Abstract
In 2013 and 2014, two U.S. universities had meningococcal serogroup B outbreaks (a total of 14 cases) caused by strains from two different clonal complexes. To control the outbreaks, students were immunized with a serogroup B meningococcal vaccine (Novartis) that was not yet licensed in the United States. The vaccine (referred to as MenB-4C) contains four components capable of eliciting bactericidal activity. Both outbreak strains had high expression levels of two of the vaccine antigens (subfamily B factor H binding protein [FHbp] and neisserial heparin binding antigen [NHba]); the university B outbreak strain also had moderate expression of a third antigen, NadA. We investigated the bactericidal activity of sera from mice immunized with FHbp, NHba, or NadA and sera from MenB-4C-immunized infant macaques and an adult human. The postimmunization bactericidal activity of the macaque or human serum against isolates from university B with FHbp identification (ID) 1 that exactly matched the vaccine FHbp sequence variant was 8- to 21-fold higher than that against isolates from university A with FHbp ID 276 (96% identity to the vaccine antigen). Based on the bactericidal activity of mouse antisera to FHbp, NadA, or NHba and macaque or human postimmunization serum that had been depleted of anti-FHbp antibody, the bactericidal activity against both outbreak strains largely or entirely resulted from antibodies to FHbp. Thus, despite the high level of strain expression of FHbp from a subfamily that matched the vaccine antigen, there can be large differences in anti-FHbp bactericidal activity induced by MenB-4C vaccination. Further, strains with moderate to high NadA and/or NHba expression can be resistant to anti-NadA or anti-NHba bactericidal activity elicited by MenB-4C vaccination.
INTRODUCTION
Meningococcal serogroup B outbreaks involving a total of 14 cases occurred on two university campuses in the United States in 2013 and 2014 (1). The outbreak at university A, which is located in New Jersey, started in March 2013 with a total of 9 cases documented in the campus population or in close contacts of the students (2). The outbreak at university B, which is located in California, started in November 2013 with a cluster of four cases (1). These cases were later connected to a fifth case in a student enrolled at university B, which had occurred 7 months earlier. The strains from the two outbreaks were from different clonal complexes and therefore were not epidemiologically related.
In response to these campus outbreaks, the U.S. Food and Drug Administration approved the immunization of the students with a serogroup B meningococcal vaccine (Bexsero; Novartis Vaccines and Diagnostics) (2), which at the time was licensed in Europe, Canada, and Australia. The vaccine contains four primary components, each capable of eliciting complement-mediated serum bactericidal activity (3, 4), and is referred to here as MenB-4C. The four antigens are factor H binding protein (FHbp), neisserial heparin binding antigen (NHba), neisserial adhesin A (NadA), and a porin protein (PorA) with variable region (VR) sequence type P1.7-2,4 contained in outer membrane vesicles (OMV) (3, 4). Since most of the anti-PorA bactericidal activity is directed to VR2, a match between the vaccine PorA and strain PorA is usually described as sharing the P1.4 VR2 antigen (5).
The purpose of the present study was to investigate the expression of MenB-4C vaccine antigens in representative isolates from each of the outbreaks and the susceptibility of the isolates to serum bactericidal activity induced by MenB-4C vaccination. We also assessed the contribution of anti-FHbp antibodies in eliciting serum bactericidal activity.
MATERIALS AND METHODS
Meningococcal outbreak strains.
We received 15 case isolates from the Meningitis Laboratory, Meningitis and Vaccine Preventable Diseases Branch, Centers for Disease Control and Prevention (CDC), Atlanta, GA. Of these, 10 isolates were from the university A outbreak, and 5 were from the university B outbreak. The CDC provided data on the date of isolation, source (blood or cerebrospinal fluid [CSF] sample), multilocus sequence type (MLST), PorB amino acid sequence type, and amino acid sequence variants of the FHbp, NadA, NHba, and PorA vaccine antigens. The isolates from university A were from sequence type 409 (ST-409), which is uncommon in the United States but part of the more common ST-41/44 clonal complex. The isolates from university B were all from ST-32, which is a common ST/clonal complex among invasive serogroup B case isolates from the west coast of the United States (6).
Control strains.
The strains used as controls are summarized in Table 1 and were previously described. Neisseria meningitidis strain NZ98/254 is a relatively low expresser of FHbp identification (ID) 14 (subfamily B) (7) and was used in flow cytometric studies as a control to measure FHbp expression. The remaining control strains were each mismatched for three of the four antigens in MenB-4C known to elicit serum bactericidal activity (8–10). Thus, the serum bactericidal activity elicited by the MenB-4C vaccine against N. meningitidis strain H44/76 is specific for antibodies to FHbp, N. meningitidis strain 5/99 is specific for antibodies to NadA, and N. meningitidis strain SK016 is specific for antibodies to PorA P1.4. In past studies, finding a control strain that is specific for anti-NHba bactericidal activity has been problematic (9, 11, 12). For our study, we used N. meningitidis strain M4407, which is mismatched for all of the vaccine antigens except NHba (i.e., lacks a NadA gene, has a PorA heterologous to that in the OMV in MenB-4C, and expresses subfamily A FHbp) (Table 1). Strain M4407 is a high expresser of NHba (100% amino acid identity to vaccine [10]). The strain is susceptible to mouse anti-NHba serum bactericidal activity but resistant to bactericidal activity by mouse antisera to subfamily B FHbp ID 1 or NadA (see Results).
TABLE 1.
Summary of control meningococcal strains
| Meningococcal strain (vaccine antigen[s])a | ST clonal complexb | MenB-4C vaccine antigens |
||||
|---|---|---|---|---|---|---|
| FHbp ID (subfamily)c | PorA VR1,2d | Antigen expressione |
||||
| NadA | FHbp | NHba | ||||
| NZ98/254 (FHbp and PorA) | 41/44 | 14 (B) | P1.7-2,4 | Absent | + | + |
| SK016 (PorA) | 103 | 25 (A) | P1.7-2,4 | Absent | + | +/− |
| H44/76 (FHbp) | 32 | 1 (B) | P1.7,16 | Absent | ++ | +/− |
| 5/99 (NadA) | 8 | 23 (A) | P1.5,2 | +++ | +/− | +/− |
| M4407 (NHba) | 41/44 | 19 (A) | P1.19,15-1 | Absent | + | ++ |
The antigens present in the MenB-4C vaccine are NadA, subfamily B FHbp, NHba, and PorA P1.4 (in the OMV). Strain NZ98/254 expresses FHbp subfamily B and PorA P1.4. The remaining four control strains are each matched for only one of the four vaccine antigens known to elicit serum bactericidal activity.
Clonal complex as defined by multilocus sequence type (ST) (46).
The MenB-4C vaccine contains subfamily B FHbp ID 1. FHbp peptide ID numbers and subfamily groups are from the FHbp database at http://pubmlst.org/neisseria/fHbp.
PorA variable region (VR) 1 and 2 inferred from gene sequences. The MenB-4C vaccine contains OMV from strain NZ98/254, which contains PorA P1.7-2,4.
