Abstract
The multispecific organic anion drug transporters OAT6 (SLC22A20) and OAT1 (SLC22A6) are expressed in nasal epithelial cells and both can bind odorants. Sequence analysis of OAT6 revealed an evolutionarily conserved 79-amino acid (AA) fragment present not only in OAT6 but also in other SLC22 transporters, such as the organic anion transporter (OAT), organic carnitine transporter (OCTN), and organic cation transporter (OCT) subfamilies. A similar fragment is also conserved in some odorant receptors (ORs) in both humans and rodents. This fragment is located in regions believed to be important for ligand/substrate preference and recognition in both classes of proteins, raising the possibility that it may be part of a potential common ligand/substrate recognition site in certain ORs and SLC22 transporters. In silico screening of an odorant database containing known OR ligands with a pharmacophore hypothesis (generated from a set of odorants known to bind OAT6 and/or OAT1), followed by in vitro uptake assays in transfected cells, identified OR ligands capable of inhibiting OAT6- and/or OAT1-mediated transport, albeit with different affinities. The conservation of the AA fragments between these two different classes of proteins, together with their coexpression in olfactory as well as other tissues, suggests the possibility that ORs and SLC22 transporters function in concert, and raises the question as to whether these transporters function in remote sensing and signaling and/or as transceptors.
Introduction
Organic anion transporters of the SLC22 transporter family are expressed in barrier tissues and mediate the cellular handling (mostly uptake/influx) of small-molecule organic solutes (Nigam et al., 2015). Together with other transporters of the ATP-binding cassette (ABC) and solute carrier (SLC) families, they enable vectorial transepithelial movement of xenobiotics, as well as endogenous metabolites and signaling molecules that are critical for homeostasis (Nigam, 2015; Nigam et al., 2015).
OAT6 (SLC22A20) is a multispecific organic anion transporter preferentially expressed in nasal epithelial cells and is also found in Sertoli cells in the testis of mice (Monte et al., 2004; Kaler et al., 2006; Schnabolk et al., 2006, 2010; Kaler et al., 2007). OAT6 is able to interact with a variety of small-molecule organic anions of physiological, pharmacological, and toxicological significance. These include estrone sulfate, para-aminohippurate (PAH), prostaglandin E2 (PG-E2), ibuprofen, and ochratoxin A (Monte et al., 2004; Kaler et al., 2006; Schnabolk et al., 2006; Truong et al., 2008; Schnabolk et al., 2010). Among the best ligands of this transporter are odorant molecules (Malnic et al., 2010; Modena et al., 2011), many of which also interact with other members of this transporter family, including OAT1 and OAT3 (Kaler et al., 2006, 2007). OAT1 [SLC22A6 or novel kidney transporter (NKT)] and OAT3 (SLC22A8) are close homologs that are highly expressed in kidney proximal tubule cells, as well as cells of the choroid plexus and other tissues, where they are responsible for the rate-limiting step in the movement of solutes crossing the blood-urine and blood-cerebrospinal fluid barriers (Lopez-Nieto et al., 1997; Brady et al., 1999; Sweet et al., 2002; Eraly et al., 2006; Ahn and Bhatnagar, 2008; Emami Riedmaier et al., 2012; Nagle et al., 2013; Cesar-Razquin et al., 2015). These two transporters have also generated significant pharmaceutical and regulatory (US Food and Drug Administration) interest because of their importance for drug disposition by the kidney and other organs (Morrissey et al., 2012; Nigam et al., 2015). Thus, from sequence and functional perspectives, OAT6 belongs to a large family of facilitative solute carriers of the SLC22 family of multispecific organic anion “drug” transporters, the best studied of which are OAT1 and OAT3. Nevertheless, the unique and rather restricted expression pattern of OAT6 in olfactory mucosa raises the interesting possibility of a role for this transporter in delivery of small-molecule xenobiotics across the nasal-epithelial barrier and potential access to the central nervous system (Genter et al., 2009; Nagashima and Touhara, 2010; Thiebaud et al., 2011; Heydel et al., 2013; Nigam et al., 2015). Moreover, in light of common odorant ligands, and because OAT1 is highly expressed in the kidney (whereas OAT6 is highly expressed in olfactory epithelium), it is possible that volatile odorants excreted via OAT1 into the urine interact with olfactory OAT6 in the nasal epithelium (Kaler et al., 2006, 2007; Wu et al., 2011; Nigam et al., 2015). A role for OAT6 in olfaction is also possible (Monte et al., 2004; Kaler et al., 2006; Ahn and Nigam, 2009; Schnabolk et al., 2010; Wu et al., 2011; Nigam et al., 2015).
We have identified a sequence motif within OAT6 that is conserved in other members of the SLC22 family of transporters, and this conserved domain shares sequence similarities with a domain found in many of the seven-transmembrane odorant receptors (ORs) of the G protein-coupled receptor family. Furthermore, using known OAT ligands that are also known odorants, pharmacophore hypotheses were generated and used to identify other odorants in a virtual screen of an odorant structural library reflecting the diversity of known and probable OR ligands, and some of these hits were then shown to interact with OAT6 and/or OAT1. Since both OATs and ORs are known to be coexpressed in olfactory as well as other tissues, these findings raise the interesting possibility that SLC22 transporters may somehow work together with signal transduction membrane receptors in organismal physiology or function in a “transceptor-like” capacity.
Materials and Methods
Materials.
Unless otherwise indicated, all chemicals, including fluorescein, 6-carboxyfluorescein (6CF), 4-bromobutyric acid, hexyl acetate, allyl heptanoate, amyl hexanoate, 6-bromohexanoic acid, and methyl octanoate were obtained from Sigma-Aldrich (St. Louis, MO). Water-soluble probenecid was obtained from Invitrogen (Carlsbad, CA).
Sequence Analysis.
For alignment, amino acid sequences in FASTA format for each individual transporter and seven-transmembrane odorant receptor were obtained from the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/) gene databases. For genes with multiple splice variants, only the full-length sequence was selected and used.
For multisequence alignment, a pairwise alignment program, ClustalW, was used (Li et al., 2015). For finding regions of similarity between protein sequences with a protein sequence database, the NCBI-blastp algorithm was used (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Boratyn et al., 2013). For finding a distant homolog of a protein, Position-Specific Iterated BLAST (PSI-BLAST) was used. The initial alignment of sequences was carried out in January, 2013; the latest sequence analysis was done in March, 2015.
Domain Localization.
For extracellular domain localization of the 79-amino acid sequence on a 12-transmembrane transporter, a transmembrane protein display software TOPO2 (http://www.sacs.ucsf.edu/TOPO2/) was used to generate a two-dimensional topology image on the basis of the sequence of the transporter. For domain localization of the 77-amino acid sequence on a seven-transmembrane odorant receptor (Malnic et al., 2010), TOPO2 and Weblogo representation (http://weblogo.berkeley.edu/logo.cgi) for odorant receptor amino acid sequences were used.
Pharmacophore Modeling.
To generate an odorant-based pharmacophore hypothesis for virtual screening purposes, the structure-data files (SDF) for those odorant molecules that have been found capable of interacting with OAT1 and/or OAT6 (i.e., have a Ki or IC50 value) (Tables 1 and 2) were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). These common chemical data file formats containing structural information were then input into the computational software ICM (Molsoft, San Diego, CA). The OAT1/OAT6-interacting odorant molecules were then superimposed and aligned using the atomic properties field (APF) superimposition method, which considers the three-dimensional representation of hydrogen bond donors, hydrogen bond acceptors, sp2 hybridization, lipophilicity, size of large atoms, and positive and negative charges (Totrov, 2008; Chen et al., 2014). On the basis of the APFs of the aligned molecules, the pharmacophore hypothesis was generated.
