Abstract
The Australian Drosophila Ecology and Evolution Resource (ADEER) collates Australian datasets on drosophilid flies, which are aimed at investigating questions around climate adaptation, species distribution limits and population genetics. Australian drosophilid species are diverse in climatic tolerance, geographic distribution and behaviour. Many species are restricted to the tropics, a few are temperate specialists, and some have broad distributions across climatic regions. Whereas some species show adaptability to climate changes through genetic and plastic changes, other species have limited adaptive capacity. This knowledge has been used to identify traits and genetic polymorphisms involved in climate change adaptation and build predictive models of responses to climate change. ADEER brings together 103 datasets from 39 studies published between 1982–2013 in a single online resource. All datasets can be downloaded freely in full, along with maps and other visualisations. These historical datasets are preserved for future studies, which will be especially useful for assessing climate-related changes over time.
Subject terms: Experimental evolution, Population genetics, Climate-change ecology, Evolutionary biology, Evolutionary genetics
Background & Summary
The Australian Drosophila Ecology and Evolution Resource (ADEER) contains three main Drosophila data collections: (1) clinal data, (2) species distribution data and (3) genomics data. The clinal and species distribution collections are described in this data descriptor, whereas the genomics data will be described elsewhere. The majority of data was generated by Ary Hoffmann’s research group at the University of Melbourne, with contributions from several other Australian researchers (see Acknowledgements).
Drosophila species have long been used as model organisms to answer fundamental questions in biology, and the most intensively studied Drosophila species (in particular Drosophila melanogaster) are Northern Hemisphere in origin1. Drosophilids as a broader taxonomic group are very diverse in Australia, with over 300 species identified in the tropical and temperate forests located on the east coast. Australia contains a disproportionately large number of species in the genus Scaptodrosophila, many of them endemic to this continent2. The ADEER collection broadens the scope of worldwide drosophilid data, by focussing on clinal patterns in traits and genes in Australian drosophilids as well as on thoroughly-studied species distributions. This collection contains data on the ecology and evolution of eleven species in the genera Drosophila and Scaptodrosophila including rainforest specialists (e.g. D. birchii), endemic species (e.g. D. bunnanda) and cosmopolitan species (e.g. D. melanogaster).
The east coast of Australia spans a gradient of climatic conditions from cool-temperate Tasmania to tropical northern Queensland. This gradient is unique as it occurs within a narrow elevation range and on a small continent with an ancient geology and a rich biodiversity with a high proportion of endemic species across several biomes3. The gradient provides a model system for studying equivalent climatic gradients on other continents, and represents an outstanding natural laboratory for the study of traits and genes that are associated with climatic adaptation4. Changes in traits and genes along this gradient (i.e. clines) can arise by natural selection, producing continuous patterns over geographic space. The eastern Australian gradient has been used to investigate the involvement of numerous phenotypic traits and genetic markers in climate adaptation4. The clinal data collection contains data from eight species from studies published between 1982–2013 and includes morphological, life-history, stress resistance traits as well as genetic markers. Most of these studies used common garden experiments to test for clinal variation, but some studies were performed in the field.
The species distribution collection includes five species from the melanogaster species group (montium subgroup) and two species from the repleta species group within the genus Drosophila, as well as two Scaptodrosophila species. These datasets contain presence records from field collections between 1924 and 2013 which are based on previously-published records in the literature, collections made by the dataset authors, and specimens in the Australian Museum5–7. Many species are restricted to the tropics, a few are temperate specialists, and some are broadly distributed across climatic regions. The varied distributions of drosophilid species along the temperate–tropical cline provide a powerful tool for studying climate adaptation and species distribution limits.
Previous work on Drosophila species using the Australian cline has demonstrated that monitoring biological changes along geographic climate gradients is a powerful approach for detecting evolutionary shifts under climate change8,9. Ongoing data collections from the eastern Australia cline provide an opportunity to monitor phenotypic traits and genetic markers by comparison to historical data, as climate change proceeds. Such temporal studies are particularly useful for tracking continuing evolutionary responses to climate change as well as dynamically projecting species distributions under ongoing climate change scenarios.
Methods
Clinal data collection
The clinal data collection contains data on morphological, life-history and stress resistance traits, as well as genetic marker data (Tables 1 and 2 (available online only), Fig. 1). All datasets of this kind involve flies collected at multiple locations within their geographic distribution, usually along a north-south gradient on the east coast of Australia. Material and methods for each dataset appear in detail in the original publication; here we provide a general summary of the approaches used.
Table 1. Overview of all datasets included in ADEER.
| Data file name | Collection | Species | Trait | Trait group | Data type | Publication | DOI / PMID for original publication | Data record accession on ADEER | DOI for ADEER | Data repository | DOI for Dryad |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Collection, species and traits studied, data type, the original publication, data depository, data record accession, DOI for ADEER and Dryad, and DOI (or PMID) for the original publication are listed for each dataset. | |||||||||||
| 1 Azevedo et al. 20 Egg size & ovariole number | Clinal | Drosophila melanogaster | Ovariole number and egg size | Life history and morphology | Population means | Azevedo et al. 20 | 10.2307/2410702 | http://adeer.pearg.com/biogs/DR00273b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 2 Azevedo et al. 11 Wing traits | Clinal | Drosophila melanogaster | Wing to aspect ratio | Morphology | Population means | Azevedo et al. 11 | 10.2307/2410702 | http://adeer.pearg.com/biogs/DR00277b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 3 Gockel et al. Microsatellite markers | Clinal | Drosophila melanogaster | Microsatellite markers | Genetic markers | Individual categories | Gockel et al. 22 | 11333239 | http://adeer.pearg.com/biogs/DR00279b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 4 Gockel et al. 22 Wing area | Clinal | Drosophila melanogaster | Wing area | Morphology | Population means | Gockel et al. 22 | 11333239 | http://adeer.pearg.com/biogs/DR00280b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 5 Griffiths et al. 23 Development time | Clinal | Drosophila birchii | Development time | Life history | Population means | Griffiths et al. 23 | 10.1111/j.1420-9101.2004.00782.x | http://adeer.pearg.com/biogs/DR00283b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 6 Griffiths et al. 23 Stress resistance & wing size | Clinal | Drosophila birchii | Cold-, desiccation-, heat-, starvation resistance and wing centroid size | Stress resistance and morphology | Population means | Griffiths et al. 23 | 10.1111/j.1420-9101.2004.00782.x | http://adeer.pearg.com/biogs/DR00282b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 7 Hallas et al. 19 Stress resistance and size | Clinal | Drosophila serrata | Cold-, desiccation- and starvation resistance, mass and wing length | Stress resistance and morphology | Population means | Hallas et al. 19 | 10.1017/S0016672301005523 | http://adeer.pearg.com/biogs/DR00315b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 8 Hoffmann & Shirriffs 24 Wing traits | Clinal | Drosophila serrata | Wing landmarks | Morphology | Individual measurements | Hoffmann & Shirriffs 24 | 10.1111/j.0014-3820.2002.tb01418.x | http://adeer.pearg.com/biogs/DR00257b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 9 Hoffmann et al. 10 Desiccation resistance | Clinal | Drosophila melanogaster | Desiccation resistance | Stress resistance | Individual measurements | Hoffmann et al. 10 | 10.1111/j.0014-3820.2001.tb00681.x | http://adeer.pearg.com/biogs/DR00259b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 10 Hoffmann et al. 10 Starvation resistance | Clinal | Drosophila melanogaster | Starvation resistance | Stress resistance | Individual measurements | Hoffmann et al. 10 | 10.1111/j.0014-3820.2001.tb00681.x | http://adeer.pearg.com/biogs/DR00261b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 11 Hoffmann et al. 10 Line means | Clinal | Drosophila melanogaster | Cold resistance, desiccation resistance, starvation resistance, lipid content and thorax length | Stress resistance