Abstract
Harmful algal blooms caused by Phaeocystis globosa have resulted in staggering losses to coastal countries because of their world-wide distribution. Bacteria have been studied for years to control the blooms of harmful alga, however, the action mechanism of them against harmful algal cells is still not well defined. Here, a previously isolated algicidal bacterium Bacillus sp. LP-10 was used to elucidate the potential mechanism involved in the dysfunction of P. globosa algal cells at physiological and molecular levels. Our results showed Bacillus sp. LP-10 induced an obvious rise of reactive oxygen species (ROS), which was supposed to be major reason for algal cell death. Meanwhile, the results revealed a significant decrease of photosynthetic physiological indexes and apparent down-regulated of photosynthesis-related genes (psbA and rbcS) and protein (PSII reaction center protein D1), after treated by Bacillus sp. LP-10 filtrates, suggesting photoinhibition occurred in the algal cells. Furthermore, our results indicated that light played important roles in the algal cell death. Our work demonstrated that the major lethal reason of P. globosa cells treated by the algicidal bacterium was the photoinhibition resulted from oxidative stress induced by Bacillus sp. LP-10.
Microalgae play an important role in carbon sink during the transportation of the carbon to deep waters. However, many of them lead to harmful algal blooms (HABs), causing adverse effects on marine ecosystems and economics1,2, even harm people by producing toxins3. Many methods were developed to manage the HABs4,5. However, physical and chemical methods are not supposed to be optimal choices in controlling HABs, because of their obvious defects of high costs and unavailability for large-scale application, and more importantly, bringing about secondary pollution. Hence, biological methods are regarded as more promising approaches to regulate HABs6,7.
Marine bacteria are important in regulating the growth dynamics of microalga8,9. Among them, a group of bacteria, now called algicidal bacteria, are suggested to show inhibitory effects on algal growth, which are considered as excellent candidates to control HABs10. The action mode of algicidal bacteria on target algal cells were divided into two types, one is lysing the algal cells by contacting with the cells surface directly, named direct attack mode, the other is lysing the algal cells by releasing active substances/allelochemicals, named indirect attack mode11,12. Most algicidal bacteria reported so far belongs to the latter action mode13,14.
Despite a lot of algicidal bacteria were isolated, the action mechanisms of them against harmful algal cells were still not well defined. Recently, some researches revealed that reactive oxygen species (ROS) played significant roles in the inhibitory effects of algicidal bacteria on algal growth15,16. Some allelochemicals were documented to trigger the producing of ROS, leading to algal cells damage17,18.
As is known, environmental stresses usually induce algal cell to produce ROS, which change the oxidative level and breaks the redox balance of the algal cells19,20. Normally, antioxidant systems of cells, including superoxide dismutase (SOD), peroxidase (POD), catalase (CAT) and glutathione (GSH) etc., could protect themselves from oxidative damage by clearing the numerous ROS to keep cells at a stable redox level21,22.
Photosynthetic systems are fundamental for the growth of plant and algal cells. Generally, performing oxygenic photosynthesis will cause damage to the photosynthetic apparatus23. The reaction center of photosystem II (PSII) was one of the main damage target sites, of which the protein D1 is sensitive and vulnerable to stresses24. Photosynthetic organisms execute self-repair program to replace the impaired protein D1 via de novo synthesis. Normally, the protein D1 content is in a dynamic balance. However, when the protein D1 damage rate exceeds the repair rate, photoinhibition occurs in photosynthetic organisms24. Recent research revealed high light induced lower expression of protein D1 in diatom Phaeodactylum tricornutum than low light exposure25. Recent researches showed that environmental factors, such as NaCl, heat and low temperature can also lead to photoinhibition by interrupting the de novo synthesis of protein D123,26.
Phaeocystis globosa Scherffel (Prymnesiophyceae) is a HABs causing species with world-wide distribution, which caused huge economic loses to coastal countries27. Bacillus sp. LP-10, a marine bacterium obtained in our laboratory before, was proved to be effective in controlling the growth of P. globosa28. However, few work were performed to illustrate the action mode of the bacterium on P. globosa. To better understand the action mechanism of the algicidal mechanism, a systematic investigation of physiological and molecular alterations in algal cells was carried out. And the physiological data coupled with transcript and protein analysis provided new insights into the algicidal mechanism of the marine bacterium on target algal cells.
Materials and Methods
Strains and culture conditions
Cultures of P. globosa PG03 were supplied by the State Key Laboratory of Marine Environmental Sciences, Xiamen University, China and f/2 (-Si) medium was used to prepare algal cultures for the experiments29. The light conditions were set to a 12 h/12 h light-dark cycle under cool-white fluorescent light with an intensity of 50 μmol photons m−2s−1 at 20 ± 1 °C.
The Bacillus sp. LP-10 strain was previously isolated in surface water samples from the East China Sea in our laboratory, and the GenBank accession number is KF411459. The marine bacterial strain was prepared in autoclaved Zobell 2216E medium and cultured at 28 °C for 48 h with 150 rpm rotation speed30. The bacterium cultures at stationary phase were collected and centrifuged at 8000 g for 10 min. The cell-free filtrates of Bacillus sp. LP-10 were prepared by filtering the supernatant through 0.22 μm size pore Millipore membrane filters. The cell-free filtrates of Bacillus sp. LP-10 was used in the whole study unless specially declaration. To ensure that the Bacillus sp. LP-10 is safe for the environment, the toxicity of the strain were tested. The test results indicated Bacillus sp. LP-10 were environmentally friendly to the aquatic environment (Supplementary Information).
