Abstract
UDP-Glucuronosyltransferases (UGTs) conjugate a glucuronyl group from glucuronic acid to a wide range of lipophilic substrates to form a hydrophilic glucuronide conjugate. The glucuronide generally has decreased bioactivity and increased water solubility to facilitate excretion. Glucuronidation represents an important detoxification pathway for both endogenous waste products and xenobiotics, including drugs and harmful industrial chemicals. Two clinically significant families of UGT enzymes are present in mammals: UGT1s and UGT2s. Although the two families are distinct in gene structure, studies using recombinant enzymes have shown considerable overlap in their ability to glucuronidate many substrates, often obscuring the relative importance of the two families in the clearance of particular substrates in vivo. To address this limitation, we have generated a mouse line, termed ΔUgt2, in which the entire Ugt2 gene family, extending over 609 kilobase pairs, is excised. This mouse line provides a means to determine the contributions of the two UGT families in vivo. We demonstrate the utility of these animals by defining for the first time the in vivo contributions of the UGT1 and UGT2 families to glucuronidation of the environmental estrogenic agent bisphenol A (BPA). The highest activity toward this chemical is reported for human and rodent UGT2 enzymes. Surprisingly, our studies using the ΔUgt2 mice demonstrate that, while both UGT1 and UGT2 isoforms can conjugate BPA, clearance is largely dependent on UGT1s.
Introduction
UDP-Glucuronosyltransferases (UGTs) are membrane-bound phase II enzymes that conjugate a wide array of lipophilic substrates with a glucuronyl group from UDP-glucuronic acid (UDPGA). The highly polar glucuronide conjugate generally decreases the bioactivity of the substrate and increases its water solubility, facilitating excretion through bile or urine. Glucuronidation is important in the metabolism of endogenous wastes and a wide range of drugs, including many chemotherapeutics, nonsteroidal anti-inflammatory drugs, and opioids, and in protecting against harmful environmental agents such as bisphenol A (BPA) and benzo-[a]-pyrene (Fang et al., 2002; Hanioka et al., 2008; Kutsuno et al., 2013). Based on evolutionary divergence and homology, mammalian UGTs have been organized into two major families: The UGT1 and UGT2 families. The mammalian UGT1 family currently contains only the UGT1A subfamily, while the mammalian UGT2 family contains the UGT2A and UGT2B subfamilies (Mackenzie et al., 2005; Rowland et al., 2013).
The mouse UGT1 family is encoded by the Ugt1a gene. It consists of nine functional isoforms, each encoded by a unique 5′ exon spliced to a shared set of downstream exons (Mackenzie et al., 2005). The first exon specifies a UGT1A isoform and its substrate specificity, while the downstream exons encode common domains such as a UDPGA binding site and a transmembrane region (Owens et al., 2005). The three Ugt2a and seven Ugt2b genes in mouse are located in a single cluster spanning approximately 600 kilobase pairs (kbp) on chromosome 5 (Mackenzie et al., 2005). Ugt2a1 and Ugt2a2, similar to Ugt1a, have unique first exons and a shared set of downstream exons (Jedlitschky et al., 1999). All other Ugt2 genes have a typical structure, with unique exons encoding both the N-terminal substrate-binding domain and a C-terminal UDPGA-binding domain and transmembrane sequence (Hum et al., 1999). Similar to the UGT1 family, the C-terminal domains of the UGT2 family are conserved, while the highly variable N-terminal domains determine substrate specificity (Radominska-Pandya et al., 1999). Many UGT isoforms exhibit activity toward a broad range of substrates, and the specificities of different isoforms often overlap (de Wildt et al., 1999).
Various approaches have been used to determine which UGTs are involved in glucuronidation of particular compounds (Rowland et al., 2013). Assignment using computational approaches is limited by the lack of established structure-activity relations for most UGTs (Kaivosaari, 2010). The majority of current knowledge comes from assessment of metabolism of substrates by microsomes reconstituted with recombinant enzymes. Such studies have shown that UGT2s are involved in conjugation of a variety of endogenous metabolites, such as retinoic acid and steroid hormones, as well as xenobiotics, such as opioid analgesics (Hum et al., 1999; Radominska-Pandya et al., 2001; Chouinard et al., 2007). While information from these studies can be used to predict the relative importance of various enzymes in vivo, extrapolation of in vitro results to in vivo systems remains an imperfect practice (Lin and Wong, 2002; Miners et al., 2006). Such modeling has to take into account not only the activities of the enzymes toward the substrate under study but also the fact that there are quantitative and qualitative differences in the expression of these enzymes in the tissues exposed to the chemical. Ultimately, in vivo metabolism is required to confirm the contribution of particular UGT enzymes to the metabolism of a compound. However, due to the functional overlap of many isoforms, it is often difficult to measure the contributions of one isoform or even one UGT family in vivo.
These challenges have been encountered in defining the contributions of different UGTs to metabolism and clearance of environmental pollutants such as BPA, a monomer commonly used in polycarbonate materials with widespread human exposure. BPA’s reported toxicity in mice at low doses (Richter et al., 2007), possibly through interference in both genomic and nongenomic estrogen responses (Luconi et al., 2002), has made it the subject of intense research (Krishnan et al., 1993). The ability of BPA to impact physiologic processes is likely limited by the rapid clearance of the inactive bisphenol A glucuronide (BPAG) (Matthews et al., 2001; Völkel et al., 2002), and studies with recombinant enzymes indicate that human UGT2B15 and rodent UGT2B1 likely play the primary role in clearance (Yokota et al., 1999; Hanioka et al., 2008). This has raised concerns regarding exposure of neonates to BPA (Ginsberg and Rice, 2009), since expression of UGT2 genes during the neonatal period is much lower than in children and adults (Coughtrie et al., 1988; de Wildt et al., 1999; Divakaran et al., 2014). Despite extensive efforts toward understanding the health impact of BPA exposure, remarkably little is known regarding its metabolic pathways in human or in the model organisms used in assessing its adverse effects.
To address these limitations, we have generated a mouse line homozygous for a targeted deletion of the entire Ugt2 family. We show that loss of these enzymes does not adversely affect the development of these animals, and that microsomes from mice lacking all Ugt2 genes can be used to establish the role of the UGT2 enzymes in glucuronidation of xenobiotics. Most importantly, this mouse line can be used to investigate the contribution of UGT2 enzymes to the metabolism of drugs and xenobiotics in vivo.
Materials and Methods
Reagents and Materials.
Alamethicin and (S)-naproxen were purchased from Cayman Chemical (Ann Arbor, MI). BPA, BPAG, UDPGA, and β-glucuronidase were purchased from Sigma-Aldrich (St. Louis, MO). Naproxen glucuronide (napG) [(S)-naproxen acyl β-D glucuronide] was purchased from Toronto Research Chemicals (Toronto). All solvents and other reagents were of analytical grade or better. Zorbax Eclipse XDB-C18 and Tosoh TSKGel 80TM columns were purchased from Agilent Technologies (Santa Clara, CA) and Tosoh Bioscience (Tokyo), respectively. Primer sequences can be found in Supplemental Material.
Generation of Mouse Lines.
A replacement-type targeting vector was assembled from 129 bacterial artificial chromosomes bMQ357m23 and bMQ37h06. DNA corresponding to the Ugt2 gene cluster was replaced with a neomycin-resistance gene driven by the phosphoglycerate kinase promoter. Homologous recombination of the targeting vector with the endogenous Ugt2 locus creates a 609 kbp deletion extending from 580 base pairs downstream from the Ugt2b34 gene to 7.2 kbp upstream of the Ugt2a1/2 gene. 129S6-derived embryonic stem (ES) cells were cultured using standard methods, and DNA was introduced by electroporation. Cells were selected in Geneticin and were evaluated by polymerase chain reaction (PCR) and Southern blot analysis. Correctly targeted ES cells were introduced into C56BL/6 blastocysts and the blastocysts introduced into B6D2F1/J foster mothers to complete their development. All mice were maintained in specific pathogen–free housing in ventilated caging. 129S6 mice purchased from Taconic (Hudson, NY) were bred to chimeras to maintain the deleted locus on this genetic background. All mice used in these studies are therefore 129S6, coisogenic for the deleted Ugt2 locus. For all studies of reproductive function, wild-type (WT) and null mice were generated by the intercross of heterozygous animals. Serum testosterone levels were measured by enzyme-linked immunosorbent assay (Endocrine Technologies, Newark, CA). For some experiments mutant mice were age and sex matched at weaning and housed together until used in an experiment. All studies were conducted in accordance with the National institutes of Health Guide for the Care and Use of Laboratory Animals (National Research Council, 2011) as well as the Institutional Animal Care and Use Committee guidelines for the University of North Carolina at Chapel Hill.