and morphology | Subgroup means | Hoffmann et al. 10 | 10.1111/j.0014-3820.2001.tb00681.x | http://adeer.pearg.com/biogs/DR00260b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 12 Hoffmann et al. 31 Cold recovery time | Clinal | Drosophila melanogaster | Cold recovery time | Stress resistance | Individual measurements | Hoffmann et al. 31 | 10.1046/j.1461-0248.2002.00367.x | http://adeer.pearg.com/biogs/DR00223b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 13 Hoffmann et al. 31 Cold resistance survival | Clinal | Drosophila melanogaster | Cold resistance survival | Stress resistance | Individual measurements | Hoffmann et al. 31 | 10.1046/j.1461-0248.2002.00367.x | http://adeer.pearg.com/biogs/DR00224b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 14 Hoffmann et al. 31 Heat knockdown time | Clinal | Drosophila melanogaster | Heat knockdown time | Stress resistance | Individual measurements | Hoffmann et al. 31 | 10.1046/j.1461-0248.2002.00367.x | http://adeer.pearg.com/biogs/DR00225b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 15 Hoffmann et al. 14 Overwinter fecundity | Clinal | Drosophila melanogaster | Overwinter mortality, overwinter fecundity | Life history | Population means | Hoffmann et al. 14 | 10.1046/j.1420-9101.2003.00561.x | http://adeer.pearg.com/biogs/DR00263b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 16 Hoffmann et al. 38 Frost locusA | Clinal | Drosophila melanogaster | Frost locus | Genetic markers | Population frequencies | Hoffmann et al. 38 | 10.1111/j.1365-2583.2012.01149.x | http://adeer.pearg.com/biogs/DR00265b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 17 Hoffmann et al. 38 Frost locusB | Clinal | Drosophila melanogaster | Frost locus | Genetic markers | Population frequencies | Hoffmann et al. 38 | 10.1111/j.1365-2583.2012.01149.x | http://adeer.pearg.com/biogs/DR00266b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 18 James & Partridge 26 Development time | Clinal | Drosophila melanogaster | Development time | Life history | Population means | James & Partridge 26 | 10.1046/j.1420-9101.1995.8030315.x | http://adeer.pearg.com/biogs/DR00286b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 19 James & Partridge 26 Time to pupation | Clinal | Drosophila melanogaster | Time to pupation | Life history | Population means | James & Partridge 26 | 10.1046/j.1420-9101.1995.8030315.x | http://adeer.pearg.com/biogs/DR00285b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 20 James et al. 17 Thorax length & wing traits | Clinal | Drosophila melanogaster | Thorax length and wing traits | Morphology | Population means | James et al. 17 | 7498744 | http://adeer.pearg.com/biogs/DR00288b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 21 Kennington & Hoffmann 34 Molecular markers & In(2L)t inversion | Clinal | Drosophila melanogaster | Microsatellite markers, Alcohol dehydrogenase (Adh) locus and in(2L)t inversion | Genetic markers | Individual categories | Kennington & Hoffmann 34 | 10.1186/1471-2148-13-100 | http://adeer.pearg.com/biogs/DR00290b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 22 Kennington et al. 41 Molecular markers & In(3R)Payne inversion | Clinal | Drosophila melanogaster | Microsatellite markers, Hsr-omega locus and In(3R)Payne inversion | Genetic markers | Individual measurements | Kennington et al. | 10.1534/genetics.105.053173 | http://adeer.pearg.com/biogs/DR00292b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 23 Knibb et al. 46 Inversion frequencies | Clinal | Drosophila melanogaster | Inversions | Genetic markers | Population frequencies | Knibb et al. 46 | 17249108 | http://adeer.pearg.com/biogs/DR00296b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 24 Kriesner et al. 43 Wolbachia infection frequencies & mtDNA haplotypes | Clinal | Drosophila simulans | mtDNA haplotype and wolbachia infection | Genetic markers and endosymbionts | Population counts | Kriesner et al. 43 | 10.1371/journal.ppat.1003607 | http://adeer.pearg.com/biogs/DR00065b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 25 Lee et al. 42 Neurofibromin gene & In(3R)Payne inversion | Clinal | Drosophila melanogaster | Neurofibromin (Nf1) locus and In(3R)Payne inversion | Genetic markers | Individual categories | Lee et al. 42 | 10.1111/mec.12301 | http://adeer.pearg.com/biogs/DR00247b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 26 Mitrovski & Hoffmann 13 Mean overwinter egg counts and longevity | Clinal | Drosophila melanogaster | Overwinter longevity and fecundity | Life history | Subgroup means | Mitrovski & Hoffmann 13 | 10.1098/rspb.2001.1787 | http://adeer.pearg.com/biogs/DR00229b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 27 Mitrovski & Hoffmann 13 Overwinter temperature, egg laying and mortality rates | Clinal | Drosophila melanogaster | Overwinter mortality and fecundity | Life history | Subgroup means | Mitrovski & Hoffmann 13 | 10.1098/rspb.2001.1787 | http://adeer.pearg.com/biogs/DR00228b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 28 Mitrovski & Hoffmann 13 Overwinter raw egg counts | Clinal | Drosophila melanogaster | Overwinter egg count | Life history | Subgroup measurements | Mitrovski & Hoffmann 13 | 10.1098/rspb.2001.1787 | http://adeer.pearg.com/biogs/DR00233b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 29 Mitrovski & Hoffmann 13 Overwinter raw mortality | Clinal | Drosophila melanogaster | Overwinter mortality | Life history | Subgroup measurements | Mitrovski & Hoffmann 13 | 10.1098/rspb.2001.1787 | http://adeer.pearg.com/biogs/DR00232b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 30 Oakeshott et al. 36 Adh & Gpdh loci | Clinal | Drosophila melanogaster | Alcohol dehydrogenase (Adh) and Glycerol-3-phosphate dehydrogenase (Gpdh) | Genetic markers | Population frequencies | Oakeshott et al. 36 | 10.2307/2407970 | http://adeer.pearg.com/biogs/DR00298b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 31 Oakeshott et al. 35 G6pd locus | Clinal | Drosophila melanogaster | Glucose-6-phosphate dehydrogenase (G6pd) locus | Genetic markers | Population frequencies | Oakeshott et al. 35 | 10.1038/hdy.1983.7 | http://adeer.pearg.com/biogs/DR00303b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 32 Oakeshott et al. 35 Pgd locus | Clinal | Drosophila melanogaster | 6-phosphogluconate dehydrogenase (Pgd) locus | Genetic markers | Population frequencies | Oakeshott et al. 35 | 10.1038/hdy.1983.7 | http://adeer.pearg.com/biogs/DR00304b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 33 Rako et al. 15 Post winter male fertility 2006 | Clinal | Drosophila melanogaster | Post winter male fertility | Life history | Individual measurements | Rako et al. 15 | 10.1111/j.1420-9101.2009.01852.x | http://adeer.pearg.com/biogs/DR00237b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 34 Rako et al. 15 Post winter male fertility 2008 | Clinal | Drosophila melanogaster | Post winter male fertility | Life history | Individual measurements | Rako et al. 15 | 10.1111/j.1420-9101.2009.01852.x | http://adeer.pearg.com/biogs/DR00308b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 35 Rako et al. 15 Post winter male size 2006 | Clinal | Drosophila melanogaster | Post winter wing centroid size | Morphology | Individual measurements | Rako et al. 15 | 10.1111/j.1420-9101.2009.01852.x | http://adeer.pearg.com/biogs/DR00307b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 36 Rako et al. 15 Male size 2008 | Clinal | Drosophila melanogaster | Thorax length and wing centroid size | Morphology | Individual measurements | Rako et al. 15 | 10.1111/j.1420-9101.2009.01852.x | http://adeer.pearg.com/biogs/DR00309b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 37 Telonis Scott et al. 25 Raw pcr data | Clinal | Drosophila melanogaster | Ebony expression | Genetic markers | Subgroup measurements | Telonis-Scott et al. 25 | 10.1111/j.1365-294X.2011.05089.x | http://adeer.pearg.com/biogs/DR00069b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 38 Telonis Scott et al. 25 Raw pigmentation data | Clinal | Drosophila melanogaster | Thoracic trident pigmentation scores | Morphology | Individual measurements | Telonis-Scott et al. 25 | 10.1111/j.1365-294X.2011.05089.x | http://adeer.pearg.com/biogs/DR00068b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 39 Telonis Scott et al. 25 Average pcr pigmentation | Clinal | Drosophila melanogaster | Ebony expression and Thoracic trident pigmentation scores | Genetic markers | Population means | Telonis-Scott et al. 25 | 10.1111/j.1365-294X.2011.05089.x | http://adeer.pearg.com/biogs/DR00070b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 40 Umina et al. 8 Adh locus | Clinal | Drosophila melanogaster | Alcohol dehydrogenase (Adh) locus | Genetic markers | Population frequencies | Umina et al. 8 | 10.1126/science.1109523 | http://adeer.pearg.com/biogs/DR00235b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 41 Umina et al. 8 In(3R)Payne inversion | Clinal | Drosophila melanogaster | In(3R)Payne inversion | Genetic markers | Population frequencies | Umina et al. 8 | 10.1126/science.1109523 | http://adeer.pearg.com/biogs/DR00236b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 42 Van Heerwaarden & Sgro 16 D.melanogaster thorax length | Clinal | Drosophila melanogaster | Thorax length | Morphology | Individual measurements | Van Heerwaarden & Sgro 16 | 10.1111/j.1558-5646.2010.01196.x | http://adeer.pearg.com/biogs/DR00240b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 43 Van Heerwaarden & Sgro 16 D.melanogaster wing centroid size | Clinal | Drosophila melanogaster | Wing centroid size | Morphology | Individual measurements | Van Heerwaarden & Sgro 16 | 10.1111/j.1558-5646.2010.01196.x | http://adeer.pearg.com/biogs/DR00239b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 44 Van Heerwaarden & Sgro 16 D.melanogaster wing thorax ratio | Clinal | Drosophila melanogaster | Wing to thorax ratio | Morphology | Individual measurements | Van Heerwaarden & Sgro 16 | 10.1111/j.1558-5646.2010.01196.x | http://adeer.pearg.com/biogs/DR00241b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 45 Van Heerwaarden & Sgro 16 D.simulans thorax length | Clinal | Drosophila simulans | Thorax length | Morphology | Individual measurements | Van Heerwaarden & Sgro 16 | 10.1111/j.1558-5646.2010.01196.x | http://adeer.pearg.com/biogs/DR00243b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 46 Van Heerwaarden & Sgro 16 D.simulans wing centroid size | Clinal | Drosophila simulans | Wing centroid size | Morphology | Individual measurements | Van Heerwaarden & Sgro 16 | 10.1111/j.1558-5646.2010.01196.x | http://adeer.pearg.com/biogs/DR00242b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 47 Van Heerwaarden & Sgro 16 D.simulans wing thorax ratio | Clinal | Drosophila simulans | Wing to thorax ratio | Morphology | Individual measurements | Van Heerwaarden & Sgro 16 | 10.1111/j.1558-5646.2010.01196.x | http://adeer.pearg.com/biogs/DR00244b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 48 Weeks et al. 2005 Clock locus | Clinal | Drosophila melanogaster | Clock locus | Genetic markers | Population frequencies | Weeks et al. 2005 | 10.1111/j.1420-9101.2005.01013.x | http://adeer.pearg.com/biogs/DR00313b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 49 Weeks et al. 2005 Period locus | Clinal | Drosophila melanogaster | Period locus | Genetic markers | Population frequencies | Weeks et al. 2005 | 10.1111/j.1420-9101.2005.01013.x | http://adeer.pearg.com/biogs/DR00311b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 50 Weeks et al. 2005 ThrGly locus | Clinal | Drosophila melanogaster | Clock locus | Genetic markers | Population means | Weeks et al. 2005 | 10.1111/j.1420-9101.2005.01013.x | http://adeer.pearg.com/biogs/DR00312b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 51 Collinge et al. Cold tolerance | Clinal | Drosophila melanogaster | Cold recovery time | Stress resistance | Individual measurements | Collinge et al. 21 | 10.1111/j.1420-9101.2005.01016.x | http://adeer.pearg.com/biogs/DR00320b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 52 Collinge et al. 21 Heat tolerance | Clinal | Drosophila melanogaster | Heat knockdown time | Stress resistance | Individual measurements | Collinge et al. 21 | 10.1111/j.1420-9101.2005.01016.x | http://adeer.pearg.com/biogs/DR00321b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 53 Collinge et al. 21 Ovariole number | Clinal | Drosophila melanogaster | Ovariole number | Life history | Individual measurements | Collinge et al. 21 | 10.1111/j.1420-9101.2005.01016.x | http://adeer.pearg.com/biogs/DR00322b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 54 Collinge et al. 21 Development time | Clinal | Drosophila melanogaster | Development time | Life history | Subgroup means | Collinge et al. 21 | 10.1111/j.1420-9101.2005.01016.x | http://adeer.pearg.com/biogs/DR00323b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 55 Collinge et al. 21 Wing area | Clinal | Drosophila melanogaster | Wing area | Morphology | Individual measurements | Collinge et al. 21 | 10.1111/j.1420-9101.2005.01016.x | http://adeer.pearg.com/biogs/DR00324b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 56 Collinge et al. 21 Egg viability | Clinal | Drosophila melanogaster | Egg viability | Life history | Subgroup measurements | Collinge et al. 21 | 10.1111/j.1420-9101.2005.01016.x | http://adeer.pearg.com/biogs/DR00325b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 57 Collinge et al. 21 Genetic markers summary | Clinal | Drosophila melanogaster | AC008193, DMTRXIII, DMU25686, Hsp70, Hsr-omega | Genetic markers | Population frequencies | Collinge et al. 21 | 10.1111/j.1420-9101.2005.01016.x | http://adeer.pearg.com/biogs/DR00326b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 58 Collinge et al. 21 Hsp70 locus | Clinal | Drosophila melanogaster | Hsp70 locus | Genetic markers | Population frequencies | Collinge et al. 21 | 10.1111/j.1420-9101.2005.01016.x | http://adeer.pearg.com/biogs/DR00327b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 59 Collinge et al. 21 DMTRXIII locus | Clinal | Drosophila melanogaster | DMTRXIII locus | Genetic markers | Population frequencies | Collinge et al. 21 | 10.1111/j.1420-9101.2005.01016.x | http://adeer.pearg.com/biogs/DR00328b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 60 Collinge et al. 21 Hsr-omega locus | Clinal | Drosophila melanogaster | Hsr-omega locus | Genetic markers | Population frequencies | Collinge et al. 21 | 10.1111/j.1420-9101.2005.01016.x | http://adeer.pearg.com/biogs/DR00329b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 61 Collinge et al. 21 DMU25686 locus | Clinal | Drosophila melanogaster | DMU25686 locus | Genetic markers | Population frequencies | Collinge et al. 21 | 10.1111/j.1420-9101.2005.01016.x | http://adeer.pearg.com/biogs/DR00330b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 62 Collinge et al. 21 AC008193 locus | Clinal | Drosophila melanogaster | AC008193 locus | Genetic markers | Population frequencies | Collinge et al. 21 | 10.1111/j.1420-9101.2005.01016.x | http://adeer.pearg.com/biogs/DR00331b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 63 McKechnie et al. 39 Dca MCA | Clinal | Drosophila melanogaster | Drosophila cold acclimation (Dca) locus | Genetic markers | Population frequencies | McKechnie et al. 39 | 10.1111/j.1365-294X.2009.04509.x | http://adeer.pearg.com/biogs/DR00333b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 64 McKechnie et al. 39 Dca locus | Clinal | Drosophila melanogaster | Drosophila cold acclimation (Dca) locus | Genetic markers | Population counts | McKechnie et al. 39 | 10.1111/j.1365-294X.2009.04509.x | http://adeer.pearg.com/biogs/DR00334b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 65 Lee et al. 29 Egg stage 2008 | Clinal | Drosophila melanogaster | Egg stage | Life history | Individual measurements | Lee et al. 29 | 10.1111/j.1365-294X.2009.04509.x | http://adeer.pearg.com/biogs/DR00335b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 66 Lee et al. 29 Egg stage 2009 | Clinal | Drosophila melanogaster | Egg stage | Life history | Individual measurements | Lee et al. 29 | 10.1111/j.1365-294X.2009.04509.x | http://adeer.pearg.com/biogs/DR00336b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 67 Lee et al. 29 Egg stage 2010 | Clinal | Drosophila melanogaster | Egg stage | Life history | Individual measurements | Lee et al. 29 | 10.1111/j.1365-294X.2009.04509.x | http://adeer.pearg.com/biogs/DR00337b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 68 Lee et al. 29 Association analysis | Clinal | Drosophila melanogaster | Egg stage and Couch potato (Cpo) locus | Life history and genetic markers | Individual measurements | Lee et al. 29 | 10.1111/j.1365-294X.2009.04509.x | http://adeer.pearg.com/biogs/DR00338b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 69 Lee et al. 29 Couch potato locus | Clinal | Drosophila melanogaster | Couch potato (Cpo) locus | Genetic markers | Population frequencies | Lee et al. 29 | 10.1111/j.1365-294X.2009.04509.x | http://adeer.pearg.com/biogs/DR00339b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 70 Lee et al. 29 Couch potato expression | Clinal | Drosophila melanogaster | Couch potato (Cpo) locus expression | Genetic markers | Population means | Lee et al. 29 | 10.1111/j.1365-294X.2009.04509.x | http://adeer.pearg.com/biogs/DR00340b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 71 James et al. 12 Thorax length & wing traits | Clinal | Drosophila melanogaster | Thorax length, wing area, wing cell area and wing cell number | Morphology | Population means | James et al. 12 | 9215894 | http://adeer.pearg.com/biogs/DR00344b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 72 Magiafoglou et al. 27 Development time | Clinal | Drosophila serrata | Development time | Life history | Population means | Magiafoglou et al. 27 | 10.1046/j.1420-9101.2002.00439.x | http://adeer.pearg.com/biogs/DR00347b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 73 Magiafoglou et al. 27 Viability and cold resistance | Clinal | Drosophila serrata | Cold resistance and viability | Stress resistance and life history | Population means | Magiafoglou et al. 27 | 10.1046/j.1420-9101.2002.00439.x | http://adeer.pearg.com/biogs/DR00348b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 74 Sgrò et al. 30 Longevity | Clinal | Drosophila melanogaster | Longevity | Life history | Subgroup means | Sgrò et al. 30 | 10.1111/mec.12353 | http://adeer.pearg.com/biogs/DR00255b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 75 Sgrò et al. 30 Methuselah expression | Clinal | Drosophila melanogaster | Methuselah (mth) locus expression | Genetic markers | Subgroup measurements | Sgrò et al. 30 | 10.1111/mec.12353 | http://adeer.pearg.com/biogs/DR00253b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 76 Sgrò et al. 30 Methuselah locus | Clinal | Drosophila melanogaster | Methuselah (mth) locus | Genetic markers | Individual