ROS assays
The fluorescence probe 2′, 7′-dichlorofluorescein diacetate (DCFH-DA) is used to measure the ROS level in algal cells treated with cell-free filtrates of Bacillus sp. LP-1031. The fluorescence intensity was measured at an excitation wavelength of 488 nm and an emission wavelength of 525 nm. For ROS detection, 20 mL algal cultures treated with strain LP-10 filtrates were centrifuged at 5000 g for 5 min, the pellets were washed twice with sterile f/2 medium, then the algal cells were incubated with DCFH-DA (final concentration 10 μM) at 37 °C for 30 min in the dark. The superfluous DCFH-DA probe was removed by centrifugation and the pellets were re-suspended in 500 μL sterile f/2 medium after washing twice. The fluorescence intensity was measured using a microplate reader and the fluorescence photos of 5% LP-10 treatment was taken by an epifluorescence microscope.
N-acetylcysteine (NAC), a common ROS scavenger, was used to clear the ROS in algal cells. 20 mL algal cultures were processed using four treatments respectively: (1) 5% (v/v) strain LP-10 filtrates (LP treatment); (2) 5% filtrates and NAC (final concentration 1 mM) (NAC + LP treatment); (3) NAC (final concentration 1 mM) (NAC control); (4) blank 2216E broth (5%) (CK). The autofluorescence of the algal cells was measured in 96-well microplates at an excitation wavelength of 440 nm and an emission wavelength of 680 nm. Similar operations were performed with glutathione (GSH) and ascorbic acid (AsA), two other ROS scavengers. The ROS contents were determined at 0, 4, 8, 12, 24 h after addition of ROS scavengers.
The MDA content and antioxidant activities assays
Algal cultures of P. globosa were treated by cell-free filtrates of Bacillus sp. LP-10 in 3, 4, 5% concentration, followed by sampling at different time course with an interval of 12 h. Algal cells were collected at 5000 g for 5 min, and the pellets were washed twice with sterile seawater. The cells were then re-suspended in 1 mL PBS solution (0.1 M, pH 7.4), followed by sonication at 100 W for 60 times. The cell debris was removed by centrifugation at 12000 g for 10 min, and the supernatants were transferred to new Eppendorf tubes for malondialdehyde (MDA) and antioxidants analysis. The MDA content and activities of the antioxidants, including superoxide dismutase (SOD), catalase (CAT), peroxidase (POD) and GSH, were measured using MDA kits and corresponding antioxidant assay kits (Nanjing Jiancheng Bioengineering Institute) following the manufacturer’s instructions. The contents of MDA and antioxidant were expressed on a protein basis.
Chlorophyll contents and variable chlorophyll fluorescence analysis
Chlorophyll contents were measured based on previously described methods with slight modifications32. 20 mL axenic exponential phase cell cultures of P. globosa were collected at 5000 g for 10 min and the pellets were re-suspended in 5 mL 90% ethanol overnight to extract the chlorophyll. After extraction, the pellets were removed using centrifugation, the optical density of the ethanol extract was measured at wavelengths of 645 and 664 nm using a VARY-50 spectrophotometer. The contents of chlorophyll a (Chl a) , chlorophyll b (Chl b), and the ratio of these two pigments were calculated as follows:
 |
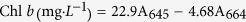 |
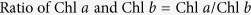 |
where A645 and A664 were the optical densities of ethanol extracts of algal cells at wavelengths of 645 and 664 nm.
The variable chlorophyll fluorescence was used as a proxy for the photosynthetic efficiency and capacity of the algal cell. And the variable chlorophyll fluorescence was measured using a PAM-CONTROL fluorometer (Walz, Effeltrich, Germany). The maximum quantum yield (Fv/Fm) and the maximum relative electron transport rate (rETR) were determined based on previously described methods after the algal cells were dark-adapted for 15 min33.
Light condition assays
20 mL algal cultures inoculated with 5% Bacillus sp. LP-10 filtrates, were respectivly placed under different light conditions: the normal light intensity (normal light, 50 μmol photons m−2s−1), low light intensity (low light, 20 μmol photons m−2s−1) and the dark condition (dark). The autofluorescence of the algal cells was measured in 96-well microplates every 24 h at an excitation wavelength of 440 nm and an emission wavelength of 680 nm. The algal cultures without LP-10 filtrates under different light conditions were set up as control.
Gene transcription analysis
For total RNA extraction, 40 mL algal cultures were treated for 24 h and harvested at 5000 g for 10 min, then the pellets were quickly frozen in liquid nitrogen and homogenized followed by addition of 1 mL RNAiso reagent (TaKaRa Biochemicals, China). The total RNA was extracted following the manufacturer’s instructions. Reverse transcription of RNA was performed using the PrimeScript™ RT-PCR Kit (TaKaRa Biochemicals, China). Two photosynthesis genes (psbA and rbcS) were selected for real-time qPCR analysis. The real-time PCR was carried out using SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa Biochemicals, China). The 18 S rRNA gene was used to standardize the results by eliminating variation in the quantity and quality. All of the primers used to amplify these genes were designed using software Primer premier 5.0 and the primer pairs are listed in Table 1. The RT-PCR program was: denaturation at 95 °C for 30 s, 40 cycles of 95 °C for 5 s and 60 °C for 30 s, followed by a melt curve program from 60 °C to 95 °C by 0.5 °C increases each 5 s. The relative gene expression abundance was quantified using the 2−ΔΔCt method34.