TABLE 1.
OAT6- and/or OAT1-interacting odorants
| CAS No. | Odorant | Kinetics (μM) |
Reference | |||
|---|---|---|---|---|---|---|
| OAT1 |
OAT6 |
|||||
| Ki | IC50 | Ki | IC50 | |||
| 123-66-0 | 2-Ethylhexanoate | 57 | 6.6 | (Kaler et al., 2006) | ||
| 623-42-7 | 2-Methylbutyrate | 920 | 40.7 | (Kaler et al., 2006) | ||
| 65-85-0 | aBenzoate | 253 | 13.8 | (Kaler et al., 2006) | ||
| 111-14-8 | Heptanoate | 16.7 | 8.2 | (Kaler et al., 2006) | ||
| 79-09-4 | Propanoic acid | 8180 | 279 | (Kaler et al., 2006) | ||
| 107-92-6 | Butyric acid | 3500 | (Kaler et al., 2007) | |||
| 69134-53-8 | Diethyl 2-hydroxyglutarate | 369 | (Hagos et al., 2007) | |||
| 142-62-1 | Hexanoic acid | 38 | (Kaler et al., 2007) | |||
| 124-07-2 | Octanoic acid | 5.41 | (Kaler et al., 2007) | |||
Note: These odorant molecules were used for the generation of pharmacophore hypotheses.
Benzoate also showed elevated plasma concentration in the Oat1 KO mouse [Eraly et al. (2006); Wikoff et al. (2011)]; OAT1 displays similar kinetics for drugs, metabolites and toxins [UCSF Transportal, http://dbts.ucsf.edu/fdatransportal; Morrissey et al. (2012); VanWert et al. (2010)].
TABLE 2.
Inhibition of OAT1- and OAT6-mediated uptake
| Odorant/Inhibitor | OAT1 IC50 | OAT6 IC50 |
|---|---|---|
| μM | μM | |
| 4-Bromobutyric acid | 45.3 | 1346 |
| 6-Bromohexanoic acid | 1012 | 16 |
| Hexyl acetate | 998 | 1149 |
| Amyl hexanoate | 444 | >3000 |
| Methyl octanoate | >3000 | 158 |
| Allyl heptanoate | >3000 | >3000 |
| Probenecid | 4.1 | 28 |
Software: GraphPad Prism, nonlinear regression fit. Tracer: OAT1-mediated uptake, 10 microM 6-CF; OAT6-mediated uptake, 10 microM fluorescein.
Virtual Screening of an Odorant Database.
The pharmacophore model derived from OAT1/OAT6-interacting odorant molecules was then used to virtually screen the OlfactionDB database (http://molsim.sci.univr.it/OlfactionDB) using the ICM computational software. Odorant molecules with APF scores ≤–60 and steric scores ≤5 were input into ChemMine (Chemmine.ucr.edu) and bin-clustered on the basis of structural similarities. The 27 molecules within the bin containing three or more molecules were further evaluated in PubChem by Tanimoto similarity coefficient. A cluster of 17 molecules (including three known OAT-interacting odorants) was identified and four molecules from this group were randomly selected for testing in the OAT transport assay.
Transport Assay.
Chinese hamster ovary (CHO) cells constitutively expressing mouse OAT6 or OAT1 (the generous gift of Dr. Douglas Sweet) were plated out and cultured to confluence in a 96-well culture plate. Cellular uptake assays were performed for 10 minutes at room temperature as previously described [for detailed descriptions of the protocol, please see Ahn et al. (2009); Schnabolk et al. (2010); and Wu et al. (2013)]. The known substrates, fluorescein (OAT6; 10 μM in phosphate-buffered saline) and 6CF (OAT1; 10 μM in phosphate-buffered saline) were used as fluorescent tracers for uptake in the transfected CHO cells. Probenecid, the prototypical inhibitor of OAT-mediated uptake, was used as a negative control for OAT6- and OAT1-mediated uptake of fluorescent tracers in the CHO cells. Quantitative uptake of fluorescent tracers (fluorescein, OAT6; 6CF, OAT1) was determined using a fluorometric plate reader as previously described (Ahn et al., 2011; Wu et al., 2013).
Kinetics Calculation.
The ability of the small-molecule odorant molecules to inhibit the uptake of the fluorescent tracer was determined at several concentrations. The inhibitory kinetics were calculated and curve fitting (nonlinear regression) was generated using GraphPad Prism 5 (www.graphpad.com/scientific-software/prism) as described (Ahn et al., 2011; Wu et al., 2013). P values were determined by using Student’s t test.
Results
Identification of a Conserved N-Terminal Region in SLC22A20/OAT6 Orthologs.
Apart from the better known SLC22 family transporters (OAT1, OAT3, OCT1, OCT2, OCTN1, OCTN2, and URAT1), a number of others have been identified by sequence similarity and evolutionary analyses (Eraly et al., 2004; Wu et al., 2009). SLC22A20/OAT6 was identified in mouse on the basis of its sequence homology to other OATs, particularly OAT1/SLC22A6, of the SLC22 small-molecule organic solute carrier family (Monte et al., 2004; Kaler et al., 2006; Kaler et al., 2007). The full-length mouse SLC22A20/OAT6 transcript codes for a peptide of ∼556 amino acids in length with 12 predicted transmembrane helixes (Monte et al., 2004; Kaler et al., 2006). Using mouse SLC22A20/OAT6 amino acid sequence as a template, a homology search of the NCBI database revealed a set of highly conserved orthologs of this transporter, with at least 50% sequence identity at the amino acid level in many mammalian species, including human, chimp, rat, dog, elephant, and others (Supplemental Fig. 1).
Alignment of the SLC22A20/OAT6 amino acid sequences from humans and rodents, using a multiple-sequence alignment tool, initially revealed conserved segments largely corresponding to transmembrane helixes. However, a large and highly conserved area located closer to the N-terminal region of the transporter that does not correspond to a transmembrane helix was also identified (Fig. 1A, Supplemental Fig. 1). To evaluate this N-terminal region of SLC22A20 in more detail, the available amino acid sequences of SLC22A20 from several vertebrate species were obtained and aligned. A high degree of sequence identity, as well as sequence similarity, was readily apparent within this N-terminal region of SLC22A20, suggesting that it was highly conserved during evolution (Fig. 1A, Supplemental Fig. 1). For example, this conservation was not only particularly evident in mammals (both placental and nonplacental), the conserved sequence of the N-terminal region was found in vertebrates as far down as frog (Xenopus laevis) and fish (Danio rerio) (Supplemental Fig. 1).
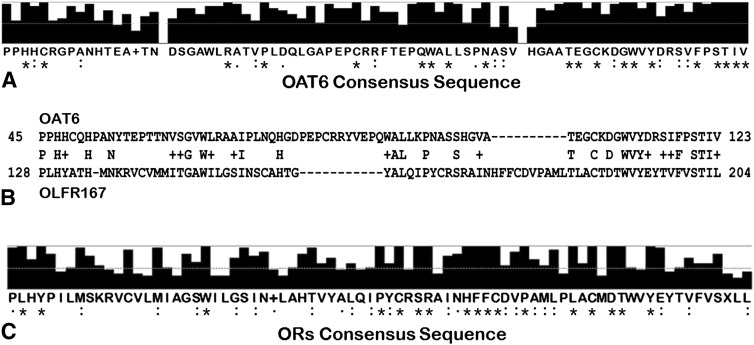
Fig. 1.