categories | Sgrò et al. 30 | 10.1111/mec.12353 | http://adeer.pearg.com/biogs/DR00254b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 77 Arthur et al. 28 Cold resistance | Clinal | Drosophila simulans | Cold recovery time | Stress resistance | Individual measurements | Arthur et al. 28 | 10.1111/j.1420-9101.2008.01617.x | http://adeer.pearg.com/biogs/DR00355b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 78 Arthur et al. 28 Desiccation resistance | Clinal | Drosophila simulans | Desiccation resistance | Stress resistance | Individual measurements | Arthur et al. 28 | 10.1111/j.1420-9101.2008.01617.x | http://adeer.pearg.com/biogs/DR00356b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 79 Arthur et al. 28 Development time | Clinal | Drosophila simulans | Development time | Life history | Individual measurements | Arthur et al. 28 | 10.1111/j.1420-9101.2008.01617.x | http://adeer.pearg.com/biogs/DR00357b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 80 Loeschcke et al Thorax and wing traits natural pops | Clinal | Drosophila aldrichi and buzzatii | Thorax length, wing traits | Morphology | Individual measurements | Loeschcke et al. 18 | 10.1046/j.1365-2540.2000.00766.x | http://adeer.pearg.com/biogs/DR00374b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 81 Loeschcke et al Wing traits and asymmetry natural pops | Clinal | Drosophila aldrichi and buzzatii | Wing traits and wing assymmetry | Morphology | Individual measurements | Loeschcke et al. 18 | 10.1046/j.1365-2540.2000.00766.x | http://adeer.pearg.com/biogs/DR00375b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 82 Loeschcke et al 18 Wing asymmetry natural pops | Clinal | Drosophila aldrichi and buzzatii | Wing assymmetry | Morphology | Individual measurements | Loeschcke et al. 18 | 10.1046/j.1365-2540.2000.00766.x | http://adeer.pearg.com/biogs/DR00376b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 83 Loeschcke et al 18 Thorax and wing traits lab pops | Clinal | Drosophila aldrichi and buzzatii | Thorax length and wing traits | Morphology | Individual measurements | Loeschcke et al. 18 | 10.1046/j.1365-2540.2000.00766.x | http://adeer.pearg.com/biogs/DR00377b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 84 Loeschcke et al 18 Wing traits and asymmetry lab pops | Clinal | Drosophila aldrichi and buzzatii | Wing traits and wing assymmetry | Morphology | Individual measurements | Loeschcke et al. 18 | 10.1046/j.1365-2540.2000.00766.x | http://adeer.pearg.com/biogs/DR00378b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 85 Loeschcke et al 18 Wing asymmetry lab pops | Clinal | Drosophila aldrichi and buzzatii | Assymmetry of wing traits | Morphology | Individual measurements | Loeschcke et al. 18 | 10.1046/j.1365-2540.2000.00766.x | http://adeer.pearg.com/biogs/DR00379b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 86 Loeschcke et al 18 Development time & viabiliy lab pops | Clinal | Drosophila aldrichi and buzzatii | Development time and viability | Life history | Population means | Loeschcke et al. 18 | 10.1046/j.1365-2540.2000.00766.x | http://adeer.pearg.com/biogs/DR00380b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 87 Barker 37 Allozyme allele frequencies 67 populations | Clinal | Drosophila buzzatii | Allozymes | Genetic markers | Population frequencies | Barker 37 | 10.1111/bij.12067 | http://adeer.pearg.com/biogs/DR00361b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 88 Barker 37 Allozyme allele frequencies 195 collections | Clinal | Drosophila buzzatii | Allozymes | Genetic markers | Population frequencies | Barker 37 | 10.1111/bij.12067 | http://adeer.pearg.com/biogs/DR00362b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 89 Barker 37 GENEPOP allozyme file 195 collections | Clinal | Drosophila buzzatii | Allozymes | Genetic markers | Individual measurements | Barker 37 | 10.1111/bij.12067 | http://adeer.pearg.com/biogs/DR00363b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 90 Barker 37 Overview 67 Populations | Clinal | Drosophila buzzatii | Allozymes | Genetic markers | Population frequencies | Barker 37 | 10.1111/bij.12067 | http://adeer.pearg.com/biogs/DR00364b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 91 Barker et al. 44 Microsatellite markers | Clinal | Drosophila buzzatii | Microsatellite markers | Genetic markers | Individual categories | Barker et al. 44 | 10.1038/hdy.2008.127 | http://adeer.pearg.com/biogs/DR00370b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 92 Barker et al. 44 GENEPOP microsatellite file | Clinal | Drosophila buzzatii | Microsatellite markers | Genetic markers | Individual categories | Barker et al. 44 | 10.1038/hdy.2008.127 | http://adeer.pearg.com/biogs/DR00371b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 93 Barker et al. 44 Microsatellite allele frequencies | Clinal | Drosophila buzzatii | Microsatellite markers | Genetic markers | Population means | Barker et al. 44 | 10.1038/hdy.2008.127 | http://adeer.pearg.com/biogs/DR00372b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 94 Barker et al. 6 S. aclinata microsatellite markers | Clinal | Scaptodrosophila aclinata | Microsatellite markers | Genetic markers | Individual categories | Barker et al. 6 | 10.1038/sj.hdy.6800592 | http://adeer.pearg.com/biogs/DR00367b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 95 Barker et al. 6 S. hibisci microsatellite markers | Clinal | Scaptodrosophila hibisci | Microsatellite markers | Genetic markers | Individual categories | Barker et al. 6 | 10.1038/sj.hdy.6800592 | http://adeer.pearg.com/biogs/DR00368b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 96 Magiafoglou et al. 27 microsatellite markers | Clinal | Drosophila serrata | Microsatellite markers | Genetic markers | Individual categories | Magiafoglou et al. 27 | 10.1046/j.1420-9101.2002.00439.x | http://adeer.pearg.com/biogs/DR00397b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 97 Hoffmann et al. 32 Desiccation resistance | Clinal | Drosophila melanogaster | Desiccation resistance | Stress resistance | Subgroup measurements | Hoffmann et al. 32 | 10.1111/j.1365-2435.2005.00959.x | http://adeer.pearg.com/biogs/DR00394b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 98 Hoffmann et al. 32 Starvation resistance | Clinal | Drosophila melanogaster | Starvation resistance | Stress resistance | Subgroup measurements | Hoffmann et al. 32 | 10.1111/j.1365-2435.2005.00959.x | http://adeer.pearg.com/biogs/DR00395b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 99 Hoffmann et al. 32 Heat resistance | Clinal | Drosophila melanogaster | Heat knockdown time | Stress resistance | Individual measurements | Hoffmann et al. 32 | 10.1111/j.1365-2435.2005.00959.x | http://adeer.pearg.com/biogs/DR00396b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
| 201 Barker 5 S. aclinata collection records | Species distribution | Scaptodrosophila aclinata | Presence records | Barker 5 | 10.1038/sj.hdy.6800592 | http://adeer.pearg.com/biogs/DR00390b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 | ||
| 202 Barker 5 S. hibisci collection records | Species distribution | Scaptodrosophila hibisci | Presence records | Barker 5 | 10.1038/sj.hdy.6800592 | http://adeer.pearg.com/biogs/DR00391b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 | ||
| 203 Barker et al. 6 D. buzzatii and aldrichi collection records | Species distribution | Drosophila buzzatii and D. aldrichi | Presence records | Barker et al. 6 | 10.1111/j.1442-9993.2005.01470.x | http://adeer.pearg.com/biogs/DR00387b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 | ||
| 204 Schiffer & McEvey 7 Montium colletion records | Species distribution | Drosophila bunnanda, D. serrata, D. birchii, D. kikkawai and D. sp. cf. jambulina | Presence records | Schiffer & McEvey 7 | na | http://adeer.pearg.com/biogs/DR00342b.htm | https://dx.doi.org/10.4225/49/555C0B8D30C3E | Dryad | http://dx.doi:10.5061/dryad.k9c31 |
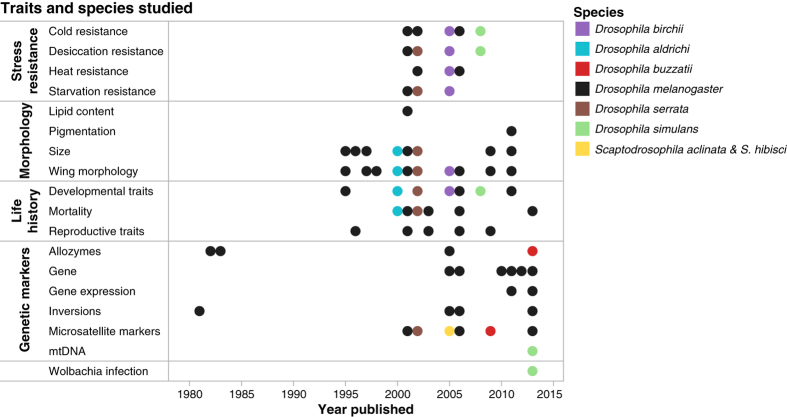
Figure 1. An overview of the trait groups and species studies in the datasets of ADEER.
All datasets were published between 1982 and 2013 in a total of 39 papers.
This clinal data collection includes data recorded at the level of the individual fly (46 datasets), the subgroup level (12 datasets) or the population level (41 datasets). The term population is here used for a group of flies collected at a single geographic location. Distinct collection sites were typically at least 40 km apart. Individual data include morphological, life-history, stress resistance traits and individual genotype at genetic marker loci. Many data were recorded as population frequencies, such as Wolbachia infection rate and genetic marker frequency (Table 1 (available online only) and Tables 2 (available online only) and 3). For other datasets the data are available as population means, including morphological, life-history and stress resistance traits and genetic markers. A few datasets report results at the subgroup level. These datasets include traits that were measured per vial (e.g. development time, longevity and mortality), per cage (e.g. mortality and fecundity) or per group of flies (desiccation and starvation resistance, gene expression). In addition, one dataset reports data on isofemale lines10. Isofemale lines are fly lines that were founded from the offspring of one single wild female (Figs 1 and 2).
Table 2. Phenotypic traits and genetic markers studied along the eastern Australian coast in drosophilid species.