Table 1. The sequences of the primer pairs for RT-PCR analysis.
| Gene name | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| 18 S | TCCGATAACGAACGAGAC | TGACGCAAACTTCCACTT |
| psbA | AGTTGCTGGTTCTCTACTTTACG | TTCCCACTCACGACCGATG |
| rbcS | AAGTCTTACTGGGAAATGTGGG | AGCAGGACGCTGAACGATG |
Protein extraction and immunodetection of protein D1
20 mL algal cultures were centrifuged at 5000 g for 10 min, the pellets were washed twice with PBS solution and then re-suspended in 1 mL PBS solution. The suspension was then sonicated at 100 W for 60 times to lyse the algal cells. The debris was removed at 12000 g for 10 min and the supernatant were transferred to new Eppendorf tubes. The protein concentrations of the supernatant were determined using Bradford assays as mentioned above.
2 μg total protein were separated by SDS-PAGE in a 12% polyacrylamide gel, the resolved protein was then transferred to PVDF membranes and probed with the antibody (Agrisera) raised in rabbits against PSII reaction center protein D1. After incubation with secondary anti-rabbit IgG antibody coupled with horseradish peroxidase for 3 h, the membranes were reacted with ECL solution. Developed films were scanned with an Epson Expression 1680 Scanner25.
Results
Effects of strain LP-10 on intracellular ROS content of algal cells
The intracellular ROS content of algal cells incubated with 3, 4 and 5% Bacillus sp. LP-10 filtrates showed an obvious variation during incubation time (Fig. 1). The fluorescence intensity of algal cells at all treatment concentrations was kept at a low level ( < 100 RFU) in the early, then increased rapidly to a high level in all three treatment concentrations in 8 h (~1200 RFU) or 12 h (~400 RFU), respectively. Subsequently, the ROS levels of all treatments decreased to a low level, but still higher than the initial levels. The results shown in the fluorescence photos of algal cells treated by 5% filtrates were consistent with the fluorescence intensity determined by microplate reader (Fig. S1).
Figure 1. The ROS fluorescence intensity of algal cells under Bacillus sp. LP-10 treatment of different concentrations (3, 4, 5%).

Effects of ROS scavengers addition on algal cell death
The impacts of addition of ROS scavengers NAC, GSH and AsA on algal cell death are shown in Fig. 2A. The addition of ROS scavengers NAC significantly influenced the algicidal activities of Bacillus sp. LP-10 on P. globosa cells. The addition of the LP-10 filtrates significantly lower the fluorescence intensity of algal cells, which was obviously less than that of the control (CK). When LP-10 treatment was added with NAC (NAC + LP), the fluorescence intensity of algal cells was much higher than that of the LP-10 treatment without NAC. And the fluorescence intensity of the NAC + LP treatment was approximate to the CK control at 24 h. However, the fluorescence intensity of the NAC + LP treatment was lower than the control at 48 h and 72 h, but still higher than the LP-10 treatment. The trends of addition of scavengers GSH and AsA showed similar pattern with the NAC.
Figure 2.
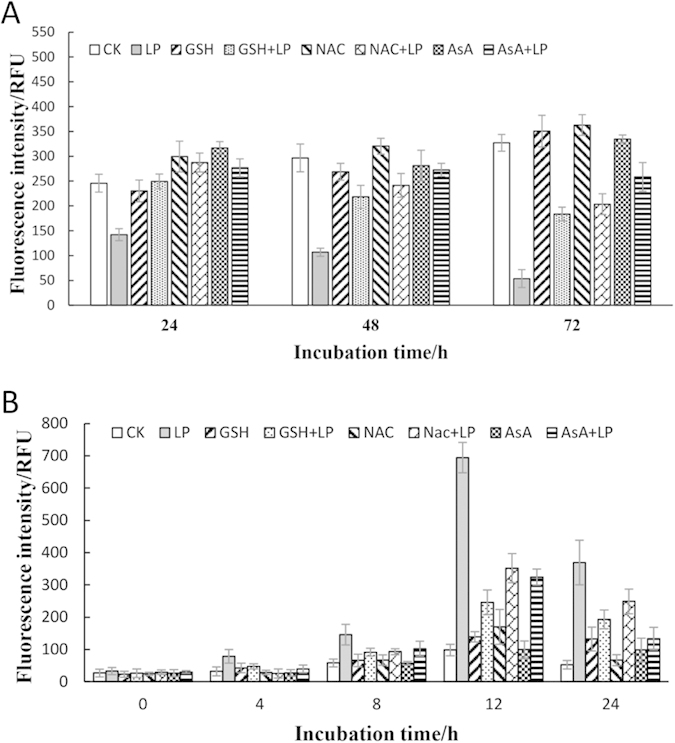
The algal autofluorescence intensity (A) and ROS fluorescence intensity (B) of algal cells under 5% Bacillus sp. LP-10 filtrates treatment with addition of ROS scavengers. CK represents algal cultures without any addition; LP represents algal cultures treated by 5% filtrates of Bacillus sp. LP-10; NAC represents algal cultures with addition of NAC; NAC + LP represents algal cultures with addition of NAC treated by 5% filtrates of Bacillus sp. LP-10. GSH and AsA are similar with those.
In the ROS scavenger assays, the ROS contents of algal cells in each groups were determined and the results were presented in Fig. 2B. The results indicated that the ROS levels of the treatment groups supplemented with ROS scavengers was apparent lower than the LP-10 treatment. NAC, GSH and AsA could effectively remove the superfluous ROS in the algal cells.