Multiple sequence alignments of SLC22A20 transporters and/or odorant receptors reveal similarity within and between these two different types of membrane proteins. (A) Full-length amino acid sequences of SLC22A20 from multiple species were aligned using ClustalW, and the consensus sequence of the 79-amino acid region is shown. Bars above the sequence and symbols below indicate the strong conservation that was observed within the N-terminal region of all species examined (asterisks indicate sequence identity, colons and periods indicate sequence conservation). (B) Alignment of N-terminal region of SLC22A20 with OLFR167 revealed the presence of a 79 amino acid fragment within the N-terminal region of OAT6 (NP_941052, starting at amino acid number 45 and ending at amino acid number 123) which had significant sequence homology to a 77-amino acid fragment in OLFR167 (NP_667146, starting at amino acid number 128 and ending at amino acid number 204) (capital letters between the two sequence fragments indicate identical amino acids at that residue, + indicates conserved residues). (C) Full-length amino acid sequences of 21 olfactory receptors from human, rat, and mouse were aligned. Strong conservation was found within the amino acid sequences of the 77-amino acid fragment (shown). Bars above the sequence and symbols below indicate the strong conservation that was observed within the N-terminal region of all species examined (asterisks indicate sequence identity, colons and periods indicate sequence conservation). Within this 77-amino acid fragment, approximately 22% of the amino acid residues were identical in all receptors examined, and an additional 30% of the residues were found to be conserved in these ORs.
Identification of a 79-Amino Acid Fragment in the N-Terminal Region of SLC22A20/OAT6 That Was Also Found in an Odorant Receptor.
In an attempt to identify additional genes possessing this conserved N-terminal region, a more detailed homology search was carried out in the mouse genome using this conserved N-terminal region sequence as a template. In addition to the SLC22A20/OAT6 and other SLC22 family transporters, a significant sequence similarity was found between the mouse SLC22A20 and the mouse olfactory receptor 167 (OLFR167) (Fig. 1B, Supplemental Fig. 2). This is interesting because: OAT6 is found in olfactory mucosa (Monte et al., 2004; Kaler et al., 2006); it has a high affinity for odorants compared with other transporters (Monte et al., 2004; Kaler et al., 2006; Kaler et al., 2007); and OAT1 (also expressed in olfactory mucosa) (Monte et al., 2004; Kaler et al., 2006; Kaler et al., 2007) was originally cloned (as an NKT) in 1996 (Lopez-Nieto and Nigam, 1996; Lopez-Nieto et al., 1997)] using degenerate primers targeting G protein-coupled receptors (Lopez-Nieto and Nigam, 1996; Lopez-Nieto et al., 1997) (among which are odorant receptors). Direct comparison of the SLC22A20 and OLFR167 peptide sequences revealed the presence of a conserved 79-amino acid fragment (amino acid 45 to amino acid 123 within the SLC22A20 protein) (Figs. 1, A and B). Within this amino acid fragment, there was an overall 28% [22 out of 79 amino acids (AA)] sequence identity and an almost 42% (33 out of 79 AA) sequence similarity between the mouse SLC22A20 and mouse OLFR167 (Fig. 1B), with a particularly high sequence conservation [50% identity (10 out 20 AA) and 70% similarity (14 out 20 AA)] found within the last 20 amino acids of the C-terminal region of the fragments (Fig. 1B).
The 79-Amino Acid Fragment of SLC22A20 Was Found in Other Transporters of the SLC22 Family.
To determine if this 79-amino acid fragment is also present in other members of the SLC22 transporter family, a BLAST search in the mouse genome was carried out using this 79-amino acid region as a query. Significant sequence similarities were found in other SLC22 family members, with SLC22A27 (Wu et al., 2009) having the highest sequence similarity at over 55% sequence identity (Supplemental Fig. 2). Overall, a higher percentage of sequence identity within this fragment was found for the OATs compared with other SLC22 family members (including OCT1/SLC22A1, a prototypical organic cation transporter; not shown). In contrast to OATs and organic cation transporters (OCTs), which include clinically important “drug” transporters, only low to very low sequence homology of this 79-AA fragment was found in the human or mouse sequences of other clinically relevant SLC drug transporters (Nigam, 2015), such as the SLCO (SLCO/SLC21) family of drug transporters or the SLC47/MATE transporters; no sequence homology for this 79-AA fragment was found in the human or mouse ABC family of drug transporters [ABCB1 (MDR1), ABCC2 (MRP2), ABCC4 (MRP4), and ABCG2 (BCRP)] (data not shown).
The Fragment Is Conserved in Some G Protein-Coupled Seven-Transmembrane Odorant Receptors in Humans and Rodents.
A homology search of the conserved fragment in the odorant receptor gene (OR) family was then conducted using the identified homology region on mouse OLFR167, which started at amino acid 128 and ended at amino acid 204 (a 77-AA fragment) (Fig. 1B, Supplemental Fig. 2). A NCBI BLAST search using this 77-AA fragment of mouse OLFR167 revealed multiple conserved homologs of olfactory receptor genes in mouse, rat, and human (Fig. 1C, Supplemental Fig. 2). (In addition, limiting the BLAST query to solute carrier family members revealed multiple SLC22 transporters from all three species, with SLC22A20 having the highest degree of homology; data not shown.) The sequence of this 77-AA fragment from mouse was 96% identical to that of rat OLFR1568, and 79% identical to the human OR2L5. In addition, there were multiple human OR genes with high levels of AA sequence identity to the mouse fragment; for example, 80% with OR2L2 and OR2L8, as well as 59% with OR2AK2. Direct comparison of 21 of these OR peptide sequences from human, mouse, and rat using a multiple alignment tool indicated the conservation of 52% of the amino acid residues with 17 out of 77 residues showing sequence identity and another 22 residues being conserved between the 21 ORs from these three species (Fig. 1C, Supplemental Fig. 3), suggesting that this fragment is highly conserved in multiple odorant receptors in humans and in rodents.
Location of this Conserved Fragment in Transporter and in Odorant Receptor.
