Abstract
Coinfections involving porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2 (PCV2) contribute to a group of disease syndromes known as porcine circovirus-associated disease (PCVAD). Presumably, PRRSV infection enhances PCV2 replication as a result of modulation of host immunity. The purpose of this study was to evaluate PCV2 replication and pathogenesis in pigs vaccinated with a PRRS modified live virus (MLV) vaccine and subsequently challenged with a combination of PRRSV and PCV2. During the early postchallenge period, the number of pigs with PRRSV-associated clinical signs was decreased, and average daily gain (ADG) was increased, in the vaccinated group, demonstrating the protective effect of PRRS vaccination. However, during the later postchallenge period, more pigs in the vaccinated group showed increased PCV2 viremia, decreased ADG, increased PCVAD clinical signs, and increased mortality. In this disease model, the early benefits of PRRSV vaccination were outweighed by the later amplification of PCVAD.
INTRODUCTION
Porcine circovirus type 2 (PCV2), a single-stranded DNA virus in the family Circoviridae, contributes to a group of syndromes collectively termed porcine circovirus-associated disease (PCVAD) (1). Two important clinical syndromes associated with PCVAD are PCV2-associated pneumonia and postweaning multisystemic wasting syndrome (PMWS) (1, 2). Management of PCV2 through the use of inactivated and subunit vaccines has led to the effective control of PCVAD in North America and Europe. However, the emergence of new PCV2 strains and the lack of PCV2 vaccination programs in other countries create an uncertain future for continued disease control.
Porcine reproductive and respiratory syndrome virus (PRRSV) is a single-stranded RNA virus in the family Arteriviridae (3, 4). For the past 20 years, PRRSV has remained the most costly disease affecting swine production worldwide (5). PRRSV infection contributes to a number of immunological outcomes that increase the susceptibility of the host to secondary infections by primary and secondary pathogens (6–8). PRRSV is frequently isolated along with PCV2 (9) and is one of the major cofactors linked with increasing PCV2 replication and pathogenesis (10–12). Previous work by us and others has shown that a principal contribution of PRRSV is to increase PCV2 viremia (13). Increased PCV2 replication is likely the result of immune stimulation that results in more PCV2-permissive cells combined with PRRSV-induced immunomodulation. The complex etiology of PCVAD, including the role of PRRSV infection, has yet to be fully understood. In an extensive body of work, we identified the aberrant recognition of a nonneutralizing decoy epitope on the PCV2 capsid protein (CP) as a contributing factor in PCVAD immunopathogenesis. Natural PCV2 infection of a population produces a mixture of pigs that recognize the decoy and neutralizing epitopes, which may explain why only a subpopulation of infected pigs goes on to develop PCVAD (13–15).
In this study, we took advantage of a host genetics study to evaluate clinical and virological outcomes after experimental challenge with PCV2 and PRRSV in pigs with or without prior vaccination with a commercial PRRS modified live virus (MLV). The results demonstrate the protective properties of vaccination; however, the short-term benefit is outweighed by the longer-term impact of MLV on PCVAD.
MATERIALS AND METHODS
Animals and housing.