Effects of strain LP-10 on the MDA content and antioxidant activities of algal cells
The MDA content increased significantly after the algal cells were treated with filtrates (Fig. S2), and the MDA content of the 4 and 5% treatment groups at 72 h were 10.78 and 29.53 times higher than that of the control. The MDA content of the 3% treatment group was also slightly higher than the control at 72 h, which implied that the addition of Bacillus sp. LP-10 filtrates promoted the lipid peroxidation of algal cells obviously.
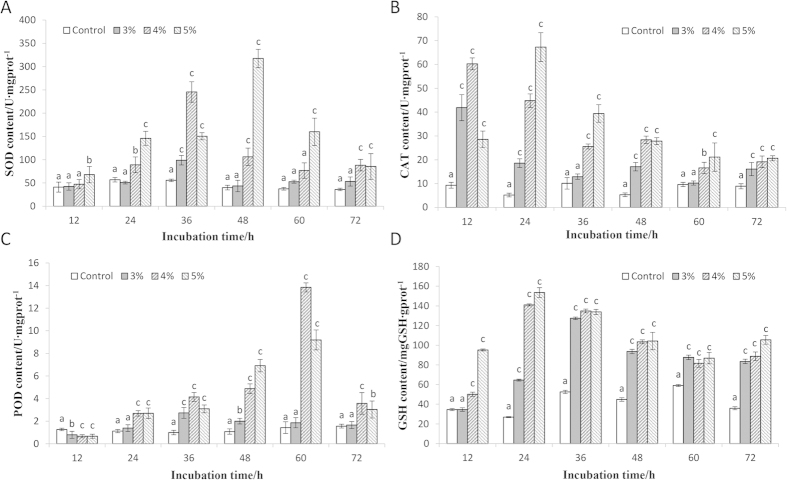
SOD activities of the different treatment groups varied based on the concentrations of the LP-10 filtrates (Fig. 3A). All three treatments significantly enhanced SOD activities and the highest SOD activities of the 3, 4 and 5% treatment groups were 1.77-, 4.41- and 7.96-fold of the control. CAT activities shared a similar pattern with the SOD activities but with a rapid increase at the early phase (Fig. 3B). All three treatment groups showed a significant increase of CAT activities after 72 h incubation with LP-10 filtrates. POD activities of algal cells changed slower than CAT activities (Fig. 3C), and the 4 and 5% treatment groups were significant higher than the control. GSH is a type of non-enzymatic antioxidant and plays an important role in protecting cells from oxidative stress. Similarly, the addition of LP-10 filtrates also induced a rapid increase of GSH content in a short time (Fig. 3D), and the GSH content of the three treatment groups was 2.32, 2.47 and 2.93 times higher than that of the control after 72 h treatment.
Figure 3. The SOD, CAT, POD and GSH activities of algal cells incubated with LP-10 filtrates of different concentrations (3, 4, and 5%).
All data represent the means ± S.D. (a) represents no significant difference, (b,c) represent statistically significant differences of p < 0.05 and p < 0.01 compared to the control respectively.
Effects of strain LP-10 on chlorophyll content and variable chlorophyll fluorescence
The contents of Chl a and b significantly decreased after algal cells were incubated with strain Bacillus sp. LP-10 filtrates (Fig. S3A,B). With the incubation time prolonged, decline of pigment contents showed the manner of concentration-dependent. The ratio of Chl a and Chl b were significantly impacted by the LP-10 filtrates (Fig. S3C). In the early phase, the ratio of the two pigments kept stable, but a rapid decrease was observed after the algal cells were incubated with filtrates for more than 36 h, and higher concentration filtrates led to more significant decreases of the ratio. In the late phase, the ratio began to increase and gradually surpassed the control group. These results suggested that the sensitivity of the two pigments to strain LP-10 filtrates was different, and the pigment structures and balance were destroyed by the filtrates.
The photosynthetic efficiency measured from maximum quantum yields (Fv/Fm) showed an apparent decrease after the algal cells were treated with LP-10 filtrates. Higher concentration filtrates and longer treatment time led to more severe damage to the algal cells (Fig. S4A). The Fv/Fm value of the 5% treatment group at 48 h was approximate to 0.3, far less than the normal level. The maximum rETR, which was used to evaluate the maximum photosynthetic capacity, showed a similar change pattern with Fv/Fm (Fig. S4B) and the rETR of all three treatment groups significantly declined after the algal cells were treated with strain LP-10 filtrates.
Effects of light conditions on algal cells
The effects of different light conditions on the algicidal activities of strain LP-10 were presented in Fig. 4. The proportion of dead cells varied based on the light conditions. The algicidal activities of 5% LP-10 filtrates treatment under normal light were 82.2% at 72 h, while those of the low light and dark treatment groups were 14.2% and 22.8% respectively, which were significantly lower than that of the normal light treatment group.
Figure 4. The autofluorescence intensity of algal cells under different light conditions after the algal cells were incubated with 5% LP-10 filtrates for 72 h.