Relative levels of antigen expression, denoted +/−, +, ++, and +++, as measured by flow cytometry using live bacteria (see Results). Absent indicates that the NadA gene is not present.
Antigen surface expression.
The expression of FHbp, NHba, and NadA on the surface of live meningococci was measured by flow cytometry, which was performed as previously described (13). FHbp was detected with an anti-FHbp monoclonal antibody (MAb), JAR 41, which recognizes all FHbp amino acid sequence variants tested from subfamily A or B (14). Additional antibodies included mouse MAbs to the capsule (SEAM 12 [15]) and polyclonal mouse antisera specific for the NHba and NadA antigens in MenB-4C vaccine (3) (see below).
Mouse sera.
Stored serum samples were available from female CD-1 mice (Charles River Laboratories) immunized with a recombinant subfamily B FHbp ID 1 vaccine, NadA, or the NHba-GNA1030 fusion protein used in the MenB-4C vaccine (3), according to previously described protocols (10). Serum pools were made from equal volumes of serum samples from 3 to 5 individual mice. The mice were housed in a facility certified by the AAALAC under a protocol approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco (UCSF) Benioff Children's Hospital Oakland Research Institute.
Rhesus macaque sera.
We tested sera from a previous study done in infant rhesus macaques immunized beginning at 3 to 4 months of age with two doses of the MenB-4C vaccine (16). We strictly adhered to the Guide for the Care and Use of Laboratory Animals (17), and the study was approved by the Institutional Animal Care and Use Committee of the University of California, Davis. For the present study, we selected postimmunization serum samples from five of the six vaccinated animals whose preimmunization serum FH bound to FHbp ID 1 with affinity similar to that of human FH; the sixth animal was excluded because of an insufficient volume of serum. For negative controls, we included serum samples from three unvaccinated animals.
Human serum.
As previously described (18), we obtained serum samples from a healthy adult 2 weeks before and 6 weeks after the subject received a third dose of the MenB-4C vaccine. The subject had been immunized approximately 5 years earlier with two doses of MenB-4C as part of a clinical trial in Europe. Informed written consent was obtained from the subject, and the studies were approved by the UCSF Benioff Children's Hospital Oakland institutional review board.
Complement-mediated serum bactericidal activity.
We measured serum bactericidal activity as described previously (19). The assay uses bacteria grown to mid-log phase in broth and exogenous human complement consisting of serum depleted of IgG via a protein G column (19). The percent survival of CFU per milliliter was calculated as a ratio of the CFU per milliliter after 60 min of incubation of dilutions of the test sera to that of the negative-control sera from the respective species.
Depletion of serum anti-FHbp antibodies.
A postimmunization serum pool from the five rhesus macaques immunized with the MenB-4C vaccine and the postimmunization serum from the immunized human were depleted of anti-FHbp antibodies via an R41S FHbp ID1 mutant recombinant protein coupled to Sepharose, as described previously (18). As a negative control, the respective serum samples were incubated with Sepharose beads that had not been coupled to a protein (referred to as mock adsorption). The efficiency of depleting serum anti-FHbp antibodies and the specificity of the depletion were measured by an enzyme-linked immunosorbent assay (ELISA) using microtiter plates (Immulon 2B; Thermo Scientific) coated with individual recombinant wild-type FHbp ID 1, NadA, or NHba, or outer membrane vesicles (OMV) prepared from a mutant of N. meningitidis strain NZ98/254 (6) in which the gene encoding FHbp had been inactivated.
RESULTS
Characteristics of outbreak isolates.
The two outbreaks were caused by epidemiologically unrelated strains that were derived from either clonal complex ST-41/44 (university A) or ST-32 (university B). The isolates from the university A outbreak had identical respective PorA, PorB, FHbp, and NHba variants, as inferred from DNA sequencing, and all of the isolates lacked the gene encoding NadA. Thus, these isolates appeared to be derived from a single clone. The isolates from university A had genes encoding two of the four MenB-4C antigens (Table 2): subfamily B FHbp (ID 276, 96% identical to FHbp ID 1 in the MenB-4C vaccine) and NHba variant 2 (100% identical to NHba in MenB-4C [20, 21]). The case isolates from the university B outbreak had genes encoding identical respective PorA, FHbp, NHba, and NadA variants but expressed one of two related PorB sequences (PorB3-24 or PorB3-461, with 97.6% amino acid identity). The slightly different PorB sequences indicated that the university B isolates were derived from two closely related but not identical strains. The university B isolates were matched for three of the four MenB-4C antigens: FHbp ID 1 (100% identical to FHbp in MenB-4C), NHba variant 5 (91% identical to NHba in MenB-4C), and NadA peptide 1 in variant group 1 (95% identical to NadA in MenB-4C and predicted to be covered by the NadA variant group 2/3 antigen in MenB-4C [20, 21]).
TABLE 2.
Characteristics of invasive isolates from two university campus outbreaksa
| Strain no. | University | ST clonal complexb | MenB-4C vaccine antigen (% identity) |
|||
|---|---|---|---|---|---|---|
| Subfamily B FHbp IDc | PorA VR1,2d | NadAe | NHbaf | |||
| CH819 | A | 41/44 | 276 (96) | P1.5,2-2 | Absent | 2 (100) |
| CH827 | A | 41/44 | 276 (96) | P1.5,2-2 | Absent | 2 (100) |
| CH838 | B | 32 | 1 (100) | P1.7,16-20 | 1.1 (95) | 5 (91) |
| CH840 | B | 32 | 1 (100) | P1.7,16-20 | 1.1 (95) | 5 (91) |
Two representative blood isolates were selected from each outbreak (see Materials and Methods).
Clonal complex as defined by multilocus sequence type (ST) (46).
The MenB-4C vaccine contains subfamily B FHbp ID 1. The percent identity between the vaccine antigen and strain antigens is given in parentheses. FHbp peptide ID numbers and subfamily groups are from the FHbp database at http://pubmlst.org/neisseria/fHbp.
Variable region (VR) type; the MenB-4C vaccine contains P1.7-2,4 (as the OMV antigen). The isolates from both outbreaks are mismatched to the PorA antigen in the vaccine.
The MenB-4C vaccine contains NadA peptide 8 in variant group 2/3. The percent identity between the vaccine NadA antigen and strain antigen is given in parentheses. Antibodies to the vaccine antigen 2/3 are reported to cover strains with NadA-1 (20).
The MenB-4C vaccine contains NHba variant 2. The percent identity between the vaccine antigen and strain antigens is given in parentheses.
We selected two representative blood isolates from each outbreak for further characterization of vaccine antigen expression and susceptibility to human complement-mediated serum bactericidal activity (Table 2). The selection of these isolates was based on including an early blood isolate and a late blood isolate from cases in each outbreak and, for university B, the inclusion of two isolates, each with a different PorB sequence type.
Strain antigen expression.
We used flow cytometry to measure the surface expression of three of the MenB-4C vaccine antigens, FHbp, NadA, and NHba. We did not test the fourth antigen, PorA P1.4, present in the OMV component of MenB-4C, since neither outbreak strain had the gene sequence encoding PorA P1.4. Representative antigen expression data for one isolate from each of the outbreaks are shown in Fig. 1. Similar data were obtained with the second isolate tested from each outbreak (data not shown).