Gene Expression Assays.
Total RNA was isolated from tissue homogenates using a Quick-RNA Miniprep Kit (Zymo Research, Irvine, CA), and reverse transcription PCR was performed using a High Capacity cDNA Reverse Transcription Kit (Life Technologies, Grand Island, NY). cDNA was amplified with the FastStart Universal Probe Master with Rox normalization (Roche Applied Science, Indiana, IN), and Ugt1 and Ugt2 expressions were measured using SYBR Green quantitative PCR primers and TaqMan quantitative PCR probe mixes (Life Technologies), respectively. Amplifications were performed in duplicate on a QuantStudio 6 Flex Real-Time PCR System (Life Technologies). Data were analyzed using the comparative threshold cycle method as described by Applied Biosystems (Waltham, MA). Relative expression was determined by normalizing samples to 18S rRNA expression.
Microsome Preparation.
Liver microsomes were prepared from 3 to 5 month old males. Median and left lobes of each liver were harvested after systemic perfusion with phosphate-buffered saline and homogenized manually in 0.05 M phosphate buffer (pH 7.4) with 0.154 M potassium chloride and 0.25 M sucrose. Homogenate was centrifuged twice at 10,000g for 10 minutes, and the postmitochondrial fraction was removed and centrifuged at 100,000g for 45 minutes. Microsomal pellets were washed with homogenization buffer and resuspended in 0.1 M phosphate buffer (pH 7.4) with 20% glycerol (v/v) and 1 mM EDTA. All preparation steps were performed at 4°C. Microsomal protein concentration was determined using Bio-Rad Protein Assay Dye (Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin as a standard. Microsomes were stored at −80°C until used and underwent no more than one freeze/thaw cycle before experimentation.
High-Performance Liquid Chromatography (HPLC).
HPLC was performed on an Agilent Technologies 1200 Series instrument. The system is comprised of a binary solvent pump with temperature-controlled autosampler, on-line degasser, variable-wavelength UV-visible detector, temperature-controlled column oven, and automated fraction collector. Chromatographic protocols are detailed in Table 1. Identities of all glucuronide peaks were verified by comparison with an authentic standard and by loss of the peak after β-glucuronidase treatment.
TABLE 1.
Chromatographic protocols
The aqueous solvent (A) was 0.1% acetic acid in water (v/v) and the organic solvent (B) was 0.1% acetic acid in acetonitrile (v/v). The autosampler chamber was held at 6°C, and columns were held at 25°C for all experiments. The protocol for BPA in vivo is based on that of Yokota et al. (1999).
| Assay |
Column |
λ |
Flow Rate |
Elution Program |
|---|---|---|---|---|
| nm | ml/min | |||
| Naproxen in vitro | Zorbax Eclipse XDB-C18 (150 × 4.6 mm, 5µm) | 225 | 1.50 | 30% B from 0 to 5 minutes |
| Linear increase to 70% B from 5 to 5.5 minutes | ||||
| Linear increase to70% B from 5.5 to 10 minutes | ||||
| BPA in vitro | Zorbax Eclipse XDB-C18 (150 × 4.6 mm, 5µm) | 280 | 0.50 | 15% B from 0 to 4.5 minutes |
| Linear increase to 60% B from 4.5 to 5 minutes | ||||
| Linear increase to 60% B from 5 to 16 minutes | ||||
| BPA in vivo | Tosoh TSKgel ODS 80TM (250 × 4.6 mm, 5 µm) | 280 | 1.00 | Isocratic 35% B |
Microsomal Glucuronidation Assays.
Glucuronidation reaction mixtures contained 0.1 M Tris buffer (pH 7.5), 4 mM magnesium chloride, substrate, and the indicated amount of microsomes in a final volume of 500 µl. Total organic solvent concentrations were less than 0.7% (v/v). Naproxen glucuronidation reactions contained 25 µg/ml alamethicin; BPA glucuronidation was performed both with and without alamethicin. After preincubation on ice and preheating at 37°C for 5 minutes, reactions were initiated by addition of UDPGA to a final concentration of 5 mM and incubated at 37°C. Reactions were terminated by addition to an equal volume of ice-cold 4% acetic acid in methanol (v/v) and vortex mixing. Microsomal protein was removed by centrifugation at 10,000g for 10 minutes at 4°C, and 30 µl of supernatant was analyzed by HPLC. Controls included baseline measurements and blanks lacking UDPGA. Glucuronide cleavage was performed by incubation of samples with 1000 units of β-glucuronidase for 4 hours at pH 5 to 6. Retention times for naproxen and its glurcuronide, napG, were 8.4 and 4.7 minutes, respectively. Consistent with previous studies (Bowalgaha et al., 2005), the napG standard was unstable, and napG was quantified based on a standard curve for naproxen, which was linear in the range of 1–100 µM (r2 > 0.999). The lower limit of detection was 1 µM for naproxen and napG. Retention times for BPA and BPAG were 15.2 and 12.8 minutes, respectively. Standard curves for BPA and BPAG were linear in the range of 0.5 µM to 1 mM (r2 > 0.999 for both), and the lower limit of quantification was 0.5 µM for BPA and BPAG. Kinetics data were analyzed using Microsoft Excel 2010 and GraphPad Prism version 4.0 software (GraphPad Software, La Jolla, CA).
In Vivo BPA Glucuronidation.
Age- and weight-matched WT and ΔUgt2 mice were anesthetized with urethane, and the bile duct was cannulated. After collection of a sample of bile prior to dosage, mice received BPA intravenously as a bolus in 10% ethanol/water (v/v). Two BPA dosage levels were examined: 2 and 20 mg BPA/kg b.wt. Bile fractions were collected in 10 minute intervals over the next 50 minutes. Samples were diluted 1:40 in deionized water, and 30 µl of each sample were analyzed by HPLC. Retention time for BPAG was 5.7 minutes. Standard curves for BPAG were linear in the range of 0.5 µM to 1 mM (r2 > 0.999) and the lower limit of quantification was 0.5 µM for BPA and BPAG.
Results
Generation of Mouse Line and Verification of ΔUgt2 Locus.
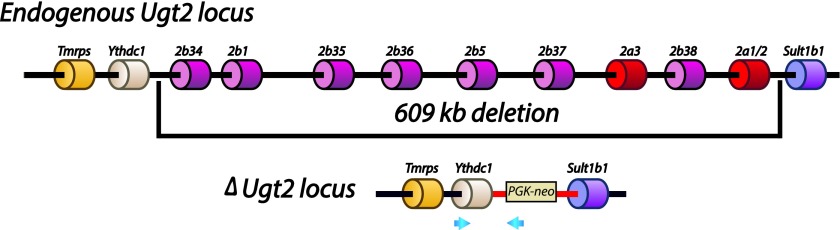
The Ugt2 genes are located in a segment of DNA on chromosome 5 between Ythdc1 and Sult1b1 (Fig. 1). While numerous polymorphisms distinguish the Ugt2 locus of the 129 strain from that of C57BL/6, no difference in the number and overall organization of the genes was identified by PCR, sequence, or Southern blot analysis. Therefore, using the organization of the published C57BL/6 locus as a guide, we generated a targeting vector designed to remove the entire gene family in a single recombination event in 129 derived ES cells. The deletion was designed to leave the Ythdc1 and Sult1b1 genes, including regulatory regions, intact (Fig. 1). Sult1b1 encodes a sulfotransferase enzyme that contributes to conjugation and excretion of endo- and xenobiotics, and therefore any compromise in the expression of this gene would complicate interpretation of the phenotypes of the UGT2-deficient animals. ES cells were identified where the targeting vector had integrated into the Ugt2 locus. These were analyzed to ensure that the recombination event had occurred at the predicted locations in the mouse genome and had resulted in loss of all Ugt2 genes. This analysis included extensive PCR, sequence, and Southern blot analyses. ES cells that met these criteria were used to generate a mouse line. For simplicity, we refer to this mutation as the ΔUgt2 allele. The ΔUgt2 allele was maintained on the 129S6 genetic background by breeding transmitting chimeras to purchased 129S6 dams. Mice heterozygous for the deletion were intercrossed to generate mice homozygous for the allele. DNA from these pups was used to further verify the loss of all Ugt2 genes by examination of both DNA and RNA from these animals. A probe was generated by PCR from exon 1 of Ugt2b38 that is expected to bind to multiple sites throughout the deleted segment, including the first exons of Ugt2b5, Ugt2b37, Ugt2b36, and Ugt2b35 as well as the intergenic region between Ugt2a3 and Ugt2b38. Analysis of DNA from pups generated by intercross of heterozygous parents showed that this probe failed to bind to DNA from the homozygous ΔUgt2 offspring (Fig. 2A). As expected, probes corresponding to regions flanking the deletion bound to the DNA from these animals and identified the predicted, novel DNA fragments generated by the recombination event.