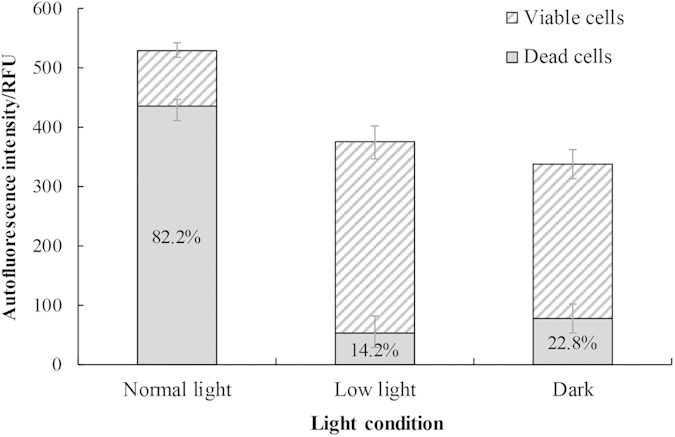
Normal light, low light and dark represent light intensities of 50, 20 and 0 μmol photons m−2s−1. The whole bar represents autofluorescence intensity of the control, while the twill part of the bar represents algal autofluorescence intensity of the 5% LP-10 treatment. And the gray part of the bar represents difference between the treatments and control, namely the dead cells under each light conditions.
Effects of strain LP-10 on the gene expression of algal cells
Genes psbA and rbcS play important roles in the photosynthesis of plant and algal cells. Gene psbA is responsible for the PSII reaction center protein D1, and gene rbcS codes for the small subunit of Rubisco, an important enzyme for CO2 fixation. Our results demonstrated that the transcriptional expression abundance of psbA was 0.29, 0.73 and 0.64-fold of that of control at 24 h when treated by LP-10 filtrates (Fig. 5). And the transcriptional abundance of rbcS at 24 h shared a similar pattern with the psbA and all three concentrations treatments suppressed the gene expression of rbcS.
Figure 5. The relative transcriptional expression abundance of two genes (psbA and rbcS) in algal cells after incubation with LP-10 filtrates for 24 h.
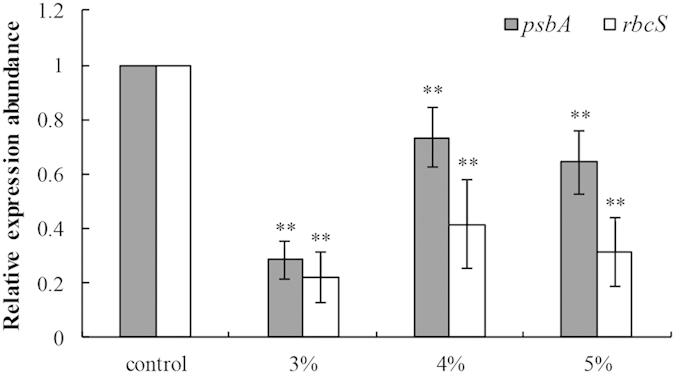
All data represent the means ± S.D. **represents statistical significance at the p < 0.01 level.
Effects of strain LP-10 on the expression of PSII reaction center protein D1
The expression of PSII reaction center protein D1 was detected in this study (Fig. 6). The D1 content of the 3% treatment group was higher than that of the control while the content of the 4 and 5% treatment groups was remarkably decreased. The D1 content of the 3, 4 and 5% treatment groups was 169, 71 and 43% of the control, based on the quantification of band intensities using densitometry with Quantity-One software.
Figure 6. Immunodetection of the PSII reaction center protein D1 after the algal cells were incubated with LP-10 filtrates for 24 h.
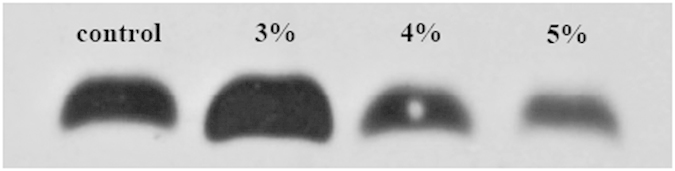
Control represents the algal cells without addition of LP-10; 3, 4, 5% represent the algal cells treated by different concentrations of LP-10 filtrates respectively.
Discussion
Marine disasters bring about huge loses to human every year, and red tides was one of the most notorious calamities. However, ocean also provide huge resources for us as a library which make it possible to utilize marine microorganisms to regulate the HABs. Marine algicidal bacteria are suggested to be excellent candidates in managing HABs8,10. Up to now, many bacteria have been isolated to be effective in removing harmful algae, but the action mechanism of algicidal bacteria on toxic algae has not been well explained. In this study, we investigated the mode of action of a marine algicidal bacterium Bacillus sp. LP-10, which was previously isolated in our laboratory from the surface seawater samples of the East China Sea.
As is known, ROS are common byproducts of aerobic metabolism, predominantly produced in chloroplasts and mitochondria of algal cells19. Normally, antioxidants system of algal cells could remove the superfluous ROS to keep the cells from oxidative damage. However, environmental stresses often induce algal cells an unusual rise of ROS level. Liu et al. found the addition of allelochemical florfenicol significantly enhanced the intracellular ROS content of Skeletonema costatum at 6 h35. Similarly, an obvious increase of ROS was observed in our study when strain LP-10 was inoculated into the algal cultures, which indicated that the algal cells were suffering from oxidative stress. And the MDA content rise also confirmed the oxidative damage in the algal cells.
NAC is a commonly used ROS scavenger, and our results demonstrated that the addition of NAC to the algal cultures significantly improved the survival rate of algal cells compared to algal cells treated by LP-10 filtrates without NAC. And the ROS content determination in ROS scavenger assays hinted that the superfluous ROS was really removed by the ROS scavengers. The tested results of two other antioxidant (GSH and AsA) showed similar consequences and confirmed that the superfluous ROS was reacted with the scavengers rather than surviving the algal cells by other means. Furthermore, the autofluorescence intensity of algal cells in NAC + LP treatment group was approximate to the CK and NAC control groups, suggesting that the ROS was the main reason for algal cell death.