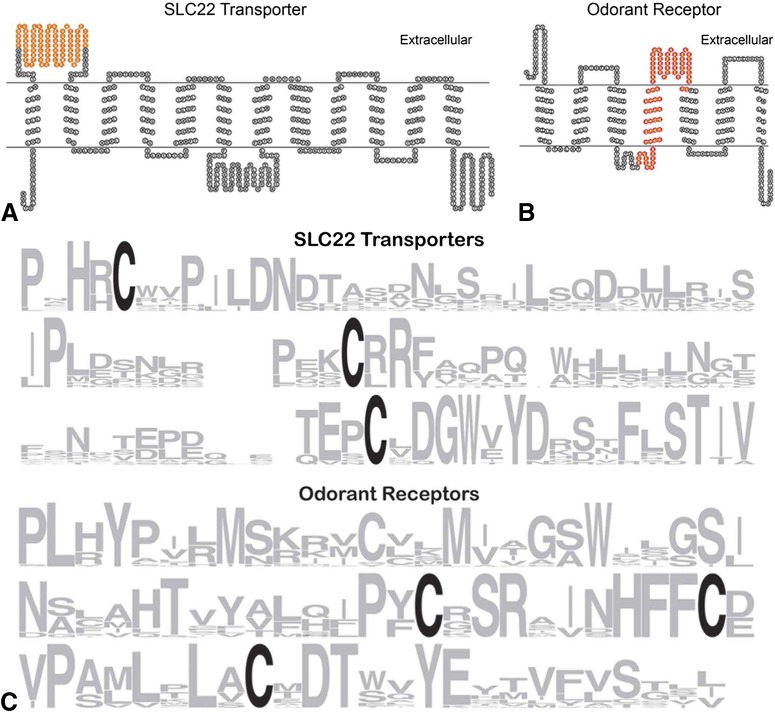
We sought to determine if this conserved fragment represented a domain of known function. However, a query of the NCBI Conserved Domains database using the 79-AA fragment of SLC22A20 did not produce any significant hits other than itself. We then sought to locate the fragments in models of the 12-transmembrane domain–containing transporters and the seven-transmembrane domain–containing odorant receptors. Although a homology-based OAT1 model and simulation has been developed, it does not include extracellular parts of the protein (Tsigelny et al., 2011). By overlaying the 79-AA fragment on a model diagram of the 12-transmembrane domain–containing organic anion and cation transporters of the SLC22 family (Pharmacogenetics.UCSF.edu. as well as the Protein Data Bank), this fragment was found to be largely situated in the first extracellular loop between transmembrane helixes 1 and 2 (Fig. 2A). This large extracellular loop (about 110 amino acids total) in the transporter has been postulated to be important for substrate recognition and selectivity (Bleasby et al., 2005; Wu et al., 2009).
Fig. 2.
Localization and conservation of the amino acid fragments in the SLC22 transporters and in the odorant receptors. The location of the conserved amino acid fragments within models of SLC22 transporters (A) and olfactory receptors (B) generated using TOPO2 (Pharmacogenetics.UCSF.edu). The 79-AA residue motif (highlighted in orange) is located in the first extracellular loop of the transporter (A), whereas the 77-AA residue motif (highlighted in red) not only encompasses the fourth transmembrane domain, but also includes the entire second (and largest) extracellular loop of the receptor (B), which corresponds to the most highly conserved portion of the motif between the transporter and the receptor (Fig. 2B). (C) Weblogo (http://weblogo.berkeley.edu/) representations of the amino acid sequences of the 79-AA fragment of transporters (top) and the 77-AA fragment of odorant receptors (bottom). A number of amino acid residues were found to be highly conserved among the transporters and OR proteins, in particular a number of conserved cysteine residues (depicted in black), which are considered critical for structural integrity of the membrane proteins (Bleasby et al., 2005).
Overlaying the corresponding 77-AA fragment of OLFR167 with a model of a typical seven-transmembrane domain–containing odorant receptor revealed that this fragment is located in the middle portion of the receptor sequence and included the second and the largest extracellular loop of the receptor between transmembrane helixes 4 and 5 (Fig. 2B). Within this region of the receptor, there were several amino acid residues that are highly conserved among the majority of OR proteins examined (Fig. 1C, Supplemental Fig. 3), including cysteine residues in extracellular loop 2 considered critical for structural integrity and for odorant recognition (Malnic et al., 2010) (Fig. 2C). Similarly, cysteine residues in the 79-AA fragment of the SLC22A20 transporter (Fig. 1, Supplemental Fig. 1) were also found to be highly conserved in the SLC22 family of transporters (Fig. 2C).
Creation of a Pharmacophore Model on the Basis of OR Ligands That Can Interact with OAT6 and/or OAT1.
Although the available functional transporter data are limited, OATs of the SLC22 family (particularly OAT6 and OAT1), which are both found in the nasal epithelium (Kaler et al., 2006), have been demonstrated to interact with a few odorant molecules (in some instances with relatively high affinity) (Table 1). We sought to examine whether small-molecule volatile odorants that are known ligands of ORs could also interact with the transporters. To further explore OAT interactions with odorant molecules known to be OR ligands, we undertook a computational in silico approach involving pharmacophore modeling followed by virtual screening and wet laboratory testing.
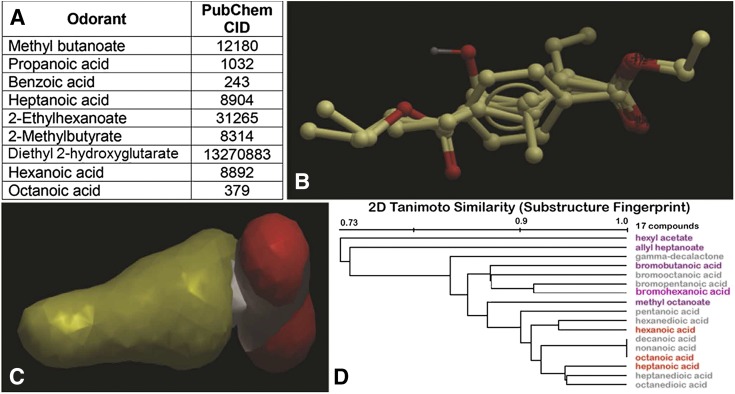
To determine the molecular features of small-molecule odorants that can also interact with the OATs, pharmacophore models were generated on the basis of those odorant molecules that can interact with OAT1 and/or OAT6 (Table 1). Since the list of known OAT-interacting odorants is small and some of the odorants can interact with both OAT1 and OAT6, a common pharmacophore model for both OAT1- and OAT6-interacting odorants was generated (Fig. 3). The structures of the OAT1/OAT6-interacting odorant molecules were superimposed and aligned using the APF superimposition method (Totrov, 2008) (Fig. 3B), and a pharmacophore hypothesis was generated. This shared pharmacophore model had a hydrophobic core that appears compatible with a ring-like structure, and two or more hydrogen bond acceptor sites on one end of the hydrophobic core (Fig. 3C).
Fig. 3.
Generation of a pharmacophore model using odorants/ligands that can also interact with OAT1 and/or OAT6. The chemical structural features of odorant molecules that can interact with OAT1 and/or OAT6 (A) were aligned (B) and used to create a consensus pharmacophore hypothesis (C) [red, hydrogen bond acceptor; yellow, encompasses the hydrophobic core probably owing to the presence of an aromatic ring structure; see (B)]. This pharmacophore hypothesis was used to query a structural library of odorants (OlfactionDB) and identified compounds were clustered in ChemMine followed by PubChem Tanimoto Similarity (D). Red font, known OAT-interacting odorants; Purple font, odorant molecules tested in in vitro OAT transport assay.
Virtual Screening of an Odorant Database with the Pharmacophore (on the Basis of OAT-Binding Odorants).
The pharmacophore model—which has as a basis OR ligands capable of interacting with OATs in transport assays—was then used in a virtual screen of a structural library of odorant molecules which have been shown to bind ORs with an EC50 of ∼1 μM to ∼1 mM. This was done by using an online odorant database, OlfactionDB (Modena et al., 2011). Although limited, this database, which is manually curated from the literature, provides information about these odorant molecules and their interactions with odorant receptors (Modena et al., 2011). Since the aim here was to identify odorants that are known to interact with ORs and which might also interact with OATs, it was expected that this database would be useful for virtual screening with the pharmacophore model.