FIG 1.
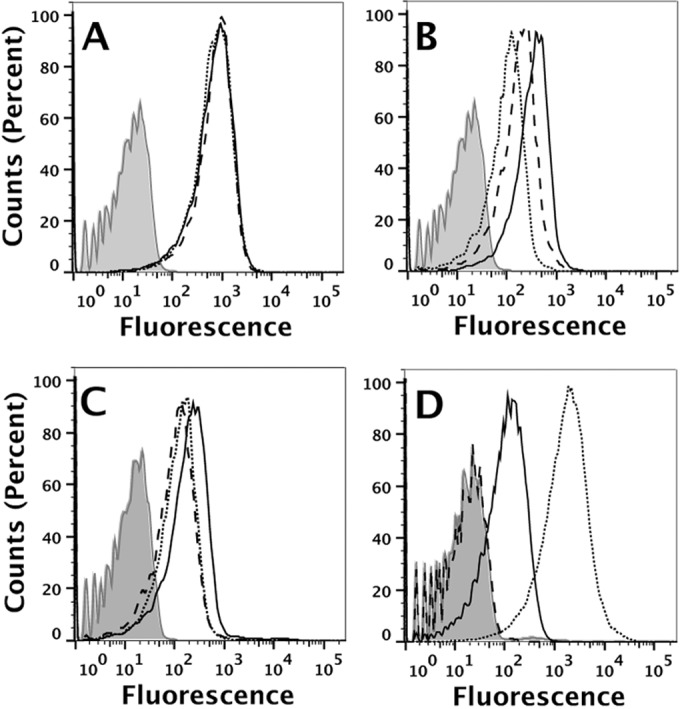
Vaccine antigen surface expression by isolates from the two outbreaks. Antigen expression was measured by flow cytometry using live bacteria and mouse MAbs or antisera to each of the individual antigens (see Materials and Methods). (A) Anti-capsular MAb. Dashed line, isolate CH819 from university A; solid black line, isolate CH840 from university B; dotted line, control strain NZ98/254. The data for the three strains are superimposed. Light gray shading, CH819 bacteria without added antibody (similar results without added antibody for the other two isolates are not shown). (B) Anti-FHbp MAb (JAR 41, which recognizes all FHbp sequence variants tested to date [14]). The line styles are the same as in panel A. (C) Mouse anti-NHba antiserum. The line styles are the same as in panel A, except the control strain is M4407, which is a naturally high expresser of NHba (10). (D) Mouse anti-NadA antiserum. The line styles are the same as in panel A, except the control strain is 5/99, which is a naturally high expresser of NadA (9, 11).
The two outbreak isolates showed similar binding by the flow assay using a control murine MAb to the serogroup B capsule (Fig. 1A). Both outbreak strains had higher FHbp expression than that of the control NZ98/254 strain (Fig. 1B, dotted line), which is known to have relatively low FHbp expression (22). The isolate from the university B outbreak had slightly higher FHbp expression than the isolate from the university A outbreak (Fig. 1B, solid and dashed lines, respectively).
The isolates from both university outbreaks also had high expression levels of NHba (Fig. 1C). The university A isolate (Fig. 1C, dashed line) had NHba expression similar to that of a high-NHba-expressing positive-control strain, M4407 (Fig. 1C, dotted line) (10). The university B isolate (Fig. 1C, solid line) had even higher NHba expression than that of the university A isolate or the high-NHba-expressing positive-control M4407 strain.
The university B isolates expressed NadA (Fig. 1D, solid line). The amount was less than that in the control 5/99 strain (Fig. 1D, dotted line), which was selected as a positive control based on high NadA expression and susceptibility to anti-NadA serum bactericidal activity (9, 11) (see also Results). As expected, there was no detectable NadA expression by the isolate from the university A outbreak (Fig. 1D, dashed line), which lacked an NadA gene (Table 2).
Susceptibility to bactericidal activity of mouse antisera.
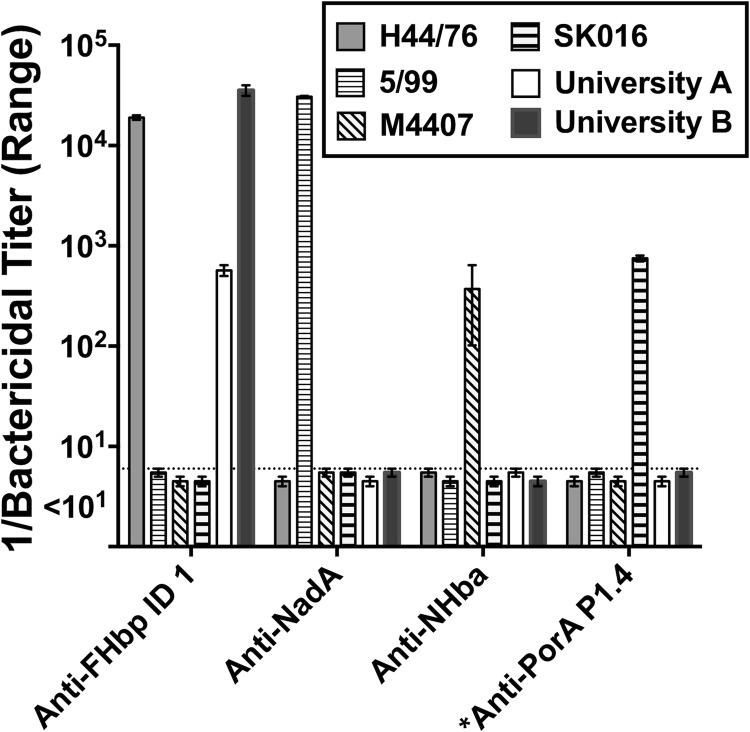
We tested the bactericidal activities of specific antisera from mice immunized with recombinant FHbp, NHba, or NadA (see Materials and Methods). For each antigen, the amino acid sequence of the recombinant protein used to immunize the mice matched the respective antigenic variants used in the MenB-4C vaccine. Similar results were obtained with two isolates tested from each outbreak. We therefore show the data for only one isolate from each outbreak. Despite high NHba expression for both isolates and moderately high NadA expression by the isolate from university B, the two isolates were susceptible only to human complement-mediated bactericidal activity elicited by antibodies to FHbp (Fig. 2). The mouse anti-FHbp bactericidal titer against the isolate from university B was ∼44-fold higher than the titer against the isolate from university A (Fig. 3B). As positive controls, the respective mouse antisera were each bactericidal against control strains (Table 1 and Fig. 2) matched for the corresponding antigen and mismatched for the other three antigens in the MenB-4C vaccine: strain SK016 (matched for PorA P1.4), H44/76 (matched for FHbp), 5/99 (matched for NadA), and M4407 (matched for NHba).
FIG 2.