MDA content was an indicator of lipid peroxidation in algal cells. Our results suggested the MDA content were keeping rising during the experiment time, and the rate of lipid peroxidation became more and more serious, implying the antioxidant system of algal cells may not clear the excessive ROS in time and the algal cells suffered from oxidative damage for long. Other reported isolates were also proved to cause the increase of MDA content in algal cells15,36.
Algal cells execute programs to clear the excess ROS via antioxidant systems21. Our results showed that addition of LP-10 enhanced SOD, CAT, POD and GSH activities of algal cells, the activities of these four antioxidants at 72 h were significantly higher than the initial levels, suggesting algal cells were kept under the oxidative stress and superfluous ROS could not be cleared in time to resume the normal status. Excess ROS triggered a series of damage processes in algal cells22, leading the dysfunction of many fundamental organelles and structures. MDA assays also revealed that algal cells were under continuous oxidative stress as discussed above, which will oxidize the lipids of membranes to MDA, thus the membrane permeability changed, and the algal cells finally disrupted for dysfunction of algal cells.
Environmental factors often affected important physiological functions of algal cells23,26. Domingues et al. investigated the response of Phaeodactylum tricornutum to high light exposure, the results showed photosynthetic activities of algal cells declined obviously25. In this study, addition of Bacillus sp. LP-10 filtrates significantly impacted the fundamental functions of algal cells. Among which, photosynthesis systems are vulnerable to oxidative stress. Photosynthesis pigments are key components of photosynthesis systems, which are responsible for light harvesting and photosynthetic reaction37. When LP-10 filtrates was inoculated into the algal cultures, an obvious decrease of Chl a and b contents was observed, and the ratio of these two pigments was significantly changed, suggesting the pigment structures were destroyed, scanning electron microscope (SEM) and transmission electron microscope (TEM) images of algal cells under treatment also confirmed the results (Figs S5, S6). The data of electron microscope indicated the thylakoid of the algal cells became sparse and structure of the chloroplast structures were changed. The unstable ratio of Chl a and b contents may be the consequence of sensitivity difference of these two pigments under strain LP-10 stress. The low ratio in the early phase indicates that photosynthetic antenna complexes are more vulnerable to oxidative stress than light-harvesting complexes17.
The Fv/Fm and maximum rETR are two important parameters used to evaluate the photosynthesis efficiency and capacity38, which were usually disturbed by environmental stresses. Domingues et al. found high light exposure will lead declination of the Fv/Fm of algal cells25. Our results revealed that the value of Fv/Fm significantly decreased when the algal cells were under strain LP-10 stress, suggesting the photosynthetic efficiency was hindered by strain LP-10. Similarly, the rETR was also significantly inhibited by strain LP-10, implying that the photosynthetic capacity of algal cells decreased after treatment with strain LP-10. These results indicated that the photosynthesis systems of algal cells were attacked by the active metabolites produced by Bacillus sp. LP-10. Combined with the pigment assays, we speculated that strain LP-10 would destroy the structures of photosynthetic system and lower the photosynthesis activities, causing the dysfunction of the photosynthetic system at last.
More than that, Bacillus sp. LP-10 also inhibited the transcriptional expression of many important genes. Wu et al. noted that allelochemical pyrogallic acid significantly affects the expression of genes psbA, grpE, fabZ, recA, cmpA, ftsZ and cyrJ39. In our study, two photosynthesis-related genes (psbA and rbcS) were investigated. At 24 h, transcriptional expression of both gene psbA and rbcS were down-regulated. We speculated that the active metabolites produced by strain LP-10 may affected the transcription processes or severe impairment occurred in the genetic molecules of the cells at the time. Gene psbA was responsible for synthesis of protein D1, one of the subunits of the reaction center of PSII. Impairment of protein D1 will decrease the photosynthetic capacity, leading to the photoinhibition of the algal cells24. The gene rbcS code for the small subunit of Rubisco, which plays a critical role in CO2 fixation and photorespiration40. The inhibition of the rbcS in the algal cells implied that the CO2 fixation cycle (Calvin cycle) was interrupted by Bacillus sp. LP-10, which would accelerate the photoinhibition of the algal cells41, because limitation of CO2 fixation will decrease the absorption capacity for solar energy of the algal cells and affect the electron transportation of photosystems. The excess energy causes the production of numerous ROS at last, which inhibits the synthesis of protein D123.
Our study also found that light was an indispensable factor in algal cell death. The algicidal activity of strain LP-10 under low light was very low (~15%) while normal light led to a higher algicidal activity (~80%), which meant the light intensity of “normal light” was too high for the stressed algal cells and the stressed algal cells could not utilize the “normal light”, so that the relative high light leads to photoinhibition of the algal cells, which indirectly revealed that strain LP-10 impacted the CO2 fixation and the utilization of solar energy. The results implied that dysfunction of the photosynthesis system (photoinhibition) was the most fundamental reason for algal cell death. Qian et al. and Yang et al. reported that the allelochemicals N-phenyl-2-naphthylamine and hydroquinone could cause photoinhibition of Chlorella vulgaris and Phaeodactylum tricornutum17,18. Though some other reasons were able to cause the algal cell death, such as destroying the cell structures, our results demonstrated the role of other factors were minor reasons. Because when the algal cultures were placed in the dark or under low light conditions, the algicidal rate was very low (~15%).