Fig. 1.
Schematic drawing showing the structure of the WT and ΔUgt2 loci. The locus includes all three Ugt2a and seven Ugt2b genes and is located between Ythdc1 and Sult1b1 on chromosome 5. The 609 kbp segment of DNA deleted during the homologous recombination event is indicated, and the structure of the ΔUgt2 locus is shown below. The locus carries marker genes, and vector sequences are shown in red. The blue arrows show primers used in the initial screening of ES cell clones to identify those in which the deletion vector is inserted by homologous recombination.
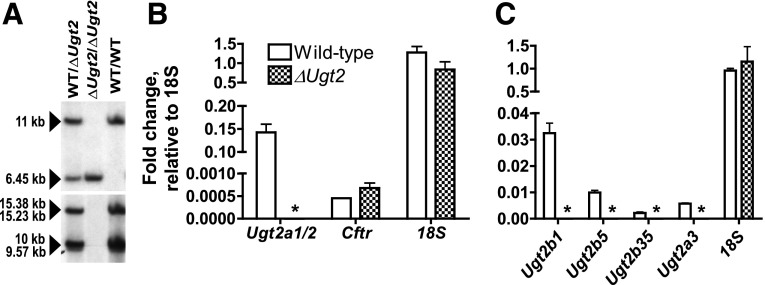
Fig. 2.
Verification of the deletion of Ugt2 genes. (A) DNA was prepared from tail biopsies of homozygous WT, homozygous ΔUgt2, and heterozygous pups, digested with BamHI, and analyzed by Southern blot. A probe corresponding to a region just outside the Ugt2 locus was used to verify the genotypes of the mice and the integrity of the DNA. As predicted, it hybridized to 11 and 6.45 kbp DNA fragments corresponding to the WT allele and ΔUgt2 locus, respectively (upper panel). The filter was then analyzed with a probe binding to BamHI fragments corresponding to a conserved region in Ugt2a3, Ugt2b1, Ugt2b5, and Ugt2b35 (lower panel). The WT locus yielded four different fragments (15.38, 15.23, 10, and 9.57 kbp), as expected. Consistent with the excision of the entire Ugt2 locus during a single recombination event, no hybridization of the probe is observed in the lanes corresponding to the mice homozygous for the ΔUgt2 locus. (B) RNA from olfactory epithelium was analyzed by quantitative PCR for Ugt2a1/2, normalized to 18S expression. As expected, robust expression is observed in WT mice, while none was detected in the ΔUgt2 animals, as indicated by the asterisks. The quality and epithelial origin of the RNA was verified by showing that Cftr expression was robust and that the expression level did not differ between the ΔUgt2 and WT animals. (C) RNA prepared from liver was analyzed by quantitative PCR for expression of Ugt2b1, Ugt2b35, Ugt2b5, and Ugt2a3 and normalized to 18S expression. N = 3 for all samples and error bars represent the S.E.M.
RNA was prepared from the nasal epithelium and liver of homozygous ΔUgt2 animals and their WT littermates. As expected, high levels of Ugt2a1/2 expression were observed in epithelium from WT animals, while expression was absent in samples collected from the null mice (Fig. 2B). Expression of members of the Ugt2b subfamily and Ugt2a3 were examined in RNA prepared from the liver. Again, expression of these genes was easily detected in the WT animals, whereas no signal was observed in ΔUgt2 animals (Fig. 2C). Taken together, these data support the DNA analysis indicating that the ΔUgt2 mice lack all Ugt2b and Ugt2a genes.
Development and Reproductive Behavior of the ΔUgt2 Mouse.
Mice lacking the Ugt2 genes were present in litters at expected Mendelian ratios. The growth and development of the mice were normal, and they could not be distinguished by observation from their WT littermates. The human UGT2B enzymes, including UGT2B4, UGT2B7, UGT2B15, and UGT2B17, have been shown to participate in the glucuronidation of endogenous steroids as well as xenobiotics (Hum et al., 1999). Alteration in steroid metabolism could dramatically alter the development of primary and secondary sex organs, and thus impair the fecundity of mice. Therefore, we intercrossed mice homozygous for the ΔUgt2 locus with WT littermates. The number of pups born to dams was within the normal size range (Supplemental Fig. 1). Furthermore, when ΔUgt2 males were presented with fertile WT females, the interval until the first mating, the number of matings, the percentage of matings resulting in pregnancies, and the size of the resulting litters were not significantly different when compared with WT males (Supplemental Fig. 2). To further examine this point, male mice were sacrificed at 8 weeks, and their body weight and the sizes of their testes and seminiferous tubules were examined. No difference in the development of these organs was observed, consistent with the normal fertility of these animals (Supplemental Fig. 3). Serum testosterone levels measured in individually housed, 5 to 6 month old male mice did not differ significantly from controls (Supplemental Fig. 4).
Gene Expression.
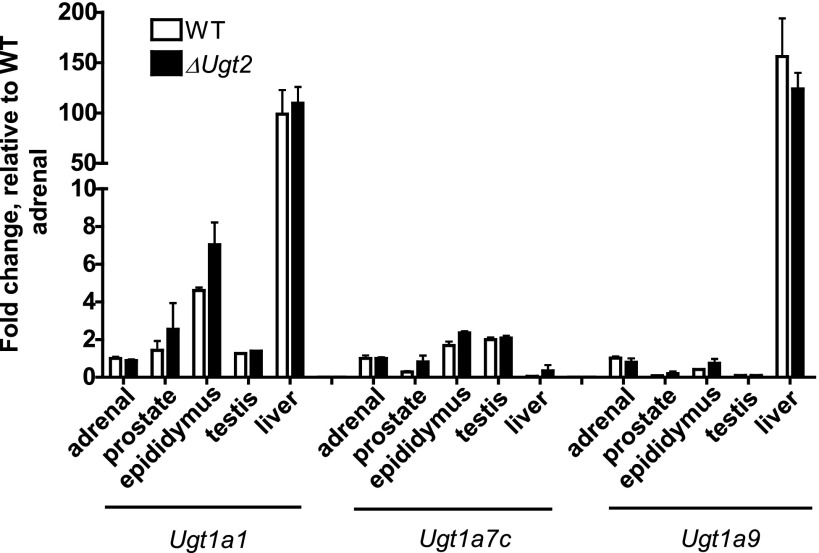
The health of the ΔUgt2 animals could reflect compensation for the loss of Ugt2 genes by increased expression of Ugt1s since substantial overlaps in the activities of these two enzyme families have been documented in both humans and rats (Radominska-Pandya et al., 1999; Lin and Wong, 2002; Bock, 2010). Increased expression of Ugt1 isoforms in response to loss of the Ugt2 genes would impact any conclusions drawn regarding observed differences in the metabolism of chemicals by the ΔUgt2 animals. As mentioned previously, the Ugt1 isoforms each have a unique 5′ exon. Primers specific for the first exons of Ugt1a1, Ugt1a7, and Ugt1a9 were generated and each was paired with a common reverse primer corresponding to the shared second exon. Quantitative PCR analysis using these primers revealed that expression patterns for these three Ugt1 mRNAs are similar to those reported previously (Buckley and Klaassen, 2007). When compared with WT mice, ΔUgt2 mice showed no significant difference in expression of any of the three mRNAs in the adrenal gland, liver, prostate, epididymis, or testes (Fig. 3). This indicates that the normal development of these mice does not reflect compensation for loss of these genes by increased expression of UGT1 isoforms. These results also support the general usefulness of this mouse line for determining the contribution of the Ugt2 genes to xenobiotic metabolism.
Fig. 3.
Expression and tissue distribution of Ugt1a mRNAs in WT and ΔUgt2 mice. Ugt1a expression levels were measured by quantitative real-time PCR with a SYBR Green quantitative PCR Master Mix. Primers were designed to amplify across the junction between the unique first exon for each isoform and the shared exons. cDNA samples were diluted to 10 pg/µl for amplification with the 18S probe and to 2.5 ng/µl for all other probes. The comparative threshold cycle method was used to quantify the relative gene expression. The amount of target was normalized to 18S as an endogenous reference and to WT adrenal gland as a calibrator. Expression of the examined isoforms did not differ significantly in any tissue examined between WT and ΔUgt2 animals. Amplifications were performed in duplicate. Error bars represent the S.E.M. (n = 3).
Naproxen Glucuronidation.
