA. (2006) Regulation of gene expression by alternative untranslated regions. Trends Genet 22:119–122. [DOI] [PubMed] [Google Scholar]
- Jackson NM, Kocarek TA. (2008) Suppression of CYP2B induction by alendronate-mediated farnesyl diphosphate synthase inhibition in primary cultured rat hepatocytes. Drug Metab Dispos 36:2030–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Grogan MJ, Jones SA, Wisely GB, Kliewer SA, Corey EJ, Mangelsdorf DJ. (1999) Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc Natl Acad Sci USA 96:266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. (1996) An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature 383:728–731. [DOI] [PubMed] [Google Scholar]
- Jiang YJ, Kim P, Elias PM, Feingold KR. (2005) LXR and PPAR activators stimulate cholesterol sulfotransferase type 2 isoform 1b in human keratinocytes. J Lipid Res 46:2657–2666. [DOI] [PubMed] [Google Scholar]
- Kocarek TA, Kraniak JM, Reddy AB. (1998) Regulation of rat hepatic cytochrome P450 expression by sterol biosynthesis inhibition: inhibitors of squalene synthase are potent inducers of CYP2B expression in primary cultured rat hepatocytes and rat liver. Mol Pharmacol 54:474–484. [DOI] [PubMed] [Google Scholar]
- Kocarek TA, Mercer-Haines NA. (2002) Squalestatin 1-inducible expression of rat CYP2B: evidence that an endogenous isoprenoid is an activator of the constitutive androstane receptor. Mol Pharmacol 62:1177–1186. [DOI] [PubMed] [Google Scholar]
- Kocarek TA, Reddy AB. (1996) Regulation of cytochrome P450 expression by inhibitors of hydroxymethylglutaryl-coenzyme A reductase in primary cultured rat hepatocytes and in rat liver. Drug Metab Dispos 24:1197–1204. [PubMed] [Google Scholar]
- Kornfeld JW, Baitzel C, Könner AC, Nicholls HT, Vogt MC, Herrmanns K, Scheja L, Haumaitre C, Wolf AM, Knippschild U, et al. (2013) Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature 494:111–115. [DOI] [PubMed] [Google Scholar]
- Kuo CJ, Conley PB, Hsieh CL, Francke U, Crabtree GR. (1990) Molecular cloning, functional expression, and chromosomal localization of mouse hepatocyte nuclear factor 1. Proc Natl Acad Sci USA 87:9838–9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffitte BA, Kast HR, Nguyen CM, Zavacki AM, Moore DD, Edwards PA. (2000) Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J Biol Chem 275:10638–10647. [DOI] [PubMed] [Google Scholar]
- Lake AD, Novak P, Fisher CD, Jackson JP, Hardwick RN, Billheimer DD, Klimecki WT, Cherrington NJ. (2011) Analysis of global and absorption, distribution, metabolism, and elimination gene expression in the progressive stages of human nonalcoholic fatty liver disease. Drug Metab Dispos 39:1954–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann JM, Kliewer SA, Moore LB, Smith-Oliver TA, Oliver BB, Su JL, Sundseth SS, Winegar DA, Blanchard DE, Spencer TA, et al. (1997) Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J Biol Chem 272:3137–3140. [DOI] [PubMed] [Google Scholar]
- Lekstrom-Himes J, Xanthopoulos KG. (1998) Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem 273:28545–28548. [DOI] [PubMed] [Google Scholar]
- Li X, Clemens DL, Anderson RJ. (2000) Sulfation of iodothyronines by human sulfotransferase 1C1 (SULT1C1)*. Biochem Pharmacol 60:1713–1716. [DOI] [PubMed] [Google Scholar]
- Lindsay J, Wang LL, Li Y, Zhou SF. (2008) Structure, function and polymorphism of human cytosolic sulfotransferases. Curr Drug Metab 9:99–105. [DOI] [PubMed] [Google Scholar]
- Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. (2002) Vitamin D receptor as an intestinal bile acid sensor. Science 296:1313–1316. [DOI] [PubMed] [Google Scholar]