| Trait/genetic marker | Species | Clinal variation | Clinal pattern | References |
|---|---|---|---|---|
| The presence and pattern of clinal variation and the publication associated with the datasets are reported | ||||
|
Morphological |
||||
| Egg size | D. melanogaster | Yes | Linear | Azevedo et al. 20 |
| Pigmentation | D. melanogaster | Yes | Linear | Telonis Scott et al. 25 |
| Thorax length | D. melanogaster | Yes | Linear | James et al. 17, 12, Rako et al. 15 |
| D. melanogaster | No | Van Heerwaarden & Sgrò 16 | ||
| D. melanogaster | Na | Hoffmann et al. 10 | ||
| D. aldrichi | Na | Loeschcke et al. 18 | ||
| D. buzzatii | Na | Loeschcke et al. 18 | ||
| D. simulans | Yes | Linear | Van Heerwaarden & Sgro 16 | |
| Mass | D. serrata | Yes | Linear and quadratic | Hallas et al. 19 |
| Wing size | D. melanogaster | Yes | Linear | Gockel et al. 22, James et al. 17, 12, Collinge et al. 21, Rako et al. 15, Van Heerwaarden & Sgrò 16 |
| D. aldrichi | Na | Loeschcke et al. 18 | ||
| D. buzzatii | Na | Loeschcke et al. 18 | ||
| D. birchii | No | Griffiths et al. 23 | ||
| D. serrata | Yes | Linear and quadratic | Hoffmann & Shirriffs 24, Hallas et al. 19 | |
| D. simulans | Yes | Linear | Van Heerwaarden & Sgro 16 | |
| Wing to aspect ratio | D. melanogaster | Yes, in the field | Linear | Azevedo et al. 11 |
| Wing to thorax ratio | D. melanogaster | Yes | Linear | Azevedo et al. 11, Van Heerwaarden & Sgrò 16 |
| D. aldrichi | Na | Loeschcke et al. 18 | ||
| D. buzzatii | Na | Loeschcke et al. 18 | ||
| D. simulans | No | Van Heerwaarden & Sgro 16 | ||
| Wing shape | D. aldrichi | Na | Loeschcke et al. 18 | |
| D. buzzatii | Na | Loeschcke et al. 18 | ||
| Fluctuating asymmetry | D. buzzatii | Na | Loeschcke et al. 18 | |
| D. aldrichi | Na | Loeschcke et al. 18 | ||
| Lipid content | D. melanogaster | Na | Hoffmann et al. 10 | |
|
Life-history |
||||
| Development time | D. melanogaster | Yes | Linear | James & Partridge 26, Collinge et al. 21 |
| D. aldrichi | Na | Loeschcke et al. 18 | ||
| D. buzzatii | Na | Loeschcke et al. 18 | ||
| D. birchii | Yes | Linear | Griffiths et al. 23 | |
| D. serrata | Yes | Linear and quadratic | Magiafoglou et al. 27 | |
| D. simulans | No | Arthur et al. 28 | ||
| Longevity | D. melanogaster | Yes | Linear and quadratic | Sgrò et al. 30 |
| Overwinter mortality | D. melanogaster | Yes | Linear and quadratic | Mitrovski & Hoffmann 13, Hoffmann et al. 14 |
| Overwinter fecundity | D. melanogaster | Yes | Linear and quadratic | Mitrovski & Hoffmann 13 |
| D. melanogaster | Linear | Hoffmann et al. 14, Rako et al. 15 | ||
| Mortality | D. aldrichi | Na | Loeschcke et al. 18 | |
| D. buzzatii | Na | Loeschcke et al. 18 | ||
| D. serrata | Yes | Linear | Magiafoglou et al. 27 | |
| Timing of overwinter fecundity | D. melanogaster | Yes | Linear | Mitrovski & Hoffmann 13, Hoffmann et al. 14 |
| Ovariole number | D. melanogaster | Yes | Quadratic | Azevedo et al. 20 |
| D. melanogaster | Linear | Collinge et al. 21 | ||
| Ovarian dormancy | D. melanogaster | Yes | Quadratic | Lee et al. 29 |
|
Stress |
||||
| Cold resistance | D. melanogaster | Yes | Linear | Hoffmann et al. 10, 31, Collinge et al. 21 |
| D. melanogaster | Na | Hoffmann et al. 10 | ||
| D. birchii | No | Griffiths et al. 23 | ||
| D. serrata | Yes | Linear | Hallas et al. 19, Magiafoglou et al. 27 | |
| D. simulans | Yes, females | Cubic | Arthur et al. 28 | |
| Desiccation resistance | D. melanogaster | No | Hoffmann et al. 10, Hoffmann et al. 32 | |
| D. melanogaster | Na | Hoffmann et al. 10 | ||
| D. birchii | Yes | Linear | Griffiths et al. 23 | |
| D. serrata | No | Hallas et al. 19 | ||
| D. simulans | No | Arthur et al. 28 | ||
| Heat resistance | D. melanogaster | Yes | Linear | Hoffmann et al. 31, Collinge et al. 21, Hoffmann et al. 10 |
| D. birchii | No | Griffiths et al. 23 | ||
| Starvation resistance | D. melanogaster | Yes, females | Linear | Hoffmann et al. 10 |
| D. melanogaster | No | Hoffmann et al. 32 | ||
| D. birchii | Yes | Linear | Griffiths et al. 23 | |
| D. serrata | Yes, males | Linear | Hallas et al. 19 | |
|
Inversions |
||||
| In(2L)t | D. melanogaster | Yes | Linear | Knibb et al. 46 |
| D. melanogaster | Na | Kennington & Hoffmann 34 | ||
| In(2R)NS | D. melanogaster | Yes | Linear | Knibb et al. 46 |
| In(3L)Payne | D. melanogaster | Yes | Linear | Knibb et al. 46 |
| In(3R)Payne | D. melanogaster | Yes | Linear | Knibb et al. 46, Lee et al. 42, Umina et al. 8, Kennington et al 41 |
| In(3R)C | D. melanogaster | Yes | Linear | Knibb et al. 46 |
|
Allozymes |
||||
| Adh | D. melanogaster | Yes | Linear | Oakeshott et al. 36, Umina et al. 8 |
| D. melanogaster | Na | Kennington & Hoffmann 34 | ||
| Gpdh | D. melanogaster | Yes | Linear | Oakeshott et al. 36 |
| G6pd | D. melanogaster | Yes | Linear | Oakeshott et al. 35 |
| Pgd | D. melanogaster | Yes | Linear | Oakeshott et al. 35 |
| Pgm, Aldox, Hex, Adh, Est1, Est2 and Lap | D. buzzatii | Yes | Linear | Barker 37 |
|
DNA sequence variation |
||||
| clock | D. melanogaster | No | Weeks et al. 2005 | |
| couch potato | D. melanogaster | Yes | Linear | Lee et al. 29 |
| drosophila cold acclimation | D. melanogaster | Yes | Linear | Mckechnie et al. 39 |
| frost | D. melanogaster | Yes | Linear | Hoffmann et al. 38 |
| hsp70 | D. melanogaster | No | Collinge et al. 21 | |
| hsr-omega | D. melanogaster | Yes | Linear | Kennington et al. 41, Collinge et al. 21 |
| methuselah | D. melanogaster | Yes | Linear and quadratic | Sgrò et al. 30 |
| neurofibromin | D. melanogaster | Yes | Linear | Lee et al. 42 |
| period | D. melanogaster | No | Weeks et al. 2005 | |
| MtDNA | D. simulans | Na | Kriesner et al. 43 | |
|
DNA repeat variation |
||||
| Microsatellite markers | D. melanogaster | Yes, 5 out of 19 | Linear | Gockel et al. 22 |
| D. melanogaster | Yes, 9 out of 24 | Linear | Kennington et al. 41 | |
| D. melanogaster | Na | Kennington & Hoffmann 34 | ||
| D. buzzatii | Yes, 6 out of 15 | Linear | Barker 2009 | |
| D. serrata | No | Magiafoglou et al. 27 | ||
| S. aclinata | Na | Barker et al. 6 | ||
| S. hibisci | Na | Barker et al. 6 | ||
|
Gene expression |
||||
| couch potato | D. melanogaster | Yes | Linear and quadratic | Lee et al. 29 |
| ebony | D. melanogaster | Yes, at 25 °C | Linear | Telonis-Scott et al. 25 |
| methuselah | D. melanogaster | Yes | Linear | Sgrò et al. 30 |
|
Others |
||||
| Wolbachia | D. simulans | Na | Kriesner et al. 43 |
Table 3. Species distribution datasets for nine drosophilid species.
| Datasets | Species | Publication | Collection years |
|---|---|---|---|
| The publication associated with the dataset and collection years are reported. | |||
| 201 Barker5 S. aclinata collection records | S. aclinata | Barker5 | 1995 |
| 202 Barker5 S. hibisci collection records | S. hibisci | Barker5 | 1998 |
| 203 Barker et al.6 D. buzzatii and aldrichi collection records | D. buzzatii | Barker et al.6 | 1971-2002 |
| 203 Barker et al.6 D. buzzatii and aldrichi collection records | D. aldrichi | Barker et al.6 | 1971-2002 |
| 204 Schiffer & McEvey7 Montium collection records | D. bunnanda | Schiffer & McEvey7 | 1924-2005 |
| 204 Schiffer & McEvey7 Montium collection records | D. serrata | Schiffer & McEvey7 | 1924-2005 |
| 204 Schiffer & McEvey7 Montium collection records | D. birchii | Schiffer & McEvey7 | 1924-2005 |
| 204 Schiffer & McEvey7 Montium collection records | D. kikkawai | Schiffer & McEvey7 | 1924-2005 |
| 204 Schiffer & McEvey7 Montium collection records | D. sp. cf. jambulina | Schiffer & McEvey7 | 1924-2005 |
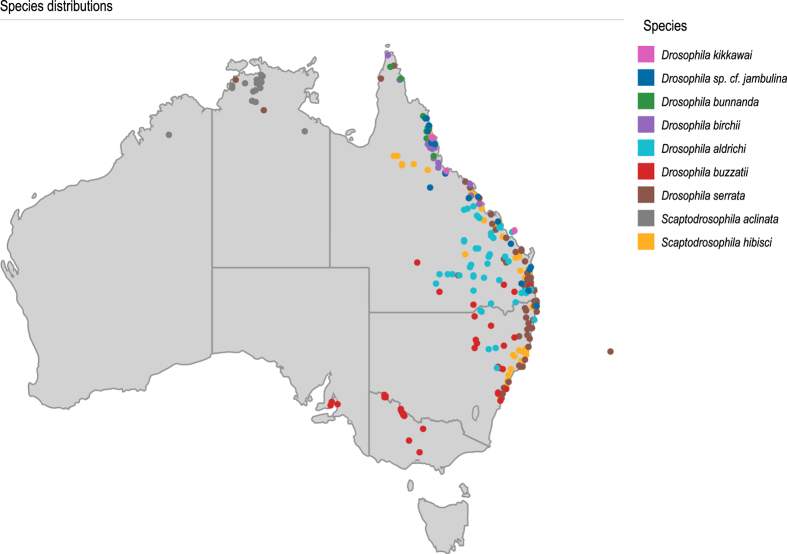
Figure 2. Collection records are shown for nine drosophilid species (D. aldrichi, D. birchii, D. bunnanda, D. buzzatii, D. kikkawai, D. serrata, D. sp. cf. jambulina, S. aclinata and S. hibisci).