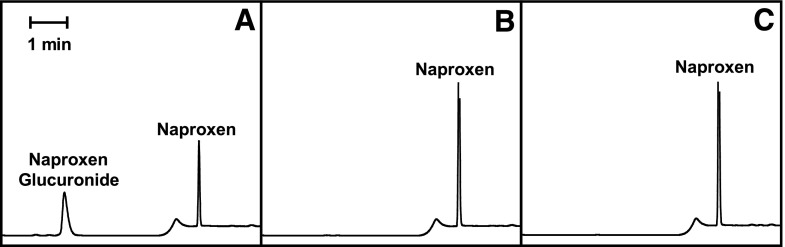
To demonstrate the usefulness of the ΔUgt2 mice in evaluation of the role of UGT2 enzymes in xenobiotic metabolism, we examined the ability of liver microsomes from these mice to metabolize naproxen. We chose this assay because a number of lines of evidence indicate that human and rat UGT2 isoforms show selectivity toward this substrate (el Mouelhi et al., 1987; Pritchard et al., 1994; Bowalgaha et al., 2005; Sanoh et al., 2012). Microsomes from ΔUgt2 and co-isogenic WT controls were incubated with naproxen for 2 hours, and supernatant was analyzed by HPLC for the presence of the glucuronide. Microsomes from WT mice formed 46.2 ± 1.9 (mean ± S.E.M., n = 3) nmol of napG per mg microsomal protein, while microsomes from ΔUgt2 mice showed no detectable amount of glucuronide formation (Fig. 4). These data indicate loss of activity against UGT2-specific substrates in the ΔUgt2 animals and support the utility of this mouse line in assigning glucuronidation of xenobiotics to the Ugt2 gene family.
Fig. 4.
Naproxen glucuronidation by liver microsomes from WT and ΔUgt2 mice. Reactions contained 50–150 µg microsomal protein, 25 µg/ml alamethicin, 100 µM naproxen, and 5 mM UDPGA. (A) Typical chromatogram of reaction products of WT microsomes. (B) Typical chromatogram of reaction products of ΔUgt2 microsomes. (C) Typical chromatogram of a control reaction lacking UDPGA. Microsomes from WT mice formed 46.2 ± 1.9 (mean ± S.E.M., n = 3) nmol NapG/mg of protein, while microsomes from ΔUgt2 mice showed no detectable amount of glucuronide formation.
BPA Glucuronidation.
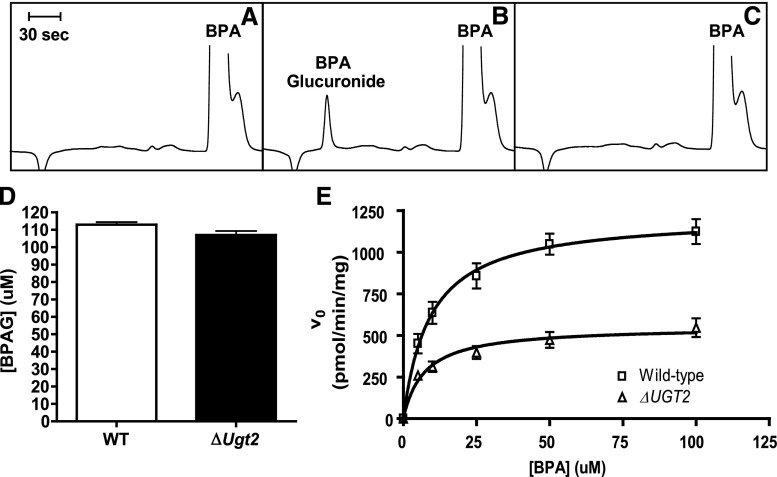
We next asked whether ΔUgt2 mice could be used to measure the contribution of UGT2 enzymes to the metabolism of xenobiotics metabolized by both UGT families. Studies using recombinant human and rat UGTs have shown that BPA is a substrate for both enzyme families, with greater activity assigned to UGT2 enzymes (Yokota et al., 1999; Hanioka et al., 2008, 2011). Therefore, we first determined whether UGT2 enzymes are exclusively responsible for the metabolism of BPA by mouse microsomes. Microsomes prepared from pairs of WT and ΔUgt mice were incubated with BPA and the cofactor UDPGA for 2 hours. The quantities of BPA and BPAG in the reaction products were determined by HPLC. Under these conditions, rapid and complete metabolism of BPA was observed in reactions carried out using both WT and mutant microsomes. No difference was observed between ΔUgt2 and WT microsomes in the total amount of BPAG produced (Fig. 5D). Thus, glucuronidation of BPA occurs in the absence of all UGT2 enzymes. Presumably, this conjugation is carried out by UGT1 enzymes. However, because the reactions were allowed to go to completion these experiments could not differentiate between the contributions of UGT1s and UGT2s to BPA glucuronidation.
Fig. 5.
BPA glucuronidation by liver microsomes from WT and ΔUgt2 mice. (A) Typical chromatogram of products of a reaction terminated immediately after addition of UDPGA. (B) Typical chromatogram of products of a reaction terminated after 10 minutes of incubation. (C) The identity of the BPAG peak was verified by its loss after treatment with β-glucuronidase. (D) BPAG formation after 2-hour incubation of BPA and 25 µg/ml alamethicin with WT and ΔUgt2 microsomes. Complete substrate conversion was observed, and BPAG concentrations were not significantly different. (E) Michaelis-Menten curves for glucuronidation of BPA by microsomes prepared from WT (squares) and ΔUgt2 (triangles) mice in the absence of alamethicin. Each data point represents microsomal preparations from four mice. Error bars represent S.D. r2 > 0.95 for WT mice and r2 > 0.85 for ΔUgt2 mice. Reactions contained 20–60 µg microsomal protein, 5–100 µM BPA, and 5 mM UDPGA.
By examining the kinetics of BPA glucuronidation in WT and ΔUgt2 microsomes, the activities of the two enzyme families can be examined. The activity of WT microsomes is expected to represent the sum of the activities of the UGT1 and UGT2 families, whereas the activity of ΔUgt2 microsomes represents that of the UGT1 family alone. Reaction conditions were optimized to allow measurement of differences in activity between WT and ΔUgt2 microsomes by increasing the substrate-to-enzyme ratio and by removal of alamethicin. Alamethicin increases the rate of glucuronidation in microsomes by facilitating the movement of the substrate and UDPGA through the microsomal lipid bilayer. Thus, removal of alamethicin from the glucuronidation reactions decreased the reaction rates and facilitated kinetic analysis of the rapidly metabolized substrate. As shown in Fig. 5, under these conditions, the ability of UGT2 enzymes to contribute to BPA metabolism is easily observed. BPA glucuronidation exhibited Michaelis-Menten kinetics in both WT and ΔUgt2 microsomes. Curves were fit to plots of glucuronidation rate (pmol/min/mg microsomal protein) versus BPA concentration (Fig. 5E). The kinetic parameters are given in Table 2. The Vmax measured for ΔUgt2 microsomes, which contain only UGT1 enzymes, is approximately 50% lower than that of WT microsomes, which contain UGT1s and UGT2s. This 50% decrease is attributed to the absence of the UGT2 enzymes in ΔUgt2 microsomes. Thus, we concluded that 50% of the activity in WT microsomes was due to UGT2s and 50% to UGT1s, suggesting the activities of the two families are approximately equal. The Km value did not differ significantly between the WT and the ΔUgt2 microsomes. This suggests that, under these conditions, the two murine UGT families have approximately equal activity toward, and affinity for, BPA. To ensure that the exclusion of alamethicin did not alter the relative activities of microsomes derived from the two mouse lines, WT and ΔUgt2 microsomes were incubated with 100 µM BPA either in the presence or absence of alamethicin. Significant differences between WT and ΔUgt2 microsomes are present under both alamethicin-activated and nonactivated conditions, and the ratio of WT activity to ΔUgt2 activity is comparable in both cases (Supplemental Fig. 5). This result is consistent with previous studies (Fisher et al., 2000) suggesting that alamethicin has an isoform-independent activating effect on UGTs.
TABLE 2.