Bactericidal activity of antisera from mice immunized with individual antigens. Data are shown for one representative isolate from each of the university outbreaks (CH819 and CH840; Table 2) and four control strains (Table 1), each matched for only one of the four antigens in MenB-4C reported to elicit bactericidal activity: H44/76 (FHbp), 5/99 (NadA), M4407 (NHba), and SK016 (PorA P1.4) (Table 1). *, the dilution of the anti-PorA P1.4 MAb that killed strain SK016 is arbitrary; the strain was killed by <0.4 μg/ml. The outbreak strains from university A and university B were killed only by the anti-FHbp antiserum. The data are reported as the means ± ranges of the results from two independent experiments. Similar results were obtained with two other tested outbreak isolates, CH827 and CH838 (from universities A and B, respectively; data not shown). The dotted line represents values of <1:10, the lowest dilution tested.
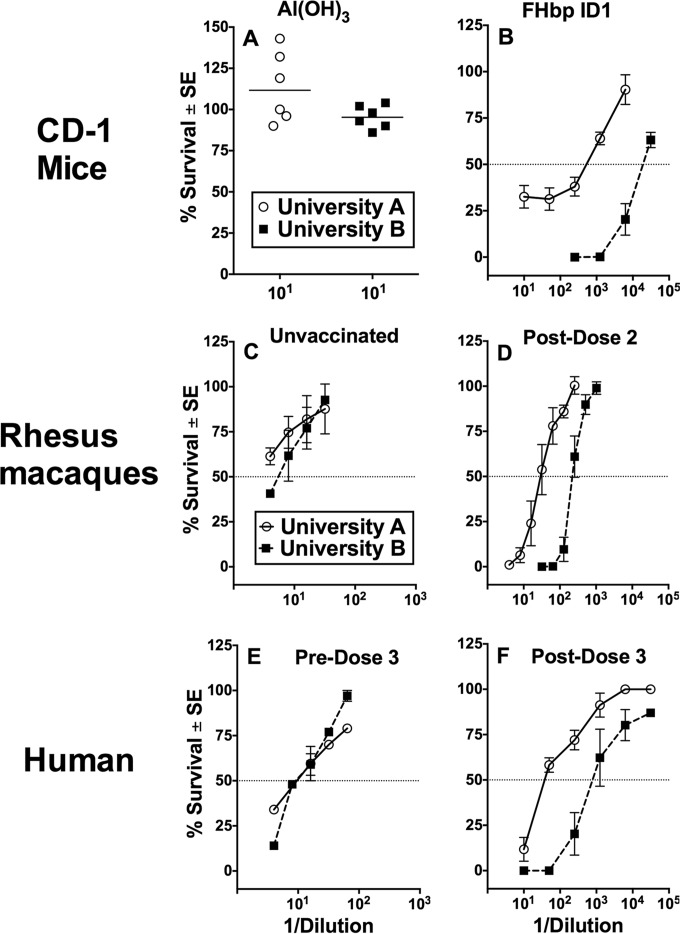
FIG 3.
Serum bactericidal antibody responses, shown as the percent survival of strain CH819 (university A, open circles) or strain CH840 (university B, filled squares) when incubated for 60 min with test serum and 20% human complement. (A) Data shown for 1:10 dilution of individual sera from negative-control mice immunized with aluminum hydroxide alone (∼100% survival, no killing). (B) Responses for sera from mice immunized with a recombinant subfamily B FHbp ID 1 (antigenic variant in MenB-4C vaccine). The mean ± standard error (SE) percent survival values of three serum pools tested at different dilutions (4 mouse sera per pool; 2 to 3 replicate assays) are shown. (C) Responses for sera from three negative-control (Neg. control) unvaccinated macaques (mean ± SE survival at different dilutions). (D) Responses for postimmunization sera from five macaques immunized with MenB-4C vaccine (mean ± SE percent survival at different dilutions tested twice). (E) Responses for serum from a healthy adult human obtained 2 weeks before a third dose of MenB-4C given 5 years after dose 2 (mean ± SE percent survival). (F) Responses for serum from the adult human obtained 6 weeks after dose 3 (mean ± SE from four replicate values). The dotted line represents 50% survival of the bacteria, which defined the endpoint titer.
Rhesus macaque sera.
We measured the bactericidal activities of sera from five infant macaques immunized with two doses of the MenB-4C vaccine. The data are expressed as the mean ± SE percent survival of each of the test isolates when incubated with human complement and different dilutions of postimmunization macaque sera. The three unvaccinated negative-control macaques had negative titers against the isolate from university A (titer, <1:4, the lowest dilution tested) and a titer of ∼1:8 against the isolate from university B (Fig. 3C). After 2 doses of MenB-4C, the serum titer was ∼1:33 against the isolate from university A and ∼1:226 against the isolate from university B (Fig. 3D), which is an 8-fold difference.
Human serum.
We also observed higher bactericidal titers against the isolates from university B outbreak than those from the university A outbreak in serum from an adult human immunized with a third dose of the MenB-4C vaccine (Fig. 3E and F). This individual had received two previous doses of MenB-4C 5 years earlier. The “preserum” for this individual was obtained 2 weeks before dose 3 and had a bactericidal titer of ∼1:10 against both isolates (Fig. 3E). The serum titers after vaccination increased to ∼1:40 against the isolate from university A and ∼1:840 against the isolate from university B (∼21-fold difference between the two isolates). Similar respective titers were obtained against the second isolates from each outbreak (data not shown).
Effect of depletion of serum anti-FHbp antibody on bactericidal activity.
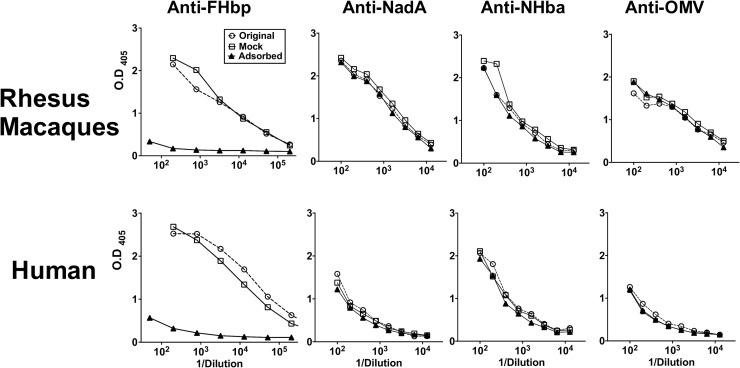
To determine the contribution of anti-FHbp antibodies to the bactericidal activity elicited by the MenB-4C vaccine, we depleted anti-FHbp antibodies from pooled sera from the five macaques immunized with MenB-4C and from the postimmunization serum of the immunized human. By ELISA, the depletion removed >99% of the serum anti-FHbp antibodies but had no significant effect on serum IgG antibody titers to NadA, NHba, or OMV (Fig. 4).
FIG 4.
Depletion of serum anti-FHbp antibodies. A post-dose 2 immunization serum pool from five rhesus macaques vaccinated with MenB-4C, and a post-dose 3 immunization serum from an immunized human were depleted of anti-FHbp antibodies by incubation with FHbp coupled with Sepharose (see Materials and Methods). The untreated sera, the FHbp-depleted sera, and the mock-adsorbed sera were tested by ELISA for IgG antibodies to FHbp, NadA, NHba, and OMV. The FHbp depletion removed >99% of the anti-FHbp antibodies but had no significant effect on the serum antibody titers to NadA, NHba, or OMV.