These data were collected between 1924 and 2005 and are based on records in the literature, collections made by the dataset authors and specimens in the Australian Museum.
Fly populations compared for clinal variation in quantitative traits have almost always been maintained in the laboratory prior to testing, for periods ranging from just one generation to several years. The effect of laboratory culture on clinal patterns was specifically investigated in two of the datasets included in this collection using D. melanogaster 11,12. Almost all clinal studies on quantitative traits in this collection involve a common garden design, where populations are reared in a common environment before they are tested for a specific trait. Flies are therefore kept under controlled temperature and day length and on standard fly medium within a study, but these conditions can vary substantially among studies. A few studies did not use common garden experiments. These include the two studies mentioned above11,12 where field flies were preserved in alcohol to measure wing traits. Other exceptions13,14 involved clinal variation in fecundity and mortality scored directly under field conditions.
Morphological traits
27 datasets from 15 publications in this collection investigated morphological traits in D. melanogaster, D. serrata, D. aldrichi, D. buzzatii, D. simulans, or D. birchii. The morphological traits include size (10 datasets), wing morphology (20 datasets), pigmentation (2 datasets) and lipid content (1 dataset).
Thorax length, measured from the anterior margin of the thorax to the posterior tip of the scutellum, is most often used as a measure of size and was investigated in four D. melanogaster studies10,15–17 in D. aldrichi and D. buzzatii18 and in D. simulans16. Mass was used as a measure of size in one D. serrata study19. In addition, egg size was measured in one D. melanogaster study20.
Wing morphology was investigated in seven D. melanogaster studies11,12,15–17,21,22, in D. aldrichi and D. buzzatii18, in D. birchii23, in D. serrata19,24 and in D. simulans16. Wings were removed from individual flies and mounted on slides, and wing traits were either directly measured under a microscope11,12,17,22 or measured from landmarked images captured under the microscope15,16,18,19,21.
Pigmentation was investigated in one D. melanogaster study and was scored by visual examination using four phenotypic classes25.
Lipid levels were scored in one D. melanogaster study where adult females were initially dried in an oven for 48 h and then soaked in ether for 24 h to extract the lipids9.
Life-history traits
There are 24 datasets in this collection from 13 publications that investigated life-history traits in D. melanogaster, D. serrata, D. aldrichi, D. buzzatii, D. simulans, or D. birchii. This includes traits related to development (11 datasets), mortality (8 datasets) and reproduction (8 datasets).
Egg-to-adult development time was investigated in two D. melanogaster studies21,26, D. aldrichi and D. buzzatii18, D. birchii23, D. serrata27 and in D. simulans28. Development time was measured from the midpoint of the egg laying period to adult eclosion (emergence from the pupal case). In addition, egg development stage was examined in female D. melanogaster after being exposed to diapause-inducing conditions for 28 days29.
Mortality was investigated in D. melanogaster13,14,30, D. aldrichi and D. buzzatii18 and D. serrata27. In D. melanogaster, mortality was recorded in field cages held at temperate winter conditions near Melbourne13 and at tropical winter conditions in Cairns14. In addition, longevity of once-mated females was scored under standard laboratory conditions30. The flies were transferred to fresh vials every day, and at each transfer, all vials were examined for dead flies30. In D. aldrichi and D. buzzatii, larvae to adult viability was scored after rearing the flies at three temperatures treatments18. Egg to adult and pupal to adult viability were scored in D. serrata collected before and after winter27. To score egg to adult and pupae to adult viability, vials were scored until no new adults emerged and the number of pupae in each vial was counted to obtain pupal viability data27.
Reproductive traits were investigated in five D. melanogaster studies13–15,20,21. Overwintering fecundity was recorded in field cages held at temperate winter conditions near Melbourne13 and at tropical winter conditions in Cairns14. Rako et al.15 tested for the maintenance of fertility in males that have survived in field cages held at temperate winter conditions near Melbourne. Males were crossed to virgin females and the number of offspring was scored for each male15. Ovariole number was scored in two studies, whereas the number of ovarioles in each ovary was counted directly after dissection of the females20,21.
Stress resistance traits
Sixteen datasets from 8 publications investigated stress traits in D. melanogaster, D. serrata, D. simulans, or D. birchii. These traits include cold resistance (8 datasets), desiccation resistance (6 datasets), heat resistance (4 datasets) and starvation resistance (5 datasets).
Cold resistance, scored as chill coma recovery time was investigated in D. melanogaster21,31, D. birchii 23, D. serrata19 and D. simulans28. Flies were placed in empty vials which were immersed in a 10% glycol solution cooled to a constant temperature of 0 °C . After 1–8 h, vials were removed from the cold bath and placed at room temperature and recovery time of flies was scored19,21,23,28,31. Cold resistance scored as mortality after chill coma was investigated in D. melanogaster10,31 and D. serrata27. Groups of females were placed into empty vials and submerged in a −2 °C waterbath for 1–3 h. Flies were allowed to recover in vials with fly medium for 24–48 h before scoring mortality10,27,31.
Desiccation resistance was investigated in D. melanogaster10,32, D. serrata19, D. simulans28 and D. birchii23. Flies were placed in empty vials covered with gauze and then transferred to a desiccator with silica gel left at 25 °C . Mortality was scored hourly until all flies had died10,19,23,28,32.
Heat resistance was investigated in D. melanogaster21,31,32, and D. birchii23. Individual flies were placed into 5 ml glass vials submerged into a glass tank with water held at 39 °C (38.5 °C for D. birchii). Resistance was scored as the time taken for flies to be knocked down21,23,31,32.
Starvation resistance was investigated in D. melanogaster10,32, D. birchii23 and D. serrata19. Flies were placed in vials/tubes containing agar and these vials were placed in a chamber with water to maintain humidity close to 100%. Chambers were held at 25 °C and mortality was scored at 6–8 h intervals until at least half the flies had died10,19,32. Griffiths et al.23 scored starvation resistance by placing flies in vials, which were then inverted over a second vial containing cotton wool and water. Flies in the vial were separated from the water with fine gauze and the two vials were sealed together with Parafilm®. The flies were scored for survival every hour until half the flies had died23.
Genetic markers
This data collection contains 41 datasets from 20 publications that investigated genetic markers in D. melanogaster, D. serrata, D. buzzatii, D. simulans, S. aclinata or S. hibisci. Genetic marker types include allozymes (9 datasets), DNA sequence polymorphism (18 datasets), DNA repeat variation (i.e. microsatellites, 8 datasets), gene expression levels (4 datasets), inversion polymorphisms (5 datasets), and mitochondrial DNA regions (1 dataset).
Allozymes are enzymes that differ in electrophoretic mobility as a result of allelic differences at a single locus33. Allozymes were investigated in D. melanogaster8,34–36 and D. buzzatii37. Allozymes were scored after electrophoresis of single fly homogenates and staining8,34–37. Adh and Pgd were scored in D. melanogaster8,34–36 and D. buzzatii37. Gpdh, G6pd and Pgd were scored in D. melanogaster35,36 and Aldox, Hex, Est1, Est2 and Lap were scored in D. buzzatii37.
DNA sequence polymorphism can be determined using polymerase chain reaction (PCR) followed by gel electrophoresis (to detect size variation) or sequencing (to detect sequence variation)33. Drosophila melanogaster has been intensively used as a model to study DNA sequence polymorphisms along the eastern Australian cline21,29,30,38–42. In addition, variation in mitochondrial DNA sequences was investigated in D. simulans43. Several genes have been investigated in D. melanogaster: clock and period40, couch potato29, drosophila cold acclimation39, frost38, hsp70 (ref. 21), hsr-omega21,41, methuselah30, and neurofibromin42. The protocols to test for clinal variation in DNA sequence polymorphism varied substantially among the studies. In short, fly DNA was most often extracted using a Chelex/Proteinase K method42 but sometimes used a modified CTAB method40. Amplification of nuclear and mitochondrial DNA was performed using standard PCR methods and variation in DNA sequences was determined by gel electrophoresis or sequencing. For further details see21,29,30,38–43.
Microsatellites are tandemly repeated sequences of 1–6 nucleotides. Microsatellite markers are highly polymorphic and are assumed to evolve neutrally33. Microsatellites were investigated in D. melanogaster22,34,41, D. buzzatii44, D. serrata27, S. aclinata and S. hibsici5. After DNA extraction, microsatellite markers were amplified by polymerase chain reaction (PCR) using the unique sequences of flanking regions as primers and then repeat length was measured either by separating radiolabelled products on a gel or separating fluorescent-labelled products on a DNA sequencer. For further details see5,22,27,34,41,44.
Gene expression assays aim to quantify the level of RNA transcript present in the cell for each gene of interest using real-time PCR or deep-sequencing technologies45. Expression of three genes was investigated in D. melanogaster: couch potato29, ebony25, and methuselah30. In each case, RNA was isolated and purified to ensure DNA removal; cDNA was then synthesised for use as template for real-time PCR on the Light-Cycler® 480 (Roche) system and normalized using housekeeping genes. Further details are available in publications25,29,30.