Kinetic parameters for BPA glucuronidation in liver microsomes from WT and ΔUgt2 mice
Values are expressed as the mean ± S.D. (n = 4). Vmax is significantly different at the P < 0.0001 level and Km is not significantly different in unpaired Student’s t tests.
|
Vmax |
Km |
|
|---|---|---|
| pmol/min/mg | µM | |
| WT | 1220.0 ± 61.4 | 9.17 ± 1.73 |
| ΔUgt2 | 553.2 ± 35.2 | 7.16 ± 1.87 |
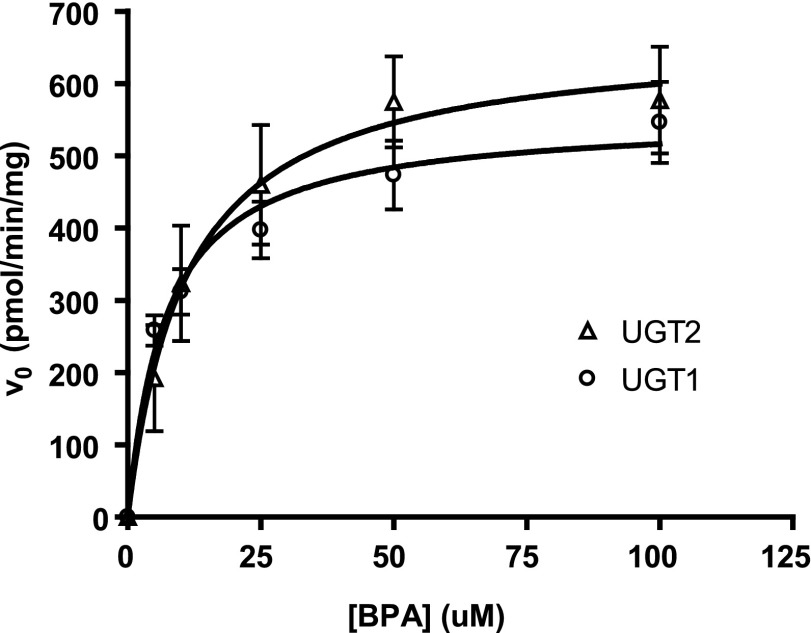
To further evaluate the relative contributions of the two UGT families to microsomal BPA glucuronidation, we used our data sets to derive theoretical kinetic parameters for the UGT1 and UGT2 enzymes, assuming that all BPAG detected was formed by these two enzyme families. Since ΔUgt2 mice express only UGT1 enzymes, the kinetic parameters of the ΔUgt2 microsomes are assigned to the UGT1 family. Glucuronidation of BPA in WT microsomes was assumed to represent the sum of the activities of both UGT2 and UGT1 enzymes, and theoretical Km and Vmax values for the UGT2 family were derived using the two-enzyme Michaelis-Menten equation (Segel, 1993):
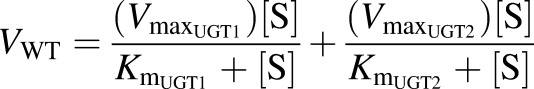 |
where V is the rate of reaction; [S] is the concentration of the substrate; and Km and Vmax are the Michaelis constant for and maximum rate attributed to each UGT family, respectively. As shown subsequently, UGT2 activity can be represented by the difference in the glucuronidation rate between WT mice and ΔUgt2 counterparts:
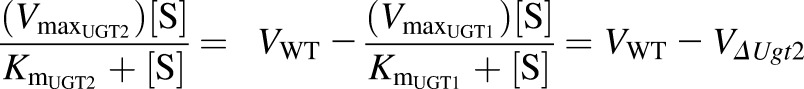 |
Thus, UGT2-mediated BPA glucuronidation was modeled using a Michaelis-Menten curve fitted to a plot of the difference in rate between paired WT and ΔUgt2 microsomes at each substrate concentration (Fig. 6). The kinetic parameters are given in Table 3. Based on this analysis, the activities and affinities of the UGT1 and UGT2 enzymes toward BPA are predicted to be very similar.
Fig. 6.
Enzyme kinetics of UGT1- and UGT2-mediated BPA glucuronidation. The glucuronidation activity remaining in mutant mice after deletion of the Ugt2s was assigned to the UGT1 family (circles). The difference in glucuronidation rate measured between microsomes from WT and ΔUgt2 animals was used to represent activity of the UGT2 family (triangles). The calculated kinetics of glucuronidation of BPA by both enzyme families was consistent with the Michaelis-Menten models. Calculations assumed equal concentrations of UGT1 and UGT2 enzymes in WT microsomes. r2 > 0.85 for UGT1s and r2 > 0.75 for UGT2s. Error bars represent the S.E.M., n = 4 microsomal preparations of each genotype.
TABLE 3.
Kinetic parameters for BPA glucuronidation by mouse UGT1s and UGT2s
Values are expressed as the mean ± S.E.M. (n = 4). Neither Vmax nor Km is significantly different in an unpaired Student’s t test.
|
Vmax |
Km |
|
|---|---|---|
| pmol/min/mg | µM | |
| UGT2 | 665.5 ± 73.1 | 10.99 ± 4.27 |
| UGT1 | 553.2 ± 35.2 | 7.16 ± 1.87 |
BPA Metabolism In Vivo.
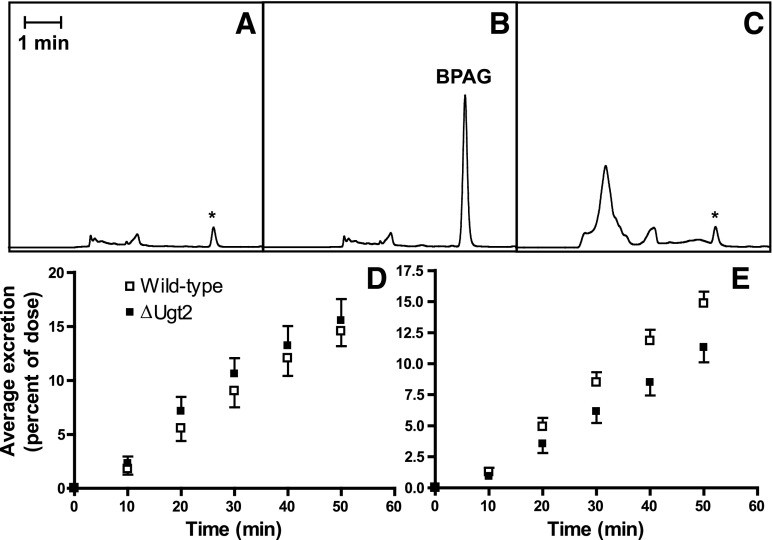
Evaluation of BPA glucuronidation by ΔUgt2 and WT microsomes indicates that UGT1 and UGT2 enzymes contribute equally to BPA metabolism. To determine whether this predicts the activities of these gene families in vivo, we examined the metabolism of BPA in WT and mutant animals. The bile ducts of paired WT and ΔUgt2 male mice matched for weight and age were cannulated. After collection of a predose bile sample for use as a baseline control, each animal received a single bolus of BPA delivered intravenously. Fractions of bile were collected over the next 50 minutes, and the quantity of BPAG in each bile fraction was determined by HLPC. No significant difference was seen in the volume or rate of bile production by the ΔUgt2 and WT animals, and BPAG accumulation in the bile was linear over the 50 minute collection in both mouse lines. The biliary excretion rate was therefore expressed as the fraction of the initial BPA dose excreted per minute.
As expected, based on the study of ΔUgt2 microsomes, BPAG was detected in the bile of the ΔUgt2 mice, clearly indicating that the UGT1 family glucuronidates BPA in vivo. More surprisingly, following a dose of 2 mg BPA/kg b.wt., no significant difference between the rate of biliary BPA excretion in WT and ΔUgt2 mice was observed (Fig. 7). This result suggests that the impact of the UGT2 family on BPA clearance is minimal at this exposure level and that the UGT1 family has significant capacity for BPA conjugation in vivo. When the BPA dose was increased to 20 mg/kg b.wt., a significant decrease between the average biliary excretion rates for WT and ΔUgt2 mice was observed. Approximately 0.31% of the BPA dose was glucuronidated and excreted per minute by WT mice. However, in mutants only 0.22% of the initial dose was glucuronidated and excreted per minute, approximately a 28% reduction in excretion rate (Fig. 7). A significant contribution of the UGT2s to BPA clearance was only observable at this extremely high dose of BPA. Male mice were used in these studies to avoid potential increases in animal-to-animal variation due to the estrous cycle in females; however, examination of hepatic BPA glucuronidation after a dosage of 2 mg BPA/kg b.wt. in female mice also failed to reveal a significant difference between WT and ΔUgt2 animals (Supplemental Fig. 6). Thus, we conclude that the UGT1 family is primarily responsible for the rapid glucuronidation and clearance of BPA in male and female mice.
Fig. 7.