Virtual screening of the database with the pharmacophore model identified 35 odorant molecules with an APF (Totrov, 2008) and steric score (Chen et al., 2014) indicative of a reasonable fit to the hypothesis (see Materials and Methods; Supplemental Table 1). These 35 odorants were input into ChemMine (Chemmine.ucr.edu), an online analytic tool for examining small molecules (Backman et al., 2011). This analysis resulted in a cluster of 27 compounds with similar chemical structures out of the 35 odorant molecules selected in the virtual screen (Supplemental Table 1). Additional similarity analysis of these 27 structures on the basis of the Tanimoto coefficient revealed a subgroup of 17 molecules that included three (hexanoic acid, octanoic acid and heptanoic acid) known to interact with OAT1 and/or OAT6 (Table 1, Fig. 3D). As described below, five of the remaining 14 compounds in the cluster, as well as one additional compound not found within this cluster, were analyzed in a well-characterized OAT transport assay (Ahn et al., 2009; Schnabolk et al., 2010; Wu et al., 2013).
Identification of Novel OAT-Interacting Odorants That Are Also Known OR Ligands.
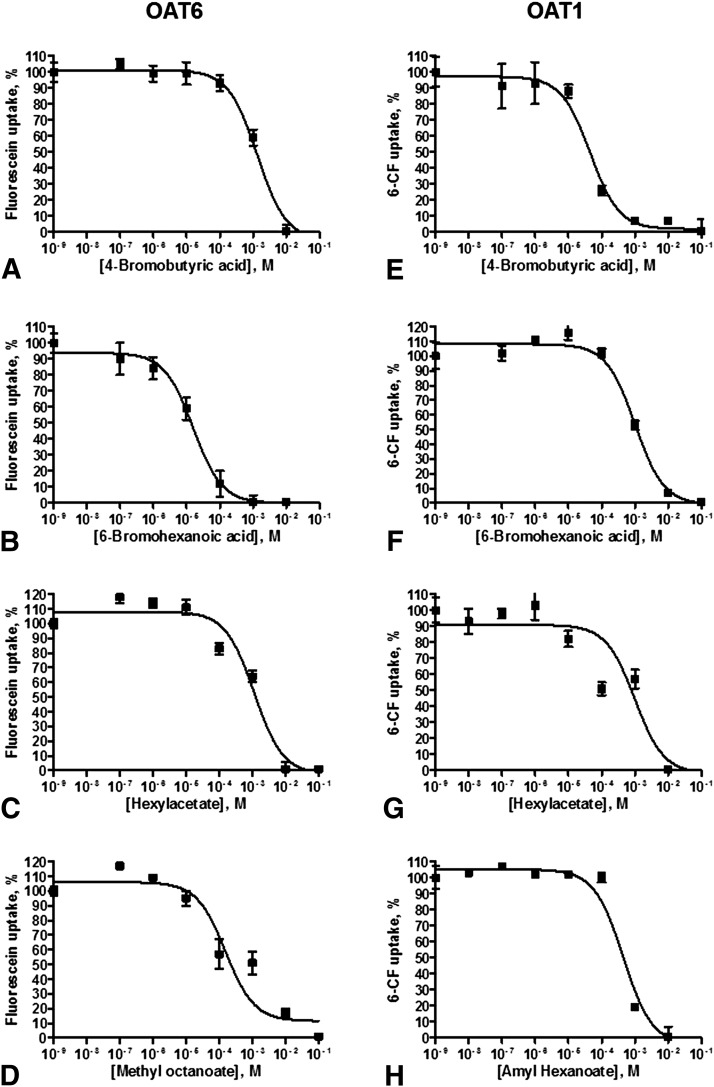
Five of the remaining 14 untested molecules (i.e., 4-bromobutyric acid, methyl octanoate, hexyl acetate, allyl heptanoate, 6-bromohexanoic acid) and one odorant not found in the cluster (i.e., amyl hexanoate) were further evaluated for the ability to inhibit OAT6- and/or OAT1-mediated uptake of a fluorescent substrate in wet laboratory assays (Fig. 4). Four of these odorant molecules (4-bromobutyric acid, 6-bromohexanoic acid, hexyl acetate and methyl ocantoate) were found to inhibit OAT6-mediated uptake of fluorescein in OAT6-expressing CHO cells, whereas allyl heptanoate and amyl hexanoate did not inhibit fluorescein uptake at a concentrations less than 3 mM (Fig. 4, Table 2). Whereas 4-bromobutyric acid, 6-bromohexanoic acid, and hexyl acetate were also found capable of inhibiting OAT1-mediated uptake of 6CF with IC50s in the range of 16–1346 μM, methyl octanoate was unable to inhibit OAT1-mediated uptake at concentrations less than 3 mM (Fig. 4, Table 2). In addition, amyl hexanoate, which did not cluster with the other five odorant molecules (Fig. 3) and was unable to inhibit OAT6-mediated uptake, was found to be capable of inhibiting OAT1-mediated uptake of 6CF (IC50 444 μM) (Fig. 4, Table 2).
Fig. 4.
Inhibition of OAT6- and/or OAT1-mediated uptake by odorants. Concentration-dependent inhibition of fluorescent substrate uptake in OAT6-expressing (A–D) and OAT1-expressing (E–H) CHO-cells (nonlinear regression fitted to curve, data are presented as Mean ± S.D., N = 4). Six odorants that are known ligands of odorant receptors (i.e., 4-bromobutyric acid, 6-bromohexanoic acid, hexyl acetate, methyl octanoate, amyl hexanoate and allyl heptanoate) were tested for their ability to inhibit OAT6- and/or OAT1-mediated uptake of a fluorescent substrate. Three of these small-molecule odorants, 4-bromobutyric acid, 6-bromohexanoic acid, and hexyl acetate, were found to inhibit both OAT6-mediated uptake of fluorescein (A–C) and OAT1-mediated uptake of 6CF (E–G). In addition, methyl octanoate was found to be capable of inhibiting OAT6-mediated transport (D), whereas amyl hexanoate inhibited OAT1-mediated transport (H). R2 values ranged from 0.89–0.99. For both OAT6 and OAT1, probenecid was used as a control.
We note that the effective dose of these odorants for interaction with OATs and ORs appear to be roughly comparable. Furthermore, comparison of the odorant inhibition data for OAT1 with that reported for drugs, metabolites, toxins, and signaling molecules revealed that the inhibitory data for the odorants fell within the range of reported data (Eraly et al., 2006; VanWert et al., 2010; Wikoff et al., 2011; Morrissey et al., 2012; Nigam et al., 2015). Although we were unable to obtain the newly identified compounds in labeled form, we note the OAT1 has been shown to transport many endogenous ligands (Nigam, 2015), and OAT6 can transport estrone sulfate, fluorescein, ochratoxin A, and PG-E2 (Monte et al., 2004; Kaler et al., 2007; Truong et al., 2008; Schnabolk et al., 2010).