Inversion polymorphism refers to the phenomenon of a chromosome region appearing in either standard or ‘reversed’ orientation in a population, which results in multiple genes being inherited together rather than assorting independently. It has been intensively investigated in D. melanogaster in Australia. The inversion In(3R)Payne is the most frequently studied inversion8,41,42,46, but In(2R)NS, In(3L)Payne, In(3R)C46 and In(2L)t34,46 have also been investigated. Two different approaches were used to test for inversion polymorphism: The BI-PASA method genotypes a SNP polymorphism shown to be in complete linkage disequilibrium with In(3R)Payne in Australia8,41,42. Alternatively, a salivary gland preparation was made from a single 3rd-instar larva and lacto-acetic orcein was used to stain the chromosome. After staining, glands were squashed under a cover slip and visualized with a light microscope to examine banding patterns and loops characteristic of inversion status34,46.
Wolbachia
Wolbachia are maternally inherited intracellular bacteria that can manipulate host reproduction43. One study in this collection investigated Wolbachia infections in D. simulans43. DNA was extracted using a standard Chelex based method and assays for Wolbachia infection status and strain type were performed with a fluorescence-based PCR assays using the Roche LightCycler® 480 system47.
Species distribution collection
The species distribution collection contains data from two Scaptodrosophila species and seven Drosophila species (Table 1 (available online only), Table 3 and Fig. 2). Schiffer and McEvey7 investigated distributions of members of the montium subgroup (Drosophila bunnanda, D. serrata, D. birchii, D. kikkawai and D. sp. cf. jambulina) along the east coast of Australia. Collection records are available for 122 locations that were sampled between 1924 and 2005 and data are based on records in the literature, collections made by the authors and specimens in the Australian Museum7. Collection records are also available for the cactophilic D. aldrichi and D. buzzatii6. These species were sampled between 1971 and 2002 in 97 locations where Opuntia cacti occur and the Opuntia species were recorded for each location. Barker5 collected distribution data of S. aclinata and S. hibisci which are both restricted to Hibiscus flowers5. Scaptodrosophila aclinata were sampled in 24 locations in 1995 and S. hibisci were sampled in 63 locations in 1998 and the Hibiscus species were recorded for all locations. For further details see the relevant publications5–7.
Data Records
All 103 datasets are freely available through the ADEER website (http://adeer.pearg.com/), where additional datasets will be added in the future. In addition to the datasets, ADEER also provides a short description and a visualisation of each dataset and a link to the publication describing the datasets (Data Citation 1). The datasets can be accessed by browsing the collections, species or traits or by using the “Search” function. All 103 datasets are listed under “Browse Datasets” or as a default using the “Search” function. The data can be downloaded by clicking on the “Data Online” icon. A static version of all datasets was also transferred to Dryad on 19.7.2015 (Data citation 2). The datasets 63–70 from the Lee et al. (2011) publication are also freely available on the Dryad repository (Data Citation 3). In addition, the dataset 25 from the Lee et al. (2013) publication (Data Citation 4), the dataset 24 from the Kriesner et al. 2013 publication (Data Citation 5), the datasets 74-76 from the Sgrò et al. (2013) publication (Data Citation 6) and the datasets 37–39 from the Telonis-Scott et al. (2011) publication (Data Citation 7) are already freely available on the Dryad repository.
Technical Validation
All datasets of this collection have been published in peer-reviewed journals confirming the technical quality of the data and the appropriate use of experimental designs. Experimental designs always included control treatments where necessary and careful replication and randomization of the experimental units. All data have also been statistically analysed, which included testing for measurement and recording errors. Furthermore, in the process of collecting this resource, each dataset was visualized and checked for potential inconsistencies. Spelling mistakes were corrected in the datasets, but only datasets where no inconsistencies were found in the data were included in this resource.
Usage Notes
The annual average daily mean temperature of Australia has risen by 0.9 °C since 1910 (CSIRO 2014) and Australian temperatures are projected to continue to increase by about 2–4 °C by 2100 following the global trend48. The increase in average and extreme temperatures presents a major challenge to biodiversity49.
The clinal and species distribution datasets will be valuable for temporal comparisons in the future to understand current and future evolutionary responses to climate change and to predict species distributions under ongoing climate change scenarios. Clinal data of phenotypic traits and genetic markers as well as species distributions can be tracked over time and tested for adaptive responses under climate change8. In addition, researchers can use the datasets for comparing shifts in species distributions and linking these to climatic variables.
There is now ample evidence that natural populations are responding to climate change by shifting their geographic distribution and phenology50–52 and an increasing number of studies have demonstrated evidence for rapid adaptive evolution in response to climate change53,54. Although plastic and genetic responses may allow some species to cope with climate change, extinction risks are predicted to be high, in particular in Australia55. One major challenge is to identify the most vulnerable species that will not be able to adapt fast enough to keep pace with climate change52,53. Collections like this one that span multiple related species with different degrees of adaptive potential and climate tolerance are important for understanding why some species are more vulnerable than others. Once this is better understood in model groups like drosophilid flies, researchers can apply general patterns to mammals, birds, plants and other groups to help prioritise conservation efforts.
Additional Information
How to cite this article: Hangartner, S. B. et al. A collection of Australian Drosophila datasets on climate adaptation and species distributions. Sci. Data 2:150067 doi: 10.1038/sdata.2015.67 (2015).
Supplementary Material
Acknowledgments
This project is supported by the Australian National Data Service (ANDS). ANDS is supported by the Australian Government through the National Collaborative Research Infrastructure Strategy Program. The project also acknowledges the support of the RDSI VicNode and NeCTAR UoM-Research Cloud Tools programs based at the University of Melbourne. We would also like to thank all the researchers that have contributed data to this collection: Professor Linda Partridge (University College London), John Oakeshott (CSIRO), Dr. Carla Sgró (Monash University), Dr. Shane McEvery (Australian Museum) and Professor James Stuart Flinton Barker (University of New England).
Footnotes
The authors declare no competing financial interests.
Data Citations
- Hoffmann A. A., Smith A., Griffin P. C., Hangartner S. B. 2015. Australian Drosophila Ecology And Evolution Resource. Adeer.pearg.com [DOI] [PMC free article] [PubMed]
- Hoffmann A. A., Smith A., Griffin P. C., Hangartner S. B. 2015. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.k9c31
- Lee S. F. 2011. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.k175g
- Lee S. F. 2013. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.n7080
- Kriesner P., Hoffmann A. A., Lee S. F., Turelli M., Weeks A. R. 2013. PLOS Pathogens. http://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1003607#s5 [DOI] [PMC free article] [PubMed]
- Sgrò C. M. 2013. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.11j35
- Telonis-Scott M., Hoffmann A. A., Sgrò C. M. 2011. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.8768
References
- Ashburner M., Golic K. G. & Hawley R. Drosophila: a laboratory handbook. Second edition. (Cold Spring Harbor Laboratory Press: Cold Spring Harbor, New York, United States, 2005). [Google Scholar]
- Bock I. R. & Parsons P. Adaptive radiation in the subgenus Scaptodrosophila of Australian. Drosophila. Nature 258, 602–602 (1975). [DOI] [PubMed] [Google Scholar]
- Steffen W. et al. Australia's biodiversity and climate change. (Csiro Publishing, 2009). [Google Scholar]
- Hoffmann A. A. & Weeks A. R. Climatic selection on genes and traits after a 100 year-old invasion: a critical look at the temperate-tropical clines in Drosophila melanogaster from eastern Australia. Genetica 129, 133–147 (2007). [DOI] [PubMed] [Google Scholar]
- Barker J. Population structure and host-plant specialization in two Scaptodrosophila flower-breeding species. Heredity 94, 129–138 (2005). [DOI] [PubMed] [Google Scholar]
- Barker J., Krebs R. A. & Davies H. Geographical distributions, relative abundance and coexistence of Drosophila aldrichi and Drosophila buzzatii in Australia. Austral. Ecol. 30, 546–557 (2005). [Google Scholar]
- Schiffer M. & McEvey S. F. Drosophila bunnanda: a new species from northern Australia with notes on other Australian members of the montium subgroup (Diptera: Drosophilidae). Zootaxa 1333, 1–23 (2006). [Google Scholar]
- Umina P., Weeks A., Kearney M., McKechnie S. & Hoffmann A. A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science 308, 691–693 (2005). [DOI] [PubMed] [Google Scholar]
- Anderson A. R., Hoffmann A. A., Mckechnie S. W., Umina P. A. & Weeks A. R. The latitudinal cline in the In (3R) Payne inversion polymorphism has shifted in the last 20 years in Australian Drosophila melanogaster populations. Mol. Ecol. 14, 851–858 (2005). [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Hallas R., Sinclair C. & Mitrovski P. Levels of variation in stress resistance in Drosophila among strains, local populations, and geographic regions: patterns for desiccation, starvation, cold resistance, and associated traits. Evolution 55, 1621–1630 (2001). [DOI] [PubMed] [Google Scholar]