In vivo BPA glucuronidation in WT and ΔUgt2 mice. (A) Typical chromatogram of bile collected from a ΔUgt2 mouse prior to dosage. (B) Typical chromatogram of a bile fraction collected from a ΔUgt2 mouse from 0 to 10 minutes after dosage. (C) Treatment with β-glucuronidase causes loss of the BPAG peak and changes in the chromatographic profile due to deconjugation of endogenous glucuronides. BPAG peak areas were corrected for the area of a small interfering peak (presumably due to a co-eluting endogenous metabolite; marked by an asterisk) in bile obtained immediately before dosage. The interfering peak remained after β-glucuronidase treatment, indicating that it does not represent BPA exposure prior to experimentation. (D) Average BPAG excretion after dosage with 2 mg/kg BPA over the collection period for WT and ΔUgt2 male mice, measured as percentage of dose/minute. Average excretion rates for WT and ΔUgt2 mice were 0.306 ± 0.030 (mean ± S.E.M., n = 3) and 0.325 ± 0.039 (mean ± S.E.M., n = 3) respectively. (E) Average BPAG excretion after dosage with 20 mg/kg BPA over the collection period for WT and ΔUgt2 mice. Average excretion rates for WT and ΔUgt2 mice were 0.313 ± 0.019 (mean ± S.E.M., n = 6) and 0.224 ± 0.024 (mean ± S.E.M., n = 7) respectively. Average percent excretion/minute was significantly different at the P < 0.05 level in an unpaired Student’s t test. BPAG excretion was linear over the time range examined (plots of percent excretion versus time had r2 > 0.93 for all animals).
Discussion
In this study, we show the generation of mice lacking the ∼600 kbp DNA segment carrying the Ugt2 gene family. Unlike mice lacking the UGT1 enzymes (Nguyen et al., 2008), the mice are healthy and indistinguishable from control animals when bred in a specific pathogen–free animal facility. While the UGT1 family is essential for the conjugation and excretion of endogenous metabolites, our data indicate that the members of the UGT2 family do not play an essential and unique role in this function. Particularly surprising is the fact that the loss of UGT2 enzymes does not significantly alter sexual development or fertility, given the ability of both rodent and human UGT2s to metabolize steroid hormones, particularly androgens (Hum et al., 1999; Chouinard et al., 2008). Studies using mice lacking the androgen receptor or mice treated with antiandrogens such as cyproterone acetate have shown that the development of the secondary sex organs is sensitive to androgen levels (Jean-Faucher et al., 1984; Yeh et al., 2002). Both of these studies reported impaired development of secondary sex organs observable at a gross anatomic level as well as infertility when androgen metabolism was impaired. This indicates that the normal development of these organs is a sensitive surrogate marker for normal androgen metabolism in mice. A possible explanation for the lack of a developmental or reproductive phenotype in the mutants is that glucuronidation is not a critical pathway for androgen excretion in mice. There is evidence that sulfonation is the predominant excretory pathway for some substrates in rodents (Sanoh et al., 2012), and a number of steroid-conjugating sulfotransferases, including the estrogen-preferring SULT1E1 and the hydroxysteroid sulfotransferases SULT2A1 and SULT2B1, could contribute to androgen metabolism (Alnouti and Klaassen, 2006). Supporting this possibility, neither the Gunn rat (Nguyen et al., 2008) nor the ΔUgt2 mouse exhibits altered fertility or sexual development, while the Sult1e1 knockout mouse exhibits alterations in both capacities (Qian et al., 2001).
The Ugt2a genes are located in the same cluster as the Ugt2b genes in both humans and rodents. Ugt2a1 and Ugt2a2 are expressed in the nasal epithelium, and studies using recombinant enzymes have shown that they are able to conjugate odorants. This is hypothesized to be important in the termination of odorant signaling and preventing receptor desensitization (Lazard et al., 1991; Sneitz et al., 2009). The ΔUgt2 mouse line should allow this hypothesis to be tested, both in vitro using cultured nasal epithelial cells and in vivo using odor-dependent behavioral paradigms (Friedrich, 2006). While the function of Ugt2a3 remains unclear (Buckley and Klaassen, 2009; Sneitz et al., 2009), its conservation across species suggests a role in excretion of endogenous metabolites (Mackenzie et al., 2005). However, our studies indicate that, at least in mice bred in a vivarium, this gene is not essential for normal development and health.
Previous studies using recombinant UGT2 enzymes indicate that the UGT2 family is important in the metabolism of many nonsteroidal anti-inflammatory drugs in both humans and rats (King et al., 2000). Using microsomes prepared from the ΔUgt2 mice, we show that glucuronidation of naproxen is dependent on the UGT2 enzymes. Even after extended incubation, only the parent compound was detected in ΔUgt2 microsomal reaction products. However, we cannot exclude the possibility that some glucuronidation may be detected at higher substrate concentrations due to low-affinity interaction with UGT1 isoforms. Such interactions have been observed using recombinant human UGT1s (Bowalgaha et al., 2005). Studies using recombinant rat UGT enzymes indicate that UGT2B1 shows high activity toward many nonsteroidal anti-inflammatory drugs, including naproxen (Ritter, 2000). While our studies only allow assignment to the UGT2 enzyme family and not individual isoforms, it is likely that naproxen metabolism in the mouse is dependent on Ugt2b1 expression.
Extensive studies have confirmed widespread human exposure to BPA (Vandenberg et al., 2007), and studies carried out in both rats and mice indicate possible adverse effects of BPA on development and health (Kabuto et al., 2004; Richter et al., 2007; Vandenberg et al., 2007; Hanioka et al., 2008). Considerable resources have been directed toward determination of the health risks associated with BPA exposure. Despite the interest in this xenobiotic, the understanding of its metabolism is incomplete. BPA is rapidly glucuronidated in both humans and rodents and excreted through urine and bile, respectively. However, less is known regarding the enzymes primarily responsible for formation of the glucuronide. Assigning this activity to the individual enzymes of the UGT1 and UGT2 families has relied largely on studies using recombinant enzymes. Studies of both human and rat enzymes indicate that the UGT2s, specifically UGT2B15 in human and UGT2B1 in rat, show the highest activity toward BPA. Therefore, it has been assumed that the UGT2 family is critical for rapid clearance of this xenobiotic. Our results are not consistent with this model and indicate that UGT1 enzymes generate a larger proportion of BPAG in vivo. One possibility is that Ugt1a9, orthologous to the BPA-conjugating human UGT1A9 (Hanioka et al., 2008), is a pseudogene in rats but functional in mice (Mackenzie et al., 2005). Thus, the UGT1s may make a larger relative contribution to BPA glucuronidation in mice than in rats.
The role of the UGT1s in BPA metabolism became apparent upon incubation of BPA with microsomes isolated from ΔUgt2 mice. When allowed to go to completion, reactions containing WT and ΔUgt2 microsomes with equal initial concentrations of BPA formed equal concentrations of BPAG. A contribution of UGT2s to BPA metabolism could be defined by examining the rate of formation of the glucuronide, and a 50% reduction was observed in the Vmax of ΔUgt2 microsomes when compared with WT controls. The approximately equal contributions of UGT1s and UGT2s to BPA metabolism in these in vitro studies did not accurately predict the impact of UGT2 deficiency on BPA metabolism in vivo. At a dose of 2 mg/kg, orders of magnitude greater than the estimated 1.5 µg/kg exposure for the general human population (European Union, 2010), there was no significant difference in biliary BPA excretion between WT and ΔUgt2 mice. Only when the dosage was increased to 20 mg/kg was a significant difference observed. These data indicate that the UGT1 family has extensive capacity to glucuronidate BPA and that this enzyme family may, in fact, be primarily responsible for BPA metabolism in vivo. Barring extreme exposures, the contribution of UGT2s to hepatic conjugation of BPA may be negligible.
This work highlights the limitations of directly extrapolating the results of in vitro studies to in vivo metabolism of xenobiotics, which have been extensively discussed previously (Lin and Wong, 2002; Soars et al., 2002; Miners et al., 2006). In general, in vitro studies are reported to underestimate in vivo enzymatic activities. In the case of the ΔUgt2 mice, the in vitro results underestimated the contribution of the UGT1 family to BPA metabolism. One possible explanation could be the differential stability of mouse UGT2s and UGT1s during microsomal preparation or incubation, as has been observed in the glucuronidation of flavonoids (Joseph et al., 2007), thereby exaggerating the contribution of one family over the other in in vitro studies. It is also possible that differential localization of UGT1 and UGT2 enzymes in the liver results in differential access to BPA in an in vivo environment. Additional experiments, including evaluation of BPA metabolism by primary mouse hepatocytes, could help to distinguish between these possibilities.