- Massé R, Huang YS, Eid K, Laliberté C, Davignon J. (1982) Plasma methyl sterol sulfates in familial hypercholesterolemia after partial ileal bypass. Can J Biochem 60:556–563. [DOI] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen CY, Chou A, Ienasescu H, et al. (2014) JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res 42:D142–D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meljon A, Watson GL, Wang Y, Shackleton CH, Griffiths WJ. (2013) Analysis by liquid chromatography-mass spectrometry of sterols and oxysterols in brain of the newborn Dhcr7(Δ3-5/T93M) mouse: a model of Smith-Lemli-Opitz syndrome. Biochem Pharmacol 86:43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel DB, Hansen LP, Graves MK, Conley PB, Crabtree GR. (1991) HNF-1 alpha and HNF-1 beta (vHNF-1) share dimerization and homeo domains, but not activation domains, and form heterodimers in vitro. Genes Dev 5:1042–1056. [DOI] [PubMed] [Google Scholar]
- Metz RP, Auyeung DJ, Kessler FK, Ritter JK. (2000) Involvement of hepatocyte nuclear factor 1 in the regulation of the UDP-glucuronosyltransferase 1A7 (UGT1A7) gene in rat hepatocytes. Mol Pharmacol 58:319–327. [DOI] [PubMed] [Google Scholar]
- Min HK, Kapoor A, Fuchs M, Mirshahi F, Zhou H, Maher J, Kellum J, Warnick R, Contos MJ, Sanyal AJ. (2012) Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab 15:665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima S, Yagyu H, Tozawa R, Tazoe F, Takahashi M, Kitamine T, Yamamuro D, Sakai K, Sekiya M, Okazaki H, et al. (2015) Plasma cholesterol-lowering and transient liver dysfunction in mice lacking squalene synthase in the liver. J Lipid Res 56:998–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Z, Jiang M, Hu B, Huang Y, Xu M, Ren S, Li S, Liu S, Xie W, Huang M. (2014) Transcriptional regulation of human hydroxysteroid sulfotransferase SULT2A1 by LXRα. Drug Metab Dispos 42:1684–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniswamy R, Teglund S, Lauth M, Zaphiropoulos PG, Shimokawa T. (2010) Genetic variations regulate alternative splicing in the 5′ untranslated regions of the mouse glioma-associated oncogene 1, Gli1. BMC Mol Biol 11:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontoglio M. (2000) Hepatocyte nuclear factor 1, a transcription factor at the crossroads of glucose homeostasis. J Am Soc Nephrol 11 (Suppl 16):S140–S143. [PubMed] [Google Scholar]
- Pontoglio M, Barra J, Hadchouel M, Doyen A, Kress C, Bach JP, Babinet C, Yaniv M. (1996) Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell 84:575–585. [DOI] [PubMed] [Google Scholar]
- Popják G, Meenan A, Parish EJ, Nes WD. (1989) Inhibition of cholesterol synthesis and cell growth by 24(R,S),25-iminolanosterol and triparanol in cultured rat hepatoma cells. J Biol Chem 264:6230–6238. [PubMed] [Google Scholar]
- Rollier A, DiPersio CM, Cereghini S, Stevens K, Tronche F, Zaret K, Weiss MC. (1993) Regulation of albumin gene expression in hepatoma cells of fetal phenotype: dominant inhibition of HNF1 function and role of ubiquitous transcription factors. Mol Biol Cell 4:59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondini EA, Fang H, Runge-Morris M, Kocarek TA. (2014) Regulation of human cytosolic sulfotransferases 1C2 and 1C3 by nuclear signaling pathways in LS180 colorectal adenocarcinoma cells. Drug Metab Dispos 42:361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge-Morris M, Kocarek TA. (2013) Expression of the sulfotransferase 1C family: implications for xenobiotic toxicity. Drug Metab Rev 45:450–459. [DOI] [PubMed] [Google Scholar]