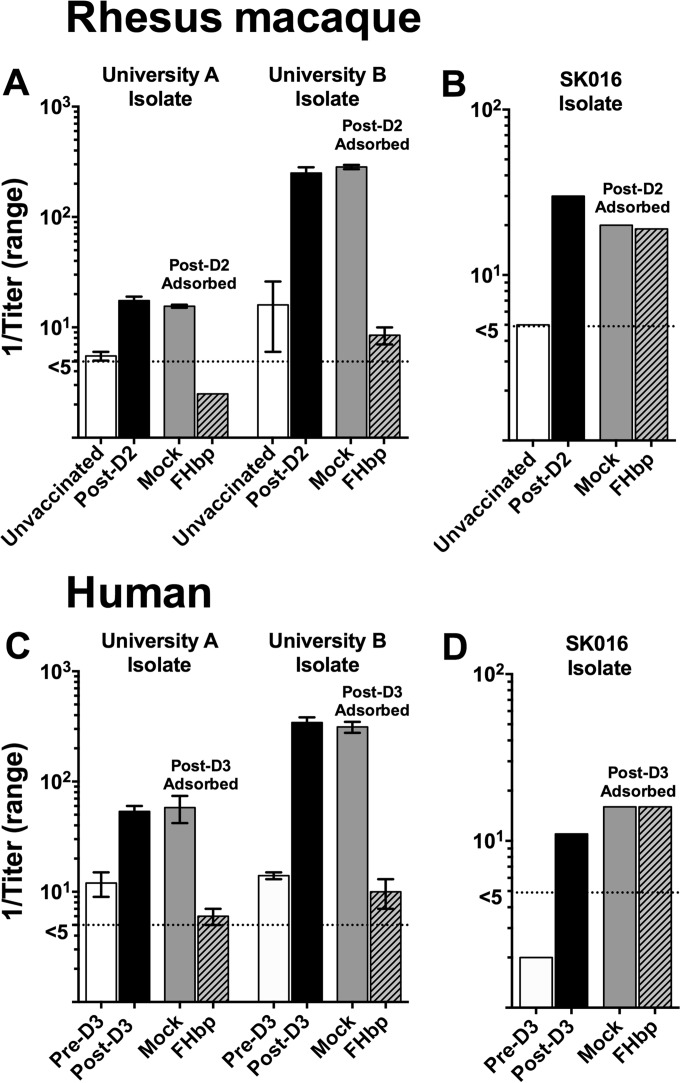
Despite the high expression of NHba by the isolates from the university A outbreak and high NHba and moderate NadA expression by the isolates from the university B outbreak, the depletion of anti-FHbp antibodies from both the postimmunization macaque and human sera removed all or nearly all of the vaccine-induced bactericidal activity (Fig. 5A and C). In contrast, the depletion of anti-FHbp antibodies had no effect on serum anti-PorA bactericidal activity against a control strain, SK016 (Fig. 5B and D), which was mismatched for all of the antigens in MenB-4C except PorA P1.4.
FIG 5.
Effect of depletion of serum anti-FHbp antibody on bactericidal activity. (A) Bactericidal activity of macaque serum depleted of anti-FHbp antibody (FHbp) against isolate CH819 (university A) or CH840 (university B). Serum pools from unvaccinated animals (n = 3) and animals immunized with two doses of MenB-4C vaccine (post-dose 2 [post-D2], n = 5) were obtained as part of a previous study (16). Serum pools were depleted using an FHbp-Sepharose column or a nonconjugated Sepharose column as a control (mock) (see Materials and Methods). Reciprocal mean bactericidal titers with ranges from two replicate assays are shown. (B) Macaque serum bactericidal activity measured against control strain SK016 (mismatched for all of the MenB-4C antigens except PorA P1.4 [8]). (C) Bactericidal activity of human serum from a subject immunized with MenB-4C 5 years after two previous doses (18). Pre-D3 refers to serum obtained 2 weeks before dose 3 (approximately 5 years after doses 1 and 2). The post-dose 3 serum was obtained 3 weeks after vaccination. (D) Human serum bactericidal activity measured against control strain SK016 (mismatched for all of the MenB-4C antigens except PorA P1.4 [8]). The dotted line represents titers of <1:5, the lowest dilution tested.
DISCUSSION
This study investigated the susceptibilities of serogroup B case isolates from two meningococcal outbreaks on U.S. university campuses to serum bactericidal antibodies elicited by the MenB-4C vaccine. Our most important finding was that the isolates from both outbreaks were susceptible to vaccine-induced serum bactericidal activity. However, the isolates from the university B outbreak were more susceptible than those from the university A outbreak. Further, for both outbreaks, despite strain expression of two or three antigens that matched antigens in the MenB-4C vaccine, the major target of the vaccine-induced serum bactericidal activity was FHbp.
FHbp can be classified into three variant groups (23) or two subfamilies (24) based on amino acid sequence similarities. In North America and Europe, approximately 50 to 60% of serogroup B case isolates have subfamily B FHbp (variant group 1), with the remaining isolates having subfamily A FHbp (variant group 2 or 3) (24, 25). Considerable data indicate that FHbp vaccines elicit complement-mediated serum bactericidal activity, primarily against strains that express an FHbp sequence variant from the same subfamily or variant group as the vaccine antigen (23, 26–28). Although the isolates from both university outbreaks expressed subfamily B FHbp, which matched the subfamily B FHbp antigen in MenB-4C FHbp, the university B isolates were much more susceptible to anti-FHbp bactericidal activity than the university A isolates.
Previous studies identified several factors that can affect anti-FHbp bactericidal activity, even when there is a match between the vaccine FHbp subfamily and the subfamily of the strain. The most important factors are the extent of amino acid identity between the strain FHbp variant and vaccine antigen (16, 22, 29) and the level of strain FHbp expression (5, 7, 30, 31). Additionally, the ability of meningococci to bind human FH using alternative ligands, such as NspA (32, 33) or PorB (34, 35), can increase resistance to anti-FHbp bactericidal activity (34) (by FH downregulation of complement activation). It is noteworthy that the more susceptible isolates from the university B outbreak expressed FHbp ID 1, which is an exact match to FHbp in the MenB-4C vaccine, whereas the less susceptible isolates from university A expressed FHbp ID 276, with 96% amino acid identity to ID 1. In immunized mice, even this small difference in amino acid sequence can have a large effect on anti-FHbp bactericidal activity, depending on the locations of the amino acid residues (29). Further, while the isolates from both outbreaks were high expressers of FHbp, the university B isolates had greater surface-accessible FHbp detected by flow cytometry than the isolates from university A. Thus, the greater anti-FHbp bactericidal titers against the university B isolates likely resulted from greater expression of an FHbp that exactly matched the variant present in the vaccine. However, we cannot rule out a role for other factors that might have rendered the isolates from the university A outbreak more resistant to complement-mediated bactericidal activity or the isolates from the university B outbreak more susceptible.