Several human UGT isoforms are capable of conjugating BPA. UGT2B15 is reported to have the highest activity, with lower activities observed for recombinant UGT1A1, UGT1A3, UGT1A9, UGT2B4, and UGT2B7 (Hanioka et al., 2008). An important issue impacted by the assignment of BPA primarily to the UGT2 family is the ability of neonates to metabolize BPA. Neonates and infants have reduced levels of UGT2 expression until 6 to 7 months of age. Therefore, they may represent a subpopulation with increased susceptibility to BPA’s adverse effects (Strassburg et al., 2002; Zaya et al., 2006; Divakaran et al., 2014). Concern regarding the ability of the newborn to metabolize BPA is the basis for restricting the use of BPA-containing plastics in infant bottles in some countries. However, if human UGT1 enzymes contribute extensively to BPA metabolism, as we have found to be the case in mice, this concern may not be well-founded. UGT1A9 reaches adult levels by 4 months, and expression of UGT1A1 increases rapidly after birth due to the necessity of bilirubin conjugation, which occurs in the maternal liver via transfer across the placenta prior to birth (Schenker et al., 1964; Onishi et al., 1979; Miyagi et al., 2012). While we cannot rule out the possibility that the predominant role of UGT1s in BPA metabolism is unique to the mouse, the UGT1 family is more closely conserved between species than the UGT2 family, suggesting that members of the UGT1 family may play a major role in BPA glucuronidation in humans.
In summary, we describe here the generation of a mouse line allowing definitive assignment of metabolism of xenobiotics to the UGT1 or UGT2 enzyme family. The normal development and good health of these animals makes them an ideal model for such studies. Reconstitution of the deleted locus with individual mouse or human UGT2 genes will provide a means of refining functional assignments to individual UGT2 enzymes in vivo.
Supplementary Material
Acknowledgments
The authors thank Dr. Brian Hogan for useful insights regarding enzyme kinetics, Dr. Linda Spremulli and Dr. Matthew Redinbo for helpful discussions, Anne Latour for ES cell work, Joseph Snouwaert for artwork, and Peter Repenning and Leonard Collins for expert technical assistance and advice.
Abbreviations
- BPA
bisphenol A
- BPAG
bisphenol A glucuronide
- ES
embryonic stem
- HPLC
high-performance liquid chromatography
- kbp
kilobase pair
- napG
naproxen glucuronide
- PCR
polymerase chain reaction
- UDPGA
UDP-glucuronic acid
- UGT
UDP-glucuronosyltransferase
- WT
wild type
Authorship Contributions
Participated in research design: Fay, Nguyen, Snouwaert, Grant, Bodnar, Koller.
Conducted experiments: Fay, Nguyen, Snouwaert, Dye, Koller.
Contributed new reagents or analytic tools: Bodnar.
Performed data analysis: Fay, Nguyen, Snouwaert, Dye, Koller.
Wrote or contributed to the writing of the manuscript: Fay, Snouwaert, Grant, Koller.
Footnotes
This work was supported by the United States Public Health Service [Grants ES021838 and HL107780]; the National Institute of Environmental Health Sciences [Grant P30ES010126]; and SURF and Taylor Honors Research Fellowship grants from UNC-Chapel Hill.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Alnouti Y, Klaassen CD. (2006) Tissue distribution and ontogeny of sulfotransferase enzymes in mice. Toxicol Sci 93:242–255. [DOI] [PubMed] [Google Scholar]
- Bock KW. (2010) Functions and transcriptional regulation of adult human hepatic UDP-glucuronosyl-transferases (UGTs): mechanisms responsible for interindividual variation of UGT levels. Biochem Pharmacol 80:771–777. [DOI] [PubMed] [Google Scholar]
- Bowalgaha K, Elliot DJ, Mackenzie PI, Knights KM, Swedmark S, Miners JO. (2005) S-Naproxen and desmethylnaproxen glucuronidation by human liver microsomes and recombinant human UDP-glucuronosyltransferases (UGT): role of UGT2B7 in the elimination of naproxen. Br J Clin Pharmacol 60:423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley DB, Klaassen CD. (2007) Tissue- and gender-specific mRNA expression of UDP-glucuronosyltransferases (UGTs) in mice. Drug Metab Dispos 35:121–127. [DOI] [PubMed] [Google Scholar]
- Buckley DB, Klaassen CD. (2009) Induction of mouse UDP-glucuronosyltransferase mRNA expression in liver and intestine by activators of aryl-hydrocarbon receptor, constitutive androstane receptor, pregnane X receptor, peroxisome proliferator-activated receptor α, and nuclear factor erythroid 2-related factor 2. Drug Metab Dispos 37:847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouinard S, Barbier O, Bélanger A. (2007) UDP-glucuronosyltransferase 2B15 (UGT2B15) and UGT2B17 enzymes are major determinants of the androgen response in prostate cancer LNCaP cells. J Biol Chem 282:33466–33474. [DOI] [PubMed] [Google Scholar]
- Chouinard S, Yueh MF, Tukey RH, Giton F, Fiet J, Pelletier G, Barbier O, Bélanger A. (2008) Inactivation by UDP-glucuronosyltransferase enzymes: the end of androgen signaling. J Steroid Biochem Mol Biol 109:247–253. [DOI] [PubMed] [Google Scholar]
- Coughtrie MW, Burchell B, Leakey JE, Hume R. (1988) The inadequacy of perinatal glucuronidation: immunoblot analysis of the developmental expression of individual UDP-glucuronosyltransferase isoenzymes in rat and human liver microsomes. Mol Pharmacol 34:729–735. [PubMed] [Google Scholar]
- de Wildt SN, Kearns GL, Leeder JS, van den Anker JN. (1999) Glucuronidation in humans. Pharmacogenetic and developmental aspects. Clin Pharmacokinet 36:439–452. [DOI] [PubMed] [Google Scholar]
- Divakaran K, Hines RN, McCarver DG. (2014) Human hepatic UGT2B15 developmental expression. Toxicol Sci 141:292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Mouelhi M, Ruelius HW, Fenselau C, Dulik DM. (1987) Species-dependent enantioselective glucuronidation of three 2-arylpropionic acids. Naproxen, ibuprofen, and benoxaprofen. Drug Metab Dispos 15:767–772. [PubMed] [Google Scholar]
- European Union (2010) European Union Risk Assessment Report: 4,4′-Isopropylidenediphenol (bisphenol-A)—Part 2 human health, Human Health Addendum of April 2008, Publications Office of the European Union, Luxembourg. [Google Scholar]
- Fang JL, Beland FA, Doerge DR, Wiener D, Guillemette C, Marques MM, Lazarus P. (2002) Characterization of benzo(a)pyrene-trans-7,8-dihydrodiol glucuronidation by human tissue microsomes and overexpressed UDP-glucuronosyltransferase enzymes. Cancer Res 62:1978–1986. [PubMed] [Google Scholar]
- Fisher MB, Campanale K, Ackermann BL, VandenBranden M, Wrighton SA. (2000) In vitro glucuronidation using human liver microsomes and the pore-forming peptide alamethicin. Drug Metab Dispos 28:560–566. [PubMed] [Google Scholar]
- Friedrich RW. (2006) Mechanisms of odor discrimination: neurophysiological and behavioral approaches. Trends Neurosci 29:40–47. [DOI] [PubMed] [Google Scholar]
- Ginsberg G, Rice DC. (2009) Does rapid metabolism ensure negligible risk from bisphenol A? Environ Health Perspect 117:1639–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanioka N, Naito T, Narimatsu S. (2008) Human UDP-glucuronosyltransferase isoforms involved in bisphenol A glucuronidation. Chemosphere 74:33–36. [DOI] [PubMed] [Google Scholar]
- Hanioka N, Oka H, Nagaoka K, Ikushiro S, Narimatsu S. (2011) Effect of UDP-glucuronosyltransferase 2B15 polymorphism on bisphenol A glucuronidation. Arch Toxicol 85:1373–1381. [DOI] [PubMed] [Google Scholar]
- Hum DW, Bélanger A, Lévesque E, Barbier O, Beaulieu M, Albert C, Vallée M, Guillemette C, Tchernof A, Turgeon D, et al. (1999) Characterization of UDP-glucuronosyltransferases active on steroid hormones. J Steroid Biochem Mol Biol 69:413–423. [DOI] [PubMed] [Google Scholar]
- Jean-Faucher C, Berger M, De Turckheim M, Veyssiere G, Jean C. (1984) Sexual maturation in male mice treated with cyproterone acetate from birth to puberty. J Endocrinol 102:103–107. [DOI] [PubMed] [Google Scholar]