The western blot analysis of D1 protein showed that the D1 protein content of algal cells at 4 and 5% treatment concentrations of strain LP-10 decreased obviously at 24 h. This results were consistent with the gene expression profiles of psbA, which confirmed that photoinhibition was taken place in the algal cells. Photoinhibition of algal cells indicated the photosynthesis would be hampered, and energy-consuming functions would be inhibited. Long-time photohibition would be unable to support the survival of the algal cells, leading to the algal cells death eventually.
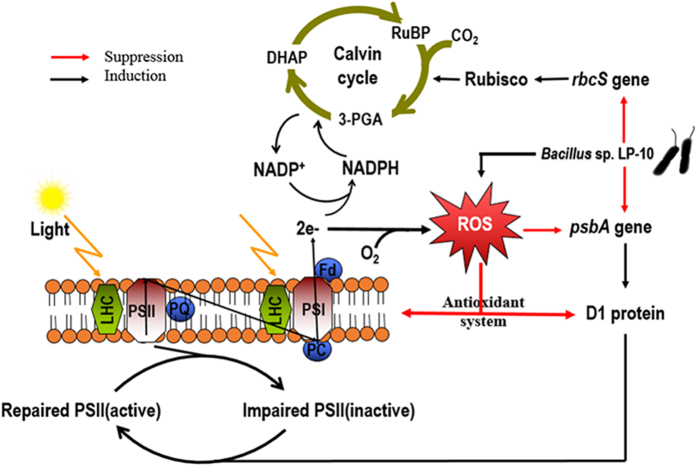
From the above, we conclude that algicidal bacterium Bacillus sp. LP-10 induce a significant increase of ROS in algal cells, and continuous stressing make the superfluous ROS not being cleared in time, which disturb the physiological functions and destruct the structures of algal cells. More important, excess ROS lead to photoinhibiton of algal cells by influencing the expression of the photosynthesis-related genes and protein, and impaired photosystem apparatus cannot be repaired, leading to the dysfunction and causing disruption of algal cells eventually (Fig. 7).
Figure 7. A hypothetical scheme for the algicidal mechanism of the algal cells upon Bacillus sp. LP-10.
Additional Information
How to cite this article: Guan, C. et al. Photoinhibition of Phaeocystis globosa resulting from oxidative stress induced by a marine algicidal bacterium Bacillus sp. LP-10. Sci. Rep. 5, 17002; doi: 10.1038/srep17002 (2015).
Supplementary Material
Acknowledgments
This work was financially supported by the Joint Project of the National Natural Science Foundation of China and Shandong Province: Marine ecology and environmental sciences (U1406403), the National Natural Science Foundation of China (41376119, 41576109, 40930847), Mr. An Tao is thanked for his help with the western blot techniques. We would like to thank Prof. I. J. Hodgkiss from The University of Hong Kong for help with English.
Footnotes
Author Contributions Conceived and designed the experiments: C.G., X.G., Z.Y. and T.Z. Performed the experiments: C.G.,X.G., Y.L., H.Z., X.L., G.C., J.G. and Z.Y. Analyzed the data: C.G., X.G. and T.Z. Wrote the paper: C.G., X.G. and T.Z.
References
- Landsberg J. H. The effects of harmful algal blooms on aquatic organisms. Reviews in Fisheries Science 10, 113–390 (2002). [Google Scholar]
- Hoagland P. & Scatasta S. Ecology of harmful algae. 391–402 (Springer, 2006). [Google Scholar]
- Van Dolah F. M. Marine algal toxins: origins, health effects, and their increased occurrence. Environmental health perspectives 108, 133 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. M. Turning back the harmful red tide. Nature 388, 513–514 (1997). [Google Scholar]
- Zingone A. & Oksfeldt Enevoldsen H. The diversity of harmful algal blooms: a challenge for science and management. Ocean & Coastal Management 43, 725–748 (2000). [Google Scholar]
- Sigee D. et al. The Ecological Bases for Lake and Reservoir Management. 161–172 (Springer, 1999). [Google Scholar]
- Gumbo R. J., Ross G. & Cloete E. T. Biological control of Microcystis dominated harmful algal blooms. African Journal of Biotechnology 7, 4765–4773 (2008). [Google Scholar]
- Cole J. J. Interactions between bacteria and algae in aquatic ecosystems. Annual Review of Ecology and Systematics 13, 291–314 (1982). [Google Scholar]
- Doucette G. J. Interactions between bacteria and harmful algae: a review. Natural toxins 3, 65–74 (1995). [DOI] [PubMed] [Google Scholar]
- Mayali X. & Azam F. Algicidal bacteria in the sea and their impact on algal blooms. Journal of Eukaryotic Microbiology 51, 139–144 (2004). [DOI] [PubMed] [Google Scholar]
- Ma H. et al. Mode of action of membrane-disruptive lytic compounds from the marine dinoflagellate Alexandrium tamarense. Toxicon 58, 247–258 (2011). [DOI] [PubMed] [Google Scholar]
- Lee S.O. et al. Involvement of an Extracellular Protease in Algicidal Activity of the Marine Bacterium Pseudoalteromonas sp. Strain A28. Applied and Environmental Microbiology 66, 4334–4339 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince E. K., Poulson K. L., Myers T. L., Sieg R. D. & Kubanek J. Characterization of allelopathic compounds from the red tide dinoflagellate Karenia brevis. Harmful Algae 10, 39–48 (2010). [Google Scholar]
- Kim D. et al. Red to red—the marine bacterium Hahella chejuensis and its product prodigiosin for mitigation of harmful algal blooms. Journal of microbiology and biotechnology 18, 1621–1629 (2008). [PubMed] [Google Scholar]