- Runge-Morris M, Kocarek TA, Falany CN. (2013) Regulation of the cytosolic sulfotransferases by nuclear receptors. Drug Metab Rev 45:15–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara Y, Yanagisawa K, Katafuchi J, Ringer DP, Takami Y, Nakayama T, Suiko M, Liu MC. (1998) Molecular cloning, expression, and characterization of novel human SULT1C sulfotransferases that catalyze the sulfonation of N-hydroxy-2-acetylaminofluorene. J Biol Chem 273:33929–33935. [DOI] [PubMed] [Google Scholar]
- Sánchez-Wandelmer J, Dávalos A, Herrera E, Giera M, Cano S, de la Peña G, Lasunción MA, Busto R. (2009) Inhibition of cholesterol biosynthesis disrupts lipid raft/caveolae and affects insulin receptor activation in 3T3-L1 preadipocytes. Biochim Biophys Acta 1788:1731–1739. [DOI] [PubMed] [Google Scholar]
- Santori FR, Huang P, van de Pavert SA, Douglass EF, Jr, Leaver DJ, Haubrich BA, Keber R, Lorbek G, Konijn T, Rosales BN, et al. (2015) Identification of natural RORγ ligands that regulate the development of lymphoid cells. Cell Metab 21:286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servitja JM, Pignatelli M, Maestro MA, Cardalda C, Boj SF, Lozano J, Blanco E, Lafuente A, McCarthy MI, Sumoy L, et al. (2009) Hnf1alpha (MODY3) controls tissue-specific transcriptional programs and exerts opposed effects on cell growth in pancreatic islets and liver. Mol Cell Biol 29:2945–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih DQ, Bussen M, Sehayek E, Ananthanarayanan M, Shneider BL, Suchy FJ, Shefer S, Bollileni JS, Gonzalez FJ, Breslow JL, et al. (2001) Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet 27:375–382. [DOI] [PubMed] [Google Scholar]
- Stanley EL, Hume R, Coughtrie MWH. (2005) Expression profiling of human fetal cytosolic sulfotransferases involved in steroid and thyroid hormone metabolism and in detoxification. Mol Cell Endocrinol 240:32–42. [DOI] [PubMed] [Google Scholar]
- Strott CA. (2002) Sulfonation and molecular action. Endocr Rev 23:703–732. [DOI] [PubMed] [Google Scholar]
- Sugimura K, Tanaka T, Tanaka Y, Takano H, Kanagawa K, Sakamoto N, Ikemoto S, Kawashima H, Nakatani T. (2002) Decreased sulfotransferase SULT1C2 gene expression in DPT-induced polycystic kidney. Kidney Int 62:757–762. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Kawada T, Goto T, Yamamoto T, Taimatsu A, Matsui N, Kimura K, Saito M, Hosokawa M, Miyashita K, et al. (2002) Dual action of isoprenols from herbal medicines on both PPARγ and PPARα in 3T3-L1 adipocytes and HepG2 hepatocytes. FEBS Lett 514:315–322. [DOI] [PubMed] [Google Scholar]
- Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, Darlington GJ. (1995) Impaired energy homeostasis in C/EBP alpha knockout mice. Science 269:1108–1112. [DOI] [PubMed] [Google Scholar]
- Wong J, Quinn CM, Gelissen IC, Brown AJ. (2008) Endogenous 24(S),25-epoxycholesterol fine-tunes acute control of cellular cholesterol homeostasis. J Biol Chem 283:700–707. [DOI] [PubMed] [Google Scholar]
- Xiangrong L, Jöhnk C, Hartmann D, Schestag F, Krömer W, Gieselmann V. (2000) Enzymatic properties, tissue-specific expression, and lysosomal location of two highly homologous rat SULT1C2 sulfotransferases. Biochem Biophys Res Commun 272:242–250. [DOI] [PubMed] [Google Scholar]
- Yang C, McDonald JG, Patel A, Zhang Y, Umetani M, Xu F, Westover EJ, Covey DF, Mangelsdorf DJ, Cohen JC, et al. (2006) Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J Biol Chem 281:27816–27826. [DOI] [PubMed] [Google Scholar]
- Zhao C, Dahlman-Wright K. (2010) Liver X receptor in cholesterol metabolism. J Endocrinol 204:233–240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.