We anticipated that NHba (36–38), which was abundantly expressed by the isolates from both outbreaks, would elicit serum bactericidal antibodies (5). However, the isolates from both outbreaks were resistant to the bactericidal activity of mouse anti-NHba antiserum. Further, the depletion of anti-FHbp antibodies from postimmunization sera from the MenB-4C-vaccinated macaques removed all of the bactericidal activity against the isolates from university A and most of the bactericidal activity against isolates from university B. The depletion of anti-FHbp antibodies from the post-dose 3 immunization human serum decreased bactericidal titers against both university A and B isolates to those obtained before dose 3 (titers of approximately 1:10; Fig. 5). This subject had received two MenB-4C doses 5 years earlier and was a microbiologist working with N. meningitidis on nearly a daily basis. Thus, the low levels of serum bactericidal activity before dose 3 may have arisen from the earlier vaccination, naturally acquired antibodies, and/or inadvertent laboratory exposure despite working in a biosafety level 2+ (BSL2+) facility.
In our previous study, the depletion of either anti-FHbp or anti-NHBA antibodies from the sera of adults who had been immunized with the MenB-3C vaccine (i.e., similar to the MenB-4C vaccine but without the OMV component) resulted in a loss of serum bactericidal activity against test strains mismatched for all of the MenB-4C antigens except NHba (10). We interpreted these results as showing “cooperative” bactericidal activity from vaccine-induced antibodies to NHba and cross-reacting antibodies to subfamily A FHbp, which individually were not bactericidal. In the present study, the isolates from both outbreaks had subfamily B FHbp, which matched the subfamily of FHbp in the MenB-4C vaccine. While we did not deplete anti-NHba antibodies, the data from tests of the antisera from mice immunized with individual antigens showed that subfamily B anti-FHbp antibodies alone were sufficient for bactericidal activity, while the isolates were resistant to anti-NHba bactericidal activity. The basis for the resistance of the isolates from both university outbreaks to bactericidal activity by the mouse anti-NHba antiserum, and the postimmunization sera from macaques or human immunized with MenB-4C, which had been depleted to remove anti-FHbp antibodies, is unknown and will require further investigation.
Preliminary data presented at the June 2015 meeting of the U.S. Advisory Committee on Immunization Practices (39) are consistent with the present findings showing that the isolates from the university A outbreak are relatively resistant to serum bactericidal activity elicited by MenB-4C vaccination.
The measurement of antigen expression and/or cross-reactivity by a meningococcal antigen typing system (MATS) has been used extensively to predict MenB-4C vaccine coverage (40–44). Further, the results have been reported to be a conservative predictor of coverage (45). However, the results of the preliminary analysis of the serum bactericidal antibody responses of the vaccinated students from university A, together with the present data showing large differences in susceptibility to MenB-4C-induced bactericidal activity by isolates from the two college outbreaks, underscore that our understanding of the factors affecting strain susceptibility to bactericidal activity remains incomplete. As a result, the prediction of vaccine strain coverage by antibodies to different MenB-4C antigens will require additional study.
ACKNOWLEDGMENTS
The work was supported by grants R01 AI046464 (to D.M.G.), R01 AI099125 (to P.T.B.) and R01 AI114701 (to D.M.G. and P.T.B.) from the National Institute of Allergy and Infectious Diseases, NIH. The work was performed in a facility funded by the Research Facilities Improvement Program grant C06 RR016226 from the National Center for Research Resources, NIH.
We thank the Meningitis Laboratory, the Meningitis and Vaccine Preventable Diseases Branch, Centers for Disease Control and Prevention (CDC), Atlanta, GA, for providing the isolates from both outbreaks and for the information on PorB and PorA, FHbp, NadA, and NHba vaccine variants inferred from DNA sequencing studies. The isolates were provided to the CDC by the California and New Jersey Departments of Health.
REFERENCES
- 1.Whelan J, Bambini S, Biolchi A, Brunelli B, Robert-Du Ry Van Beest Holle M. 2015. Outbreaks of meningococcal B infection and the 4CMenB vaccine: historical and future perspectives. Expert Rev Vaccines 14:713–736. doi: 10.1586/14760584.2015.1004317. [DOI] [PubMed] [Google Scholar]
- 2.McNamara LA, Shumate AM, Johnsen P, MacNeil JR, Patel M, Bhavsar T, Cohn AC, Dinitz-Sklar J, Duffy J, Finnie J, Garon D, Hary R, Hu F, Kamiya H, Kim HJ, Kolligian J Jr, Neglia J, Oakley J, Wagner J, Wagner K, Wang X, Yu Y, Montana B, Tan C, Izzo R, Clark TA. 2015. First use of a serogroup B meningococcal vaccine in the U.S. in response to a university outbreak. Pediatrics 135:798–804. doi: 10.1542/peds.2014-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giuliani MM, Adu-Bobie J, Comanducci M, Arico B, Savino S, Santini L, Brunelli B, Bambini S, Biolchi A, Capecchi B, Cartocci E, Ciucchi L, Di Marcello F, Ferlicca F, Galli B, Luzzi E, Masignani V, Serruto D, Veggi D, Contorni M, Morandi M, Bartalesi A, Cinotti V, Mannucci D, Titta F, Ovidi E, Welsch JA, Granoff D, Rappuoli R, Pizza M. 2006. A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci U S A 103:10834–10839. doi: 10.1073/pnas.0603940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serruto D, Bottomley MJ, Ram S, Giuliani MM, Rappuoli R. 2012. The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: immunological, functional and structural characterization of the antigens. Vaccine 30(Suppl 2):B87–B97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnelly J, Medini D, Boccadifuoco G, Biolchi A, Ward J, Frasch C, Moxon ER, Stella M, Comanducci M, Bambini S, Muzzi A, Andrews W, Chen J, Santos G, Santini L, Boucher P, Serruto D, Pizza M, Rappuoli R, Giuliani MM. 2010. Qualitative and quantitative assessment of meningococcal antigens to evaluate the potential strain coverage of protein-based vaccines. Proc Natl Acad Sci U S A 107:19490–19495. doi: 10.1073/pnas.1013758107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Shutt KA, Vuong JT, Cohn A, MacNeil J, Schmink S, Plikaytis B, Messonnier NE, Harrison LH, Clark TA, Mayer LW. 2015. Changes in the population structure of invasive Neisseria meningitidis in the United States after quadrivalent meningococcal conjugate vaccine licensure. J Infect Dis 211:1887–1894. [DOI] [PubMed] [Google Scholar]