Experiments involving animals and viruses were performed in accordance with the Guide for the Care and Use of Agricultural Animals in Research and Teaching by the Federation of Animal Science Societies (FASS) (16) and with the USDA Animal Welfare Act and Animal Welfare Regulations and were approved by the Kansas State University Institutional Animal Care and Use Committees and Institutional Biosafety Committees. The study was conducted as part of the evaluation of a previously described genomic marker, WUR (17). The population used in this study was composed of pigs with two genotypes: 50% had WUR genotype AA, and 50% had WUR genotype AB or BB. The AB and BB genotypes were predicted to have beneficial effects on the response to PRRSV infection. Both the vaccine group and the nonvaccine group were balanced according to WUR genotype; therefore, WUR was not a factor in the comparison of the outcomes of the vaccine and nonvaccine groups. Three-week-old barrows (n = 226; average age, 19.4 ± 1.8 days) were obtained from a high-health commercial source negative for PRRSV. While the pigs were derived from a sow herd previously vaccinated with a PCV2 capsid subunit vaccine, the piglets were not vaccinated for PCV2 and were obtained after weaning without regard to maternal antibody levels. All pigs were housed in two environmentally controlled rooms at the Kansas State University Large Animal Research Center and were maintained under biosafety level 2 (BSL-2) conditions. Rooms were chemically disinfected, cleaned with a high-heat pressure washer, and gas decontaminated with vaporized hydrogen peroxide prior to use. Both rooms were empty for at least 19 days prior to the start of the study. Pigs were housed in 20 pens, each with an area of 144 ft2, with 11 to 12 pigs per pen. Pigs were given access to food and water ad libitum.
Experimental design.
A total of 226 pigs were randomly allocated to two identical rooms by use of a random number assignment protocol and were housed in groups of 11 to 12 pigs per pen. After acclimation for 4 days, 115 pigs in one room were vaccinated with a 2-ml dose of a commercial PRRS MLV vaccine (Ingelvac PRRS MLV; Boehringer Ingelheim Animal Health; GenBank accession no. AF159149) administered intramuscularly according to the vaccine label instructions. At 28 days postvaccination (dpv), all pigs in both rooms were challenged with a combination of PRRSV and PCV2b. Individual body weights were determined on days −3, 0, 7, 14, 21, 28, 35, 42, 49, 56, 63, and 70 postvaccination. Blood samples were collected from all pigs at 0, 4, 7, 11, 14, 21, 28, 35, and 42 days postinfection (dpi). Blood was also collected from the vaccinated group at 0, 4, 7, 11, 14, and 21 dpv. At 11 dpi, 10 vaccinated and 10 nonvaccinated pigs were randomly selected for humane euthanasia, and complete necropsies were performed. Between days 32 and 42 postinfection, 11 pigs showing clinical signs of PCVAD and 7 pigs not showing such signs were humanely euthanized, and complete necropsies were performed. These pigs were selected on the basis of clinical disease without regard to vaccine status.
Challenge inoculum.
The PRRSV and PCV2b isolates used to prepare the inoculum were originally derived from the lymph node of a pig with severe PMWS, as described previously (13, 14). PRRSV (isolate KS62; GenBank accession no. KM035803) was isolated by propagation on MARC-145 cells. The PRRSV component of the challenge inoculum, KS62, shared 88.06% identity with the MLV (GenBank accession no. AF159149) at the peptide sequence level of GP5. Since wild-type PCV2b (GenBank accession no. JQ692110) does not propagate to high levels in cell culture, we took advantage of the heat stability of PCV2 to make a virus preparation from a lymph node suspension enriched for PCV2. The suspension was heat treated at 55°C for 30 min to remove PRRSV, bacteria, and other heat-labile agents. The treated homogenate was recombined with the isolated PRRSV in order to infect cesarean-derived, colostrum-deprived (CD/CD) pigs. A combination lung/lymph node homogenate was prepared from the CD/CD pigs, and PRRSV and PCV2 were isolated from the homogenate by the methods described above. Analysis of the heat-treated preparation for common agents showed that the preparation was negative for most heat-stable agents, such as parvovirus, but still positive for torque teno sus virus (TTSuV) and porcine oncovirus (PCOV), which are ubiquitous.
The titers of PRRSV were determined on MARC-145 cells. Briefly, the virus was serially diluted 1:10 in minimal essential medium (MEM; Corning) supplemented with 7% fetal bovine serum (FBS; Sigma-Aldrich), penicillin-streptomycin (Pen Strep; 80 U/ml and 80 μg/ml, respectively; Gibco), 3 μg/ml amphotericin B (Fungizone) (Gibco), and 25 mM HEPES (Life Technologies). The dilutions were then added in