- Azevedo R. B., James A. C., McCabe J. & Partridge L. Latitudinal variation of wing: thorax size ratio and wing-aspect ratio in Drosophila melanogaster . Evolution 1353–1362 (1998). [DOI] [PubMed] [Google Scholar]
- James A. C., Azevedo R. B. & Partridge L. Genetic and environmental responses to temperature of Drosophila melanogaster from a latitudinal cline. Genetics 146, 881–890 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovski P. & Hoffmann A. A. Postponed reproduction as an adaptation to winter conditions in Drosophila melanogaster: evidence for clinal variation under semi-natural conditions. Proc. R. Soc. B 268, 2163–2168 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Scott M., Partridge L. & Hallas R. Overwintering in Drosophila melanogaster: outdoor field cage experiments on clinal and laboratory selected populations help to elucidate traits under selection. J. Evol. Biol 16, 614–623 (2003). [DOI] [PubMed] [Google Scholar]
- Rako L., Poulsen N., Shirriffs J. & Hoffmann A. Clinal variation in post‐winter male fertility retention; an adaptive overwintering strategy in Drosophila melanogaster . J. Evol. Biol 22, 2438–2444 (2009). [DOI] [PubMed] [Google Scholar]
- Van Heerwaarden B. & Sgrò C. M. The effect of developmental temperature on the genetic architecture underlying size and thermal clines in Drosophila melanogaster and D. simulans from the east coast of Australia. Evolution 65, 1048–1067 (2011). [DOI] [PubMed] [Google Scholar]
- James A. C., Azevedo R. & Partridge L. Cellular basis and developmental timing in a size cline of Drosophila melanogaster . Genetics 140, 659–666 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeschcke V., Bundgaard J. & Barker J. Variation in body size and life history traits in Drosophila aldrichi and D. buzzatii from a latitudinal cline in eastern Australia. Heredity 85, 423–433 (2000). [DOI] [PubMed] [Google Scholar]
- Hallas R., Schiffer M. & Hoffmann A. A. Clinal variation in Drosophila serrata for stress resistance and body size. Genet. Res. 79, 141–148 (2002). [DOI] [PubMed] [Google Scholar]
- Azevedo R. B., French V. & Partridge L. Thermal evolution of egg size in Drosophila melanogaster . Evolution 2338–2345 (1996). [DOI] [PubMed] [Google Scholar]
- Collinge J., Hoffmann A. & McKechnie S. Altitudinal patterns for latitudinally varying traits and polymorphic markers in Drosophila melanogaster from eastern Australia. J. Evol. Biol 19, 473–482 (2006). [DOI] [PubMed] [Google Scholar]
- Gockel J., Kennington W. J., Hoffmann A., Goldstein D. B. & Partridge L. Nonclinality of molecular variation implicates selection in maintaining a morphological cline of Drosophila melanogaster . Genetics 158, 319–323 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J., Schiffer M. & Hoffmann A. Clinal variation and laboratory adaptation in the rainforest species Drosophila birchii for stress resistance, wing size, wing shape and development time. J. Evol. Biol 18, 213–222 (2005). [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A. & Shirriffs J. Geographic variation for wing shape in Drosophila serrata . Evolution 56, 1068–1073 (2002). [DOI] [PubMed] [Google Scholar]
- Telonis-Scott M., Hoffmann A. & Sgrò C. M. The molecular genetics of clinal variation: a case study of ebony and thoracic trident pigmentation in Drosophila melanogaster from eastern Australia. Mol. Ecol. 20, 2100–2110 (2011). [DOI] [PubMed] [Google Scholar]
- James A. C. & Partridge L. Thermal evolution of rate of larval development in Drosophila melanogaster in laboratory and field populations. J. Evol. Biol 8, 315–330 (1995). [Google Scholar]
- Magiafoglou A., Carew M. & Hoffmann A. Shifting clinal patterns and microsatellite variation in Drosophila serrata populations: a comparison of populations near the southern border of the species range. J. Evol. Biol 15, 763–774 (2002). [Google Scholar]
- Arthur A., Weeks A. & Sgro C. Investigating latitudinal clines for life history and stress resistance traits in Drosophila simulans from eastern Australia. J. Evol. Biol 21, 1470–1479 (2008). [DOI] [PubMed] [Google Scholar]
- Lee S. F. et al. Polymorphism in the couch potato gene clines in eastern Australia but is not associated with ovarian dormancy in Drosophila melanogaster . Mol. Ecol. 20, 2973–2984 (2011). [DOI] [PubMed] [Google Scholar]
- Sgrò C. M. et al. Complexity of the genetic basis of ageing in nature revealed by a clinal study of lifespan and methuselah, a gene for ageing, in Drosophila from eastern Australia. Mol. Ecol. 22, 3539–3551 (2013). [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Anderson A. & Hallas R. Opposing clines for high and low temperature resistance in Drosophila melanogaster . Ecol. Lett. 5, 614–618 (2002). [Google Scholar]
- Hoffmann A. A., Shirriffs J. & Scott M. Relative importance of plastic vs genetic factors in adaptive differentiation: geographical variation for stress resistance in Drosophila melanogaster from eastern Australia. Funct. Ecol. 19, 222–227 (2005). [Google Scholar]
- Schlötterer C. The evolution of molecular markers–just a matter of fashion? Nat. Rev. Genet. 5, 63–69 (2004). [DOI] [PubMed] [Google Scholar]
- Kennington W. J. & Hoffmann A. A. Patterns of genetic variation across inversions: geographic variation in the In (2L) t inversion in populations of Drosophila melanogaster from eastern Australia. BMC Evol. Biol. 13, 100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeshott J., Chambers G., Gibson J., Eanes W. & Willcocks D. Geographic variation in G6pd and Pgd allele frequencies in Drosophila melanogaster . Heredity 50, 67–72 (1983). [DOI] [PubMed] [Google Scholar]
- Oakeshott J. et al. Alcohol dehydrogenase and glycerol-3-phosphate dehydrogenase clines in Drosophila melanogaster on different continents. Evolution 86–96 (1982). [DOI] [PubMed] [Google Scholar]
- Barker J. S. F. Genetic history of a colonizing population: Drosophila buzzatii (Diptera: Drosophilidae) in Australia. Biol. J. Linn. Soc. 109, 682–698 (2013). [Google Scholar]
- Hoffmann A. et al. A proline repeat polymorphism of the Frost gene of Drosophila melanogaster showing clinal variation but not associated with cold resistance. Insect Mol. Biol. 21, 437–445 (2012). [DOI] [PubMed] [Google Scholar]
- McKechnie S. et al. A clinally varying promoter polymorphism associated with adaptive variation in wing size in Drosophila . Mol. Ecol. 19, 775–784 (2010). [DOI] [PubMed] [Google Scholar]
- Weeks A., McKechnie S. & Hoffmann A. In search of clinal variation in the period and clock timing genes in Australian Drosophila melanogaster populations. J. Evol. Biol 19, 551–557 (2006). [DOI] [PubMed] [Google Scholar]
- Kennington W. J., Partridge L. & Hoffmann A. A. Patterns of diversity and linkage disequilibrium within the cosmopolitan inversion In (3R) Payne in Drosophila melanogaster are indicative of coadaptation. Genetics 172, 1655–1663 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. F. et al. Polymorphism in the neurofibromin gene, Nf1, is associated with antagonistic selection on wing size and development time in Drosophila melanogaster . Mol. Ecol. 22, 2716–2725 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriesner P., Hoffmann A. A., Lee S. F., Turelli M. & Weeks A. R. Rapid sequential spread of two Wolbachia variants in Drosophila simulans . PLoS Pathog. 9, e1003607 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker J. et al. Bottlenecks, population differentiation and apparent selection at microsatellite loci in Australian Drosophila buzzatii . Heredity 102, 389–401 (2009). [DOI] [PubMed] [Google Scholar]
- Wang Z., Gerstein M. & Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibb W., Oakeshott J. & Gibson J. Chromosome inversion polymorphisms in Drosophila melanogaster. I. Latitudinal clines and associations between inversions in Australasian populations. Genetics 98, 833–847 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. F., White V. L., Weeks A. R., Hoffmann A. A. & Endersby N. M. High-throughput PCR assays to monitor Wolbachia infection in the dengue mosquito (Aedes aegypti) and Drosophila simulans . Appl. Environ. Microbiol. 78, 4740–4743 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker T. F. et al. IPCC, 2013: Climate Change 2013: The Physical Science Basis Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. (Cambridge University Press: Cambridge, United Kingdom and New York, United States, 1535, 2013). [Google Scholar]
- Pereira H. M. et al. Scenarios for Global Biodiversity in the 21st Century. Science 330, 1496–1501 (2010). [DOI] [PubMed] [Google Scholar]
- Walther G.-R. et al. Ecological responses to recent climate change. Nature 416, 389–395 (2002). [DOI] [PubMed] [Google Scholar]
- Parmesan C. & Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature 421, 37–42 (2003). [DOI] [PubMed] [Google Scholar]
- Parmesan C. Ecological and evolutionary responses to recent climate change. Annual Review of Ecology, Evolution, and Systematics 37, 637–669 (2006). [Google Scholar]
- Hoffmann A. A. & Sgro C. M. Climate change and evolutionary adaptation. Nature 470, 479–485 (2011). [DOI] [PubMed] [Google Scholar]
- Lavergne S., Mouquet N., Thuiller W. & Ronce O. Biodiversity and climate change: integrating evolutionary and ecological responses of species and communities. Annu. Rev. Ecol. Evol. Syst. 41, 321–350 (2010). [Google Scholar]
- Urban M. C. Accelerating extinction risk from climate change. Science 348, 571–573 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Hoffmann A. A., Smith A., Griffin P. C., Hangartner S. B. 2015. Australian Drosophila Ecology And Evolution Resource. Adeer.pearg.com [DOI] [PMC free article] [PubMed]
- Hoffmann A. A., Smith A., Griffin P. C., Hangartner S. B. 2015. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.k9c31
- Lee S. F. 2011. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.k175g
- Lee S. F. 2013. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.n7080
- Kriesner P., Hoffmann A. A., Lee S. F., Turelli M., Weeks A. R. 2013. PLOS Pathogens. http://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1003607#s5 [DOI] [PMC free article] [PubMed]
- Sgrò C. M. 2013. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.11j35
- Telonis-Scott M., Hoffmann A. A., Sgrò C. M. 2011. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.8768