- 7.Welsch JA, Ram S, Koeberling O, Granoff DM. 2008. Complement-dependent synergistic bactericidal activity of antibodies against factor H-binding protein, a sparsely distributed meningococcal vaccine antigen. J Infect Dis 197:1053–1061. doi: 10.1086/528994. [DOI] [PubMed] [Google Scholar]
- 8.Costa I, Pajon R, Granoff DM. 2014. Human factor H (FH) impairs protective meningococcal anti-FHbp antibody responses and the antibodies enhance FH binding. mBio 5(5):e01625-14. doi: 10.1128/mBio.01625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snape MD, Dawson T, Oster P, Evans A, John TM, Ohene-Kena B, Findlow J, Yu LM, Borrow R, Ypma E, Toneatto D, Pollard AJ. 2010. Immunogenicity of two investigational serogroup B meningococcal vaccines in the first year of life: a randomized comparative trial. Pediatr Infect Dis J 29:e71–e79. [DOI] [PubMed] [Google Scholar]
- 10.Vu DM, Wong TT, Granoff DM. 2011. Cooperative serum bactericidal activity between human antibodies to meningococcal factor H binding protein and neisserial heparin binding antigen. Vaccine 29:1968–1973. doi: 10.1016/j.vaccine.2010.12.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Findlow J, Borrow R, Snape MD, Dawson T, Holland A, John TM, Evans A, Telford KL, Ypma E, Toneatto D, Oster P, Miller E, Pollard AJ. 2010. Multicenter, open-label, randomized phase II controlled trial of an investigational recombinant meningococcal serogroup B vaccine with and without outer membrane vesicles, administered in infancy. Clin Infect Dis 51:1127–1137. doi: 10.1086/656741. [DOI] [PubMed] [Google Scholar]
- 12.Gossger N, Snape MD, Yu LM, Finn A, Bona G, Esposito S, Principi N, Diez-Domingo J, Sokal E, Becker B, Kieninger D, Prymula R, Dull P, Ypma E, Toneatto D, Kimura A, Pollard AJ, European MenB Vaccine Study Group . 2012. Immunogenicity and tolerability of recombinant serogroup B meningococcal vaccine administered with or without routine infant vaccinations according to different immunization schedules: a randomized controlled trial. JAMA 307:573–582. [DOI] [PubMed] [Google Scholar]
- 13.Giuntini S, Reason DC, Granoff DM. 2011. Complement-mediated bactericidal activity of anti-factor H binding protein monoclonal antibodies against the meningococcus relies upon blocking factor H binding. Infect Immun 79:3751–3759. doi: 10.1128/IAI.05182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vu DM, Pajon R, Reason DC, Granoff DM. 2012. A broadly cross-reactive monoclonal antibody against an epitope on the N-terminus of meningococcal fHbp. Sci Rep 2:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granoff DM, Bartoloni A, Ricci S, Gallo E, Rosa D, Ravenscroft N, Guarnieri V, Seid RC, Shan A, Usinger WR, Tan S, Mchugh YE, Moe GR. 1998. Bactericidal monoclonal antibodies that define unique meningococcal B polysaccharide epitopes that do not cross-react with human polysialic acid. J Immunol 160:5028–5036. [PubMed] [Google Scholar]
- 16.Granoff DM, Costa I, Konar M, Giuntini S, Van Rompay KK, Beernink PT. 2015. Binding of complement factor H (FH) decreases protective anti-FH binding protein antibody responses of infant rhesus macaques immunized with a meningococcal serogroup B vaccine. J Infect Dis 212:784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Committee for the Update of the Guide for the Care and Use of Laboratory Animals. 2011. Guide for the care and use of laboratory animals, 8th ed National Research Council, National Academies Press, Washington, DC: https://grants.nih.gov/grants/olaw/Guide-for-the-Care-and-use-of-laboratory-animals.pdf. [Google Scholar]
- 18.Beernink PT, Giuntini S, Costa I, Lucas AH, Granoff DM. 2015. Functional analysis of the human antibody response to meningococcal Factor H binding protein. mBio 6(3):e00842-15. doi: 10.1128/mBio.00842-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beernink PT, Shaughnessy J, Braga EM, Liu Q, Rice PA, Ram S, Granoff DM. 2011. A meningococcal factor H binding protein mutant that eliminates factor H binding enhances protective antibody responses to vaccination. J Immunol 186:3606–3614. doi: 10.4049/jimmunol.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bambini S, De Chiara M, Muzzi A, Mora M, Lucidarme J, Brehony C, Borrow R, Masignani V, Comanducci M, Maiden MC, Rappuoli R, Pizza M, Jolley KA. 2014. Neisseria adhesin A variation and revised nomenclature scheme. Clin Vaccine Immunol 21:966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bambini S, Piet J, Muzzi A, Keijzers W, Comandi S, De Tora L, Pizza M, Rappuoli R, van de Beek D, van der Ende A, Comanducci M. 2013. An analysis of the sequence variability of meningococcal fHbp, NadA and NHBA over a 50-year period in the Netherlands. PLoS One 8:e65043. doi: 10.1371/journal.pone.0065043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pajon R, Beernink PT, Harrison LH, Granoff DM. 2010. Frequency of factor H-binding protein modular groups and susceptibility to cross-reactive bactericidal activity in invasive meningococcal isolates. Vaccine 28:2122–2129. doi: 10.1016/j.vaccine.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masignani V, Comanducci M, Giuliani MM, Bambini S, Adu-Bobie J, Arico B, Brunelli B, Pieri A, Santini L, Savino S, Serruto D, Litt D, Kroll S, Welsch JA, Granoff DM, Rappuoli R, Pizza M. 2003. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J Exp Med 197:789–799. doi: 10.1084/jem.20021911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy E, Andrew L, Lee KL, Dilts DA, Nunez L, Fink PS, Ambrose K, Borrow R, Findlow J, Taha MK, Deghmane AE, Kriz P, Musilek M, Kalmusova J, Caugant DA, Alvestad T, Mayer LW, Sacchi CT, Wang X, Martin D, von Gottberg A, du Plessis M, Klugman KP, Anderson AS, Jansen KU, Zlotnick GW, Hoiseth SK. 2009. Sequence diversity of the factor H binding protein vaccine candidate in epidemiologically relevant strains of serogroup B Neisseria meningitidis. J Infect Dis 200:379–389. doi: 10.1086/600141. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Cohn A, Comanducci M, Andrew L, Zhao X, MacNeil JR, Schmink S, Muzzi A, Bambini S, Rappuoli R, Pizza M, Murphy E, Hoiseth SK, Jansen KU, Anderson AS, Harrison LH, Clark TA, Messonnier NE, Mayer LW. 2011. Prevalence and genetic diversity of candidate vaccine antigens among invasive Neisseria meningitidis isolates in the United States. Vaccine 29:4739–4744. doi: 10.1016/j.vaccine.2011.04.092. [DOI] [PubMed] [Google Scholar]