Discussion
A combined in silico and wet-laboratory approach was used to investigate the relationship between anion transporters of the SLC22 family, odorant molecules, and odorant receptors, and resulted in the identification of novel OAT6 and OAT1 ligands that are known OR ligands. To summarize, it was found that: 1) Oat6/Slc22a20, a mouse organic anion transporter expressed in the nasal epithelium (Monte et al., 2004; Kaler et al., 2006; Kaler et al., 2007), is highly conserved in many vertebrate species, particularly in placental mammals (Fig. 1, Supplemental Fig. 1), and, importantly, a 79-AA motif, within the N-terminal region of OAT6/SLC22A20, is present in all 20 vertebrate species examined (Fig. 1, Supplemental Fig. 1); 2) This 79-AA motif is also found in other members of the SLC22 drug transporter family and in some seven-transmembrane domain–containing odorant receptors as a 77-AA motif (Fig. 1, Supplemental Fig. 2); 3) The 77-AA motif is present in many ORs (Fig. 1, Supplemental Figs. 2 and 3); 4) The location of this conserved motif overlaps postulated substrate binding domains (on the basis of published wet laboratory data discussed below), being found in the largest/main extracellular loop of both the SLC22 transporters and the odorant receptors (Fig. 2). It is perhaps also interesting to note in passing that OAT1, the prototypical OAT transporter underlying the classic organic anion transport pathway, was initially cloned (as NKT) through a homology-based approach using degenerate primers targeting seven-transmembrane G protein coupled receptors (Lopez-Nieto et al., 1996; Lopez-Nieto et al., 1997); 5) Odorants known to bind odorant receptors are also capable of interaction with OAT1 and OAT6 (Table 1); 6) Odorant molecules that are capable of interacting with OAT1 and/or OAT6 share similar structural features (Fig. 3); and 7) In silico screening of an odorant database, which includes many known and putative OR ligands, identified additional odorant molecules that had the ability, in wet laboratory assays using transfected cells, to inhibit OAT6- and/or OAT1-mediated transport (Fig. 4, and Table 2).
Collectively, this information suggests that a highly conserved motif shared by both SLC22 transporters and odorant receptors might play a role in substrate/ligand recognition and binding by these two different classes of membrane proteins. In support of this notion, it has been shown that replacing the entire large extracellular loop between transmembrane domains 1 and 2, which encompasses the 79-AA motif (Fig. 2) of OCT1/SLC22A1, with that of OAT1/SLC22A6 [amino acid 40-136, OCT1(Oat1-l)] significantly alters the uptake of tetraethylammonium (TEA, a prototypical OCT1 substrate) (Keller et al., 2011). Furthermore, an examination of available data on single-nucleotide polymorphisms (SNPs) discovered in organic anion transporter genes indicates that a number of nonsynonymous SNPs within the boundary of this 79-AA motif have functional ligand-binding and/or transport consequences in several SLC22 transporters, including OAT1 and URAT1 (Table 3). For example, in OAT1, a nonsynonymous SNP resulting in a change from arginine to histidine at residue 50 (R50H) significantly reduced the uptake of antiviral drugs (i.e., adefovir, cidofovir, and tenofovir) (Bleasby et al., 2005), whereas a nonsynonymous SNP resulting in a change from arginine to histidine at residue 90 (R90H) in URAT1 significantly reduced the uptake of urate (Ichida et al., 2004). In addition, an SNP in a conserved residue located five amino-acid residues downstream of the 79-AA motif in URAT1, V138M, also significantly reduced uptake of urate by the mutant transporter (Ichida et al., 2004).
TABLE 3.
Effects of mutations/polymorphisms in shared motif region of OATs and ORs
| OAT or OR | Variant | Substrate/Ligand with Altered Response | Reference |
|---|---|---|---|
| OAT1(SLC22A6) | R50H | Adefovir, cidofovir, tenofovir | (Bleasby et al., 2005) |
| URAT1(SLC22A12) | R90H | Urate | (Ichida et al., 2004; Cheong et al., 2005) |
| URAT1(SLC22A12) | V138M | Urate | (Ichida et al., 2004) |
| Or15 | A195X | Multiple odorants | (Hughes et al., 2014) |
| OR5A1 | N183D | β-Ionone | (Jaeger et al., 2013) |
| OR2A25 | S75N | Geranyl acetate | (Mainland et al., 2014) |
| OR2B11 | I130S+V198M | Spearmint oil | (Mainland et al., 2014) |
X, various amino acids in Hughes et al. (2014).
Interestingly, similar findings related to the function of the conserved motif have been reported for the ORs. For example, in a systematic study investigating the structural basis of ligand specificity for odorant receptors, site-directed mutagenesis targeting 37 residues individually in odorant receptor Or15 in Anopheles gambiae found that a determinant of odorant specificity resides in the large extracellular loop 2 (which is within the 77-AA motif of the OR) (Hughes et al., 2014). In particular, substitution of an alanine residue at position 195 within the motif resulted in the largest reduction in sensitivity of the receptor (Hughes et al., 2014). In addition, in humans, a nonsynonymous SNP resulting in the substitution of aspartic acid for asparagine at residue 183 (N183D) (located in the extracellular loop 2) of OR5A1 was identified as a causal variant of β-ionone sensitivity (Jaeger et al., 2013). More recently, in a comprehensive large-scale structural-functional relationship study of polymorphisms of human odorant receptors, 18 residues within the boundary of the 77-AA motif were deemed to be important for the function of human odorant receptors, and concluded that these SNPs or mutations probably alter the ligand binding and selectivity of the underlying odorant receptor (Mainland et al., 2014). For example, polymorphisms in OR2A25, whose close paralogs have the highest sequence homology with the 79-AA motif of the transporters (Supplemental Fig. 2), shifted its dose-response curve for geranyl acetate, whereas substituting serine for isoleucine at residue 130 (I130S) and methionine for valine at residue 198 (V198M) (both of which are within the boundary of the motif) of OR2B11 effectively eliminated the mutant’s ability to respond to spearmint oil (Mainland et al., 2014).
Furthermore, our findings raise the possibility that the transporters and ORs might function in a parallel fashion via their ability to respond to the same or similar ligands. For example, as described above, propionic acid (a gut microbiome–derived short-chain fatty acid) is a substrate for renal organic anion transporters (OAT1/OAT3, Table 1), and this small-molecule odorant is also able to interact with ORs (OLFR78 and OR51E) (Modena et al., 2011). Interestingly, deletion of renally expressed Olfr78, which also contains the shared motif, results in altered renin secretion and blood pressure regulation (Pluznick et al., 2013). Thus, in the kidney, OATs (OAT1/3) probably play a role in regulating the plasma concentration of propionic acid, and the OR (e.g., OLFR78) can sense changes in the concentration of this short-chain fatty acid and transduce this information to juxtaglomerular cells, leading to physiologic changes (i.e., blood pressure) (Pluznick and Caplan, 2012; Pluznick et al., 2013). This may also be interesting in the context of understanding the altered blood pressure in the Oat3 knockout mouse (Vallon et al., 2008).
The potential signaling function of SLC22 transporters also needs to be examined. Apart from odorants, other OAT ligands are also known to activate G protein-coupled receptors (Nigam, 2015). A number of mammalian SLC transporters are thought to play a role in sensing and signaling, and some may function in a manner analogous to bacterial “transceptors” (Hyde et al., 2007; Thevelein and Voordeckers, 2009; Chantranupong et al., 2015). These transporters and receptors might conceivably also be part of a proposed system for mediating remote communication between organs and/or organisms via these small-molecule odorants, according to the remote sensing and signaling hypothesis (Ahn and Nigam, 2009; Wu et al., 2011; Nigam, 2015; Nigam et al., 2015). For example, Oat6 is mainly expressed in the nasal epithelium and is able to interact with odorants such as propionic acid whose excretion into the urine is probably mediated, at least in part, by the renally expressed OATs. Thus, propionic acid not only would be able to bind to its nasally expressed OR, but it could also be taken up by an OAT (e.g., OAT6 and/or OAT1), gaining entry to the blood and the central nervous system. Although speculative at present, these possibilities could have a major impact on current paradigms in the fields of olfaction and drug transport.