- Jedlitschky G, Cassidy AJ, Sales M, Pratt N, Burchell B. (1999) Cloning and characterization of a novel human olfactory UDP-glucuronosyltransferase. Biochem J 340:837–843. [PMC free article] [PubMed] [Google Scholar]
- Joseph TB, Wang SWJ, Liu X, Kulkarni KH, Wang J, Xu H, Hu M. (2007) Disposition of flavonoids via enteric recycling: enzyme stability affects characterization of prunetin glucuronidation across species, organs, and UGT isoforms. Mol Pharm 4:883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabuto H, Amakawa M, Shishibori T. (2004) Exposure to bisphenol A during embryonic/fetal life and infancy increases oxidative injury and causes underdevelopment of the brain and testis in mice. Life Sci 74:2931–2940. [DOI] [PubMed] [Google Scholar]
- Kaivosaari S (2010) N-Glucuronidation of drugs and other xenobiotics, Academic dissertation, University of Helsinki, Helsinki, Finland. [Google Scholar]
- King CD, Rios GR, Green MD, Tephly TR. (2000) UDP-glucuronosyltransferases. Curr Drug Metab 1:143–161. [DOI] [PubMed] [Google Scholar]
- Krishnan AV, Stathis P, Permuth SF, Tokes L, Feldman D. (1993) Bisphenol-A: an estrogenic substance is released from polycarbonate flasks during autoclaving. Endocrinology 132:2279–2286. [DOI] [PubMed] [Google Scholar]
- Kutsuno Y, Sumida K, Itoh T, Tukey RH, Fujiwara R. (2013) Glucuronidation of drugs in humanized UDP-glucuronosyltransferase 1 mice: Similarity with glucuronidation in human liver microsomes. Pharmacol Res Perspect 1:e00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazard D, Zupko K, Poria Y, Nef P, Lazarovits J, Horn S, Khen M, Lancet D. (1991) Odorant signal termination by olfactory UDP glucuronosyl transferase. Nature 349:790–793. [DOI] [PubMed] [Google Scholar]
- Lin JH, Wong BK. (2002) Complexities of glucuronidation affecting in vitro in vivo extrapolation. Curr Drug Metab 3:623–646. [DOI] [PubMed] [Google Scholar]
- Luconi M, Forti G, Baldi E. (2002) Genomic and nongenomic effects of estrogens: molecular mechanisms of action and clinical implications for male reproduction. J Steroid Biochem Mol Biol 80:369–381. [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, Miners JO, Owens IS, Nebert DW. (2005) Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics 15:677–685. [DOI] [PubMed] [Google Scholar]
- Matthews JB, Twomey K, Zacharewski TR. (2001) In vitro and in vivo interactions of bisphenol A and its metabolite, bisphenol A glucuronide, with estrogen receptors α and β. Chem Res Toxicol 14:149–157. [DOI] [PubMed] [Google Scholar]
- Miners JO, Knights KM, Houston JB, Mackenzie PI. (2006) In vitro–in vivo correlation for drugs and other compounds eliminated by glucuronidation in humans: pitfalls and promises. Biochem Pharmacol 71:1531–1539. [DOI] [PubMed] [Google Scholar]
- Miyagi SJ, Milne AM, Coughtrie MW, Collier AC. (2012) Neonatal development of hepatic UGT1A9: implications of pediatric pharmacokinetics. Drug Metab Dispos 40:1321–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (2011) Guide for the Care and Use of Laboratory Animals, Eighth Edition, National Academies Press, Washington D.C.
- Nguyen N, Bonzo JA, Chen S, Chouinard S, Kelner MJ, Hardiman G, Bélanger A, Tukey RH. (2008) Disruption of the ugt1 locus in mice resembles human Crigler-Najjar type I disease. J Biol Chem 283:7901–7911. [DOI] [PubMed] [Google Scholar]
- Onishi S, Kawade N, Itoh S, Isobe K, Sugiyama S. (1979) Postnatal development of uridine diphosphate glucuronyltransferase activity towards bilirubin and 2-aminophenol in human liver. Biochem J 184:705–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens IS, Basu NK, Banerjee R. (2005) UDP-glucuronosyltransferases: gene structures of UGT1 and UGT2 families. Methods Enzymol 400:1–22. [DOI] [PubMed] [Google Scholar]
- Pritchard M, Fournel-Gigleux S, Siest G, Mackenzie P, Magdalou J. (1994) A recombinant phenobarbital-inducible rat liver UDP-glucuronosyltransferase (UDP-glucuronosyltransferase 2B1) stably expressed in V79 cells catalyzes the glucuronidation of morphine, phenols, and carboxylic acids. Mol Pharmacol 45:42–50. [PubMed] [Google Scholar]
- Qian YM, Sun XJ, Tong MH, Li XP, Richa J, Song WC. (2001) Targeted disruption of the mouse estrogen sulfotransferase gene reveals a role of estrogen metabolism in intracrine and paracrine estrogen regulation. Endocrinology 142:5342–5350. [DOI] [PubMed] [Google Scholar]
- Radominska-Pandya A, Czernik PJ, Little JM, Battaglia E, Mackenzie PI. (1999) Structural and functional studies of UDP-glucuronosyltransferases. Drug Metab Rev 31:817–899. [DOI] [PubMed] [Google Scholar]
- Radominska-Pandya A, Little JM, Czernik PJ. (2001) Human UDP-glucuronosyltransferase 2B7. Curr Drug Metab 2:283–298. [DOI] [PubMed] [Google Scholar]
- Richter CA, Birnbaum LS, Farabollini F, Newbold RR, Rubin BS, Talsness CE, Vandenbergh JG, Walser-Kuntz DR, vom Saal FS. (2007) In vivo effects of bisphenol A in laboratory rodent studies. Reprod Toxicol 24:199–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter JK. (2000) Roles of glucuronidation and UDP-glucuronosyltransferases in xenobiotic bioactivation reactions. Chem Biol Interact 129:171–193. [DOI] [PubMed] [Google Scholar]
- Rowland A, Miners JO, Mackenzie PI. (2013) The UDP-glucuronosyltransferases: their role in drug metabolism and detoxification. Int J Biochem Cell Biol 45:1121–1132. [DOI] [PubMed] [Google Scholar]
- Sanoh S, Horiguchi A, Sugihara K, Kotake Y, Tayama Y, Uramaru N, Ohshita H, Tateno C, Horie T, Kitamura S, et al. (2012) Predictability of metabolism of ibuprofen and naproxen using chimeric mice with human hepatocytes. Drug Metab Dispos 40:2267–2272. [DOI] [PubMed] [Google Scholar]
- Schenker S, Dawber NH, Schmid R. (1964) Bilirubin metabolism in the fetus. J Clin Invest 43:32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segel IH. (1993) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems, Wiley, New York. [Google Scholar]
- Sneitz N, Court MH, Zhang X, Laajanen K, Yee KK, Dalton P, Ding X, Finel M. (2009) Human UDP-glucuronosyltransferase UGT2A2: cDNA construction, expression, and functional characterization in comparison with UGT2A1 and UGT2A3. Pharmacogenet Genomics 19:923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soars MG, Burchell B, Riley RJ. (2002) In vitro analysis of human drug glucuronidation and prediction of in vivo metabolic clearance. J Pharmacol Exp Ther 301:382–390. [DOI] [PubMed] [Google Scholar]
- Strassburg CP, Strassburg A, Kneip S, Barut A, Tukey RH, Rodeck B, Manns MP. (2002) Developmental aspects of human hepatic drug glucuronidation in young children and adults. Gut 50:259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg LN, Hauser R, Marcus M, Olea N, Welshons WV. (2007) Human exposure to bisphenol A (BPA). Reprod Toxicol 24:139–177. [DOI] [PubMed] [Google Scholar]
- Völkel W, Colnot T, Csanády GA, Filser JG, Dekant W. (2002) Metabolism and kinetics of bisphenol A in humans at low doses following oral administration. Chem Res Toxicol 15:1281–1287. [DOI] [PubMed] [Google Scholar]
- Yeh S, Tsai MY, Xu Q, Mu XM, Lardy H, Huang KE, Lin H, Yeh SD, Altuwaijri S, Zhou X, et al. (2002) Generation and characterization of androgen receptor knockout (ARKO) mice: an in vivo model for the study of androgen functions in selective tissues. Proc Natl Acad Sci USA 99:13498–13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota H, Iwano H, Endo M, Kobayashi T, Inoue H, Ikushiro S, Yuasa A. (1999) Glucuronidation of the environmental oestrogen bisphenol A by an isoform of UDP-glucuronosyltransferase, UGT2B1, in the rat liver. Biochem J 340:405–409. [PMC free article] [PubMed] [Google Scholar]
- Zaya MJ, Hines RN, Stevens JC. (2006) Epirubicin glucuronidation and UGT2B7 developmental expression. Drug Metab Dispos 34:2097–2101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.