- 26.Fletcher LD, Bernfield L, Barniak V, Farley JE, Howell A, Knauf M, Ooi P, Smith RP, Weise P, Wetherell M, Xie X, Zagursky R, Zhang Y, Zlotnick GW. 2004. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect Immun 72:2088–2100. doi: 10.1128/IAI.72.4.2088-2100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seib KL, Brunelli B, Brogioni B, Palumbo E, Bambini S, Muzzi A, DiMarcello F, Marchi S, van der Ende A, Arico B, Savino S, Scarselli M, Comanducci M, Rappuoli R, Giuliani MM, Pizza M. 2011. Characterization of diverse subvariants of the meningococcal factor H (fH) binding protein for their ability to bind fH, to mediate serum resistance, and to induce bactericidal antibodies. Infect Immun 79:970–981. doi: 10.1128/IAI.00891-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beernink PT, Granoff DM. 2008. Bactericidal antibody responses induced by meningococcal recombinant chimeric factor H-binding protein vaccines. Infect Immun 76:2568–2575. doi: 10.1128/IAI.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konar M, Granoff DM, Beernink PT. 2013. Importance of inhibition of binding of complement factor H for serum bactericidal antibody responses to meningococcal factor H-binding protein vaccines. J Infect Dis 208:627–636. doi: 10.1093/infdis/jit239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pajon R, Fergus AM, Koeberling O, Caugant DA, Granoff DM. 2011. Meningococcal factor H binding proteins in epidemic strains from Africa: implications for vaccine development. PLoS Negl Trop Dis 5:e1302. doi: 10.1371/journal.pntd.0001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang HQ, Hoiseth SK, Harris SL, McNeil LK, Zhu D, Tan C, Scott AA, Alexander K, Mason K, Miller L, DaSilva I, Mack M, Zhao XJ, Pride MW, Andrew L, Murphy E, Hagen M, French R, Arora A, Jones TR, Jansen KU, Zlotnick GW, Anderson AS. 2010. Broad vaccine coverage predicted for a bivalent recombinant factor H binding protein based vaccine to prevent serogroup B meningococcal disease. Vaccine 28:6086–6093. doi: 10.1016/j.vaccine.2010.06.083. [DOI] [PubMed] [Google Scholar]
- 32.Lewis LA, Carter M, Ram S. 2012. The relative roles of factor H binding protein, neisserial surface protein A, and lipooligosaccharide sialylation in regulation of the alternative pathway of complement on meningococci. J Immunol 188:5063–5072. doi: 10.4049/jimmunol.1103748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis LA, Ngampasutadol J, Wallace R, Reid JE, Vogel U, Ram S. 2010. The meningococcal vaccine candidate neisserial surface protein A (NspA) binds to factor H and enhances meningococcal resistance to complement. PLoS Pathog 6:e1001027. doi: 10.1371/journal.ppat.1001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giuntini S, Pajon R, Ram S, Granoff DM. 2015. Binding of complement factor H to PorB3 and NspA enhances resistance of Neisseria meningitidis to anti-FHbp bactericidal activity. Infect Immun 83:1536–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis LA, Vu DM, Vasudhev S, Shaughnessy J, Granoff DM, Ram S. 2013. Factor H-dependent alternative pathway inhibition mediated by porin B contributes to virulence of Neisseria meningitidis. mBio 4(5):e00339-13. doi: 10.1128/mBio.00339-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welsch JA, Moe GR, Rossi R, Adu-Bobie J, Rappuoli R, Granoff DM. 2003. Antibody to genome-derived neisserial antigen 2132, a Neisseria meningitidis candidate vaccine, confers protection against bacteremia in the absence of complement-mediated bactericidal activity. J Infect Dis 188:1730–1740. doi: 10.1086/379375. [DOI] [PubMed] [Google Scholar]
- 37.Serruto D, Spadafina T, Ciucchi L, Lewis LA, Ram S, Tontini M, Santini L, Biolchi A, Seib KL, Giuliani MM, Donnelly JJ, Berti F, Savino S, Scarselli M, Costantino P, Kroll JS, O'Dwyer C, Qiu J, Plaut AG, Moxon R, Rappuoli R, Pizza M, Arico B. 2010. Neisseria meningitidis GNA2132, a heparin-binding protein that induces protective immunity in humans. Proc Natl Acad Sci U S A 107:3770–3775. doi: 10.1073/pnas.0915162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giuliani MM, Biolchi A, Serruto D, Ferlicca F, Vienken K, Oster P, Rappuoli R, Pizza M, Donnelly J. 2010. Measuring antigen-specific bactericidal responses to a multicomponent vaccine against serogroup B meningococcus. Vaccine 28:5023–5030. doi: 10.1016/j.vaccine.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 39.MacNeil J. 2015. Considerations for use of serogroup B meningococcal (MenB) vaccines in adolescents. Advisory Committee on Immunization Practices, Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/vaccines/acip/meetings/downloads/slides-2015-06/mening-03-macneil.pdf. [Google Scholar]
- 40.Tzanakaki G, Hong E, Kesanopoulos K, Xirogianni A, Bambini S, Orlandi L, Comanducci M, Muzzi A, Taha MK. 2014. Diversity of Greek meningococcal serogroup B isolates and estimated coverage of the 4CMenB meningococcal vaccine. BMC Microbiol 14:111. doi: 10.1186/1471-2180-14-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Křížová P, Musilek M, Vacková Z, Kozáková J, Claus H, Vogel U, Medini D. 2014. Predicted strain coverage of a new protein-based meningococcal vaccine in the Czech Republic. Epidemiol Mikrobiol Imunol 63:103–106. [PubMed] [Google Scholar]
- 42.Vogel U, Taha MK, Vazquez JA, Findlow J, Claus H, Stefanelli P, Caugant DA, Kriz P, Abad R, Bambini S, Carannante A, Deghmane AE, Fazio C, Frosch M, Frosi G, Gilchrist S, Giuliani MM, Hong E, Ledroit M, Lovaglio PG, Lucidarme J, Musilek M, Muzzi A, Oksnes J, Rigat F, Orlandi L, Stella M, Thompson D, Pizza M, Rappuoli R, Serruto D, Comanducci M, Boccadifuoco G, Donnelly JJ, Medini D, Borrow R. 2013. Predicted strain coverage of a meningococcal multicomponent vaccine (4CMenB) in Europe: a qualitative and quantitative assessment. Lancet Infect Dis 13:416–425. doi: 10.1016/S1473-3099(13)70006-9. [DOI] [PubMed] [Google Scholar]
- 43.Bettinger JA, Scheifele DW, Halperin SA, Vaudry W, Findlow J, Borrow R, Medini D, Tsang R, Members of the Canadian Immunization Monitoring Program, Active (IMPACT) . 2013. Diversity of Canadian meningococcal serogroup B isolates and estimated coverage by an investigational meningococcal serogroup B vaccine (4CMenB). Vaccine 32:124–130. doi: 10.1016/j.vaccine.2013.03.063. [DOI] [PubMed] [Google Scholar]
- 44.Vogel U, Stefanelli P, Vazquez J, Taha MK, Claus H, Donnelly J. 2012. The use of vaccine antigen characterization, for example by MATS, to guide the introduction of meningococcus B vaccines. Vaccine 30(Suppl 2):B73–B77. [DOI] [PubMed] [Google Scholar]
- 45.Frosi G, Biolchi A, Lo Sapio M, Rigat F, Gilchrist S, Lucidarme J, Findlow J, Borrow R, Pizza M, Giuliani MM, Medini D. 2013. Bactericidal antibody against a representative epidemiological meningococcal serogroup B panel confirms that MATS underestimates 4CMenB vaccine strain coverage. Vaccine 31:4968–4974. doi: 10.1016/j.vaccine.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 46.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]