Supplementary Material
Acknowledgments
The authors acknowledge the assistance of Anne Goldenberg.
Abbreviations
- AA
amino acids
- ABC
ATP-binding cassette
- APF
atomic properties field
- CHO
Chinese hamster ovary
- NKT
novel kidney transporter
- OAT
organic anion transporter
- OAT6
organic anion transporter 6/SLC22A20
- OAT1
organic anion transporter 1/SLC22A6
- OAT3
organic anion transporter 3/SLC22A8
- OR
odorant receptor
- OCT1
organic cation transporter 1/SLC22A1
- SLC
solute carrier
- SNP
single-nucleotide polymorphisms
Authorship Contributions
Participated in research design: Wu, Nigam.
Conducted experiments: Wu, Bush, Liu, Zhu.
Contributed new reagents or analytic tools: Abagyan, Nigam.
Performed data analysis: Wu, Bush.
Wrote or contributed to the writing of the manuscript: Wu, Bush, Abagyan, Nigam.
Footnotes
This work was partly supported by National Institutes of Health Grants [GM104098] and [GM098449] to S.K.N. and the Nancy Kaehr Chair in Research at UCSD.
References
- Ahn SY, Bhatnagar V. (2008) Update on the molecular physiology of organic anion transporters. Curr Opin Nephrol Hypertens 17:499–505. [DOI] [PubMed] [Google Scholar]
- Ahn SY, Eraly SA, Tsigelny I, Nigam SK. (2009) Interaction of organic cations with organic anion transporters. J Biol Chem 284:31422–31430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SY, Jamshidi N, Mo ML, Wu W, Eraly SA, Dnyanmote A, Bush KT, Gallegos TF, Sweet DH, Palsson BO, et al. (2011) Linkage of organic anion transporter-1 to metabolic pathways through integrated “omics”-driven network and functional analysis. J Biol Chem 286:31522–31531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SY, Nigam SK. (2009) Toward a systems level understanding of organic anion and other multispecific drug transporters: a remote sensing and signaling hypothesis. Mol Pharmacol 76:481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman TW, Cao Y, Girke T. (2011) ChemMine tools: an online service for analyzing and clustering small molecules. Nucleic Acids Res 39:W486–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleasby K, Hall LA, Perry JL, Mohrenweiser HW, Pritchard JB. (2005) Functional consequences of single nucleotide polymorphisms in the human organic anion transporter hOAT1 (SLC22A6). J Pharmacol Exp Ther 314:923–931. [DOI] [PubMed] [Google Scholar]
- Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, et al. (2013) BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41:W29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady KP, Dushkin H, Förnzler D, Koike T, Magner F, Her H, Gullans S, Segre GV, Green RM, Beier DR. (1999) A novel putative transporter maps to the osteosclerosis (oc) mutation and is not expressed in the oc mutant mouse. Genomics 56:254–261. [DOI] [PubMed] [Google Scholar]
- César-Razquin A, Snijder B, Frappier-Brinton T, Isserlin R, Gyimesi G, Bai X, Reithmeier RA, Hepworth D, Hediger MA, Edwards AM, et al. (2015) A Call for Systematic Research on Solute Carriers. Cell 162:478–487. [DOI] [PubMed] [Google Scholar]
- Chantranupong L, Wolfson RL, Sabatini DM. (2015) Nutrient-sensing mechanisms across evolution. Cell 161:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Totrov M, Abagyan R. (2014) Docking to multiple pockets or ligand fields for screening, activity prediction and scaffold hopping. Future Med Chem 6:1741–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong HI, Kang JH, Lee JH, Ha IS, Kim S, Komoda F, Sekine T, Igarashi T, Choi Y. (2005) Mutational analysis of idiopathic renal hypouricemia in Korea. Pediatr Nephrol 20:886–890. [DOI] [PubMed] [Google Scholar]
- Emami Riedmaier A, Nies AT, Schaeffeler E, Schwab M. (2012) Organic anion transporters and their implications in pharmacotherapy. Pharmacol Rev 64:421–449. [DOI] [PubMed] [Google Scholar]
- Eraly SA, Monte JC, Nigam SK. (2004) Novel slc22 transporter homologs in fly, worm, and human clarify the phylogeny of organic anion and cation transporters. Physiol Genomics 18:12–24. [DOI] [PubMed] [Google Scholar]
- Eraly SA, Vallon V, Vaughn DA, Gangoiti JA, Richter K, Nagle M, Monte JC, Rieg T, Truong DM, Long JM, et al. (2006) Decreased renal organic anion secretion and plasma accumulation of endogenous organic anions in OAT1 knock-out mice. J Biol Chem 281:5072–5083. [DOI] [PubMed] [Google Scholar]
- Genter MB, Kendig EL, Knutson MD. (2009) Uptake of materials from the nasal cavity into the blood and brain: are we finally beginning to understand these processes at the molecular level? Ann N Y Acad Sci 1170:623–628. [DOI] [PubMed] [Google Scholar]
- Hagos Y, Stein D, Ugele B, Burckhardt G, Bahn A. (2007) Human renal organic anion transporter 4 operates as an asymmetric urate transporter. J Am Soc Nephrol 18:430–439. [DOI] [PubMed] [Google Scholar]
- Heydel JM, Coelho A, Thiebaud N, Legendre A, Le Bon AM, Faure P, Neiers F, Artur Y, Golebiowski J, Briand L. (2013) Odorant-binding proteins and xenobiotic metabolizing enzymes: implications in olfactory perireceptor events. Anat Rec (Hoboken) 296:1333–1345. [DOI] [PubMed] [Google Scholar]
- Hughes DT, Wang G, Zwiebel LJ, Luetje CW. (2014) A determinant of odorant specificity is located at the extracellular loop 2-transmembrane domain 4 interface of an Anopheles gambiae odorant receptor subunit. Chem Senses 39:761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde R, Cwiklinski EL, MacAulay K, Taylor PM, Hundal HS. (2007) Distinct sensor pathways in the hierarchical control of SNAT2, a putative amino acid transceptor, by amino acid availability. J Biol Chem 282:19788–19798. [DOI] [PubMed] [Google Scholar]
- Ichida K, Hosoyamada M, Hisatome I, Enomoto A, Hikita M, Endou H, Hosoya T. (2004) Clinical and molecular analysis of patients with renal hypouricemia in Japan-influence of URAT1 gene on urinary urate excretion. J Am Soc Nephrol 15:164–173. [DOI] [PubMed] [Google Scholar]
- Jaeger SR, McRae JF, Bava CM, Beresford MK, Hunter D, Jia Y, Chheang SL, Jin D, Peng M, Gamble JC, et al. (2013) A Mendelian trait for olfactory sensitivity affects odor experience and food selection. Curr Biol 23:1601–1605. [DOI] [PubMed] [Google Scholar]
- Kaler G, Truong DM, Khandelwal A, Nagle M, Eraly SA, Swaan PW, Nigam SK. (2007) Structural variation governs substrate specificity for organic anion transporter (OAT) homologs. Potential remote sensing by OAT family members. J Biol Chem 282:23841–23853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaler G, Truong DM, Sweeney DE, Logan DW, Nagle M, Wu W, Eraly SA, Nigam SK. (2006) Olfactory mucosa-expressed organic anion transporter, Oat6, manifests high affinity interactions with odorant organic anions. Biochem Biophys Res Commun 351:872–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T, Egenberger B, Gorboulev V, Bernhard F, Uzelac Z, Gorbunov D, Wirth C, Koppatz S, Dötsch V, Hunte C, et al. (2011) The large extracellular loop of organic cation transporter 1 influences substrate affinity and is pivotal for oligomerization. J Biol Chem 286:37874–37886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, Lopez R. (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43 (W1):W580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Nieto CE, Nigam SK. (1996) Selective amplification of protein-coding regions of large sets of genes using statistically designed primer sets. Nat Biotechnol 14:857–861. [DOI] [PubMed] [Google Scholar]