- Zhang H. et al. Effect of Oxidative Stress Induced by Brevibacterium sp. BS01 on a HAB Causing Species-Alexandrium tamarense. PloS one 8, e63018, doi: 10.1371/journal.pone.0063018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. Towards molecular, physiological, and biochemical understanding of photosynthetic inhibition and oxidative stress in the toxic Alexandrium tamarense induced by a marine bacterium. Applied microbiology and biotechnology 98, 4637–4652 (2014). [DOI] [PubMed] [Google Scholar]
- Qian H. et al. Allelochemical stress causes oxidative damage and inhibition of photosynthesis in Chlorella vulgaris. Chemosphere 75, 368–375 (2009). [DOI] [PubMed] [Google Scholar]
- Yang C. et al. Allelochemical induces growth and photosynthesis inhibition, oxidative damage in marine diatom Phaeodactylum tricornutum. Journal of Experimental Marine Biology and Ecology 444, 16–23 (2013). [Google Scholar]
- Apel K. & Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399 (2004). [DOI] [PubMed] [Google Scholar]
- Atkinson N. J. & Urwin P. E. The interaction of plant biotic and abiotic stresses: from genes to the field. Journal of experimental botany 63, 3523–3543 (2012). [DOI] [PubMed] [Google Scholar]
- Yu B. P. Cellular defenses against damage from reactive oxygen species. Physiological reviews 74, 139 (1994). [DOI] [PubMed] [Google Scholar]
- Avery Simon V. Molecular targets of oxidative stress. Biochemical Journal 434, 201–210 (2011). [DOI] [PubMed] [Google Scholar]
- Takahashi S. & Murata N. How do environmental stresses accelerate photoinhibition. Trends in Plant Science 13, 178–182 (2008). [DOI] [PubMed] [Google Scholar]
- Takahashi S. & Badger M. R. Photoprotection in plants: a new light on photosystem II damage. Trends in Plant Science 16, 53–60 (2011). [DOI] [PubMed] [Google Scholar]
- Domingues N., Matos A. R., Marques da Silva J. & Cartaxana P. Response of the Diatom Phaeodactylum tricornutum to Photooxidative Stress Resulting from High Light Exposure. PLoS One 7, e38162, doi: 10.1371/journal.pone.0038162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata N., Takahashi S., Nishiyama Y. & Allakhverdiev S. Photoinhibition of photosystem II under environmental stress. Biochimica et Biophysica Acta (BBA) - Bioenergetics 1767, 414–421 (2007). [DOI] [PubMed] [Google Scholar]
- Schoemann V., Becquevort S., Stefels J., Rousseau V. & Lancelot C. Phaeocystis blooms in the global ocean and their controlling mechanisms: a review. Journal of Sea Research 53, 43–66 (2005). [Google Scholar]
- Guan C. et al. Novel algicidal evidence of a bacterium Bacillus sp. LP-10 killing Phaeocystis globosa, a harmful algal bloom causing species. Biological Control 76, 79–86 (2014). [Google Scholar]
- Guillard R. R. In Culture of marine invertebrate animals 29-60 (Springer, 1975). [Google Scholar]
- Schut F. et al. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Applied and environmental microbiology 59, 2150–2160 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBel C. P., Ischiropoulos H. & Bondy S. C. Evaluation of the probe 2′, 7′-dichlorofluorescin as an indicator of reactive oxygen species formation and oxidative stress. Chemical research in toxicology 5, 227–231 (1992). [DOI] [PubMed] [Google Scholar]
- Marr I. L., Suryana N., Lukulay P. & Marr M. I. Determination of chlorophyll a and b by simultaneous multi-component spectrophotometry. Fresenius’ journal of analytical chemistry 352, 456–460 (1995). [Google Scholar]
- Grebe M. et al. An Integrated Analysis of Molecular Acclimation to High Light in the Marine Diatom Phaeodactylum tricornutum. PLoS One 4, e7743 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Liu W., Ming Y., Huang Z. & Li P. Impacts of florfenicol on marine diatom Skeletonema costatum through photosynthesis inhibition and oxidative damages. Plant Physiology and Biochemistry 60, 165–170 (2012). [DOI] [PubMed] [Google Scholar]
- Kong Y., Xu X. & Zhu L. Cyanobactericidal Effect of Streptomyces sp. HJC-D1 on Microcystis auruginosa. PLoS One 8, e57654, doi: 10.1371/journal.pone.0057654 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan K. S. Photosynthetic pigments of algae (CUP Archive, 1989). [Google Scholar]
- Schreiber U., Bilger W. & Neubauer C. In Ecophysiology of photosynthesis 49–70 (Springer, 1995). [Google Scholar]
- Wu Z., Shi J. & Yang S. The effect of pyrogallic acid on growth, oxidative stress, and gene expression in Cylindrospermopsis raciborskii (Cyanobacteria). Ecotoxicology 22, 271–278 (2012). [DOI] [PubMed] [Google Scholar]
- Spreitzer R. J. & Salvucci M. E. Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annual review of plant biology 53, 449–475 (2002). [DOI] [PubMed] [Google Scholar]
- Takahashi S. & Murata N. Interruption of the Calvin cycle inhibits the repair of Photosystem II from photodamage. Biochim Biophys Acta 1708, 352–61 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.