- Lopez-Nieto CE, You G, Barros EJG, Beier DR, Nigam SK. (1996) Molecular cloning and characterization of a novel transporter protein with very high expression in the kidney. abstract J Am Soc Nephrol 7:1301. [Google Scholar]
- Lopez-Nieto CE, You G, Bush KT, Barros EJ, Beier DR, Nigam SK. (1997) Molecular cloning and characterization of NKT, a gene product related to the organic cation transporter family that is almost exclusively expressed in the kidney. J Biol Chem 272:6471–6478. [DOI] [PubMed] [Google Scholar]
- Mainland JD, Keller A, Li YR, Zhou T, Trimmer C, Snyder LL, Moberly AH, Adipietro KA, Liu WL, Zhuang H, et al. (2014) The missense of smell: functional variability in the human odorant receptor repertoire. Nat Neurosci 17:114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B, Gonzalez-Kristeller DC, and Gutiyama LM (2010) Odorant receptors, in The Neurobiology of Olfaction (Menini A ed) CRC Press, Boca Raton, FL. [PubMed] [Google Scholar]
- Modena D, Trentini M, Corsini M, Bombaci A, Giorgetti A. (2011) OlfactionDB: A database of olfactory receptors and their ligands. Adv Lif Sci 1:1–5. [Google Scholar]
- Monte JC, Nagle MA, Eraly SA, Nigam SK. (2004) Identification of a novel murine organic anion transporter family member, OAT6, expressed in olfactory mucosa. Biochem Biophys Res Commun 323:429–436. [DOI] [PubMed] [Google Scholar]
- Morrissey KM, Wen CC, Johns SJ, Zhang L, Huang SM, Giacomini KM. (2012) The UCSF-FDA TransPortal: a public drug transporter database. Clin Pharmacol Ther 92:545–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima A, Touhara K. (2010) Enzymatic conversion of odorants in nasal mucus affects olfactory glomerular activation patterns and odor perception. J Neurosci 30:16391–16398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle MA, Wu W, Eraly SA, Nigam SK. (2013) Organic anion transport pathways in antiviral handling in choroid plexus in Oat1 (Slc22a6) and Oat3 (Slc22a8) deficient tissue. Neurosci Lett 534:133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam SK. (2015) What do drug transporters really do? Nat Rev Drug Discov 14:29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam SK, Bush KT, Martovetsky G, Ahn SY, Liu HC, Richard E, Bhatnagar V, Wu W. (2015) The organic anion transporter (OAT) family: a systems biology perspective. Physiol Rev 95:83–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluznick JL, Caplan MJ. (2012) Novel sensory signaling systems in the kidney. Curr Opin Nephrol Hypertens 21:404–409. [DOI] [PubMed] [Google Scholar]
- Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan LX, Rey F, Wang T, et al. (2013) Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci USA 110:4410–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabolk GW, Gupta B, Mulgaonkar A, Kulkarni M, Sweet DH. (2010) Organic anion transporter 6 (Slc22a20) specificity and Sertoli cell-specific expression provide new insight on potential endogenous roles. J Pharmacol Exp Ther 334:927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabolk GW, Youngblood GL, Sweet DH. (2006) Transport of estrone sulfate by the novel organic anion transporter Oat6 (Slc22a20). Am J Physiol Renal Physiol 291:F314–F321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet DH, Miller DS, Pritchard JB, Fujiwara Y, Beier DR, Nigam SK. (2002) Impaired organic anion transport in kidney and choroid plexus of organic anion transporter 3 (Oat3 (Slc22a8)) knockout mice. J Biol Chem 277:26934–26943. [DOI] [PubMed] [Google Scholar]
- Thevelein JM, Voordeckers K. (2009) Functioning and evolutionary significance of nutrient transceptors. Mol Biol Evol 26:2407–2414. [DOI] [PubMed] [Google Scholar]
- Thiebaud N, Menetrier F, Belloir C, Minn AL, Neiers F, Artur Y, Le Bon AM, Heydel JM. (2011) Expression and differential localization of xenobiotic transporters in the rat olfactory neuro-epithelium. Neurosci Lett 505:180–185. [DOI] [PubMed] [Google Scholar]
- Totrov M. (2008) Atomic property fields: generalized 3D pharmacophoric potential for automated ligand superposition, pharmacophore elucidation and 3D QSAR. Chem Biol Drug Des 71:15–27. [DOI] [PubMed] [Google Scholar]
- Truong DM, Kaler G, Khandelwal A, Swaan PW, Nigam SK. (2008) Multi-level analysis of organic anion transporters 1, 3, and 6 reveals major differences in structural determinants of antiviral discrimination. J Biol Chem 283:8654–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsigelny IF, Kovalskyy D, Kouznetsova VL, Balinskyi O, Sharikov Y, Bhatnagar V, Nigam SK. (2011) Conformational changes of the multispecific transporter organic anion transporter 1 (OAT1/SLC22A6) suggests a molecular mechanism for initial stages of drug and metabolite transport. Cell Biochem Biophys 61:251–259. [DOI] [PubMed] [Google Scholar]
- Vallon V, Eraly SA, Wikoff WR, Rieg T, Kaler G, Truong DM, Ahn SY, Mahapatra NR, Mahata SK, Gangoiti JA, et al. (2008) Organic anion transporter 3 contributes to the regulation of blood pressure. J Am Soc Nephrol 19:1732–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanWert AL, Gionfriddo MR, Sweet DH. (2010) Organic anion transporters: discovery, pharmacology, regulation and roles in pathophysiology. Biopharm Drug Dispos 31:1–71. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Nagle MA, Kouznetsova VL, Tsigelny IF, Nigam SK. (2011) Untargeted metabolomics identifies enterobiome metabolites and putative uremic toxins as substrates of organic anion transporter 1 (Oat1). J Proteome Res 10:2842–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Baker ME, Eraly SA, Bush KT, Nigam SK. (2009) Analysis of a large cluster of SLC22 transporter genes, including novel USTs, reveals species-specific amplification of subsets of family members. Physiol Genomics 38:116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Dnyanmote AV, Nigam SK. (2011) Remote communication through solute carriers and ATP binding cassette drug transporter pathways: an update on the remote sensing and signaling hypothesis. Mol Pharmacol 79:795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Jamshidi N, Eraly SA, Liu HC, Bush KT, Palsson BO, Nigam SK. (2013) Multispecific drug transporter Slc22a8 (Oat3) regulates multiple metabolic and signaling pathways. Drug Metab Dispos 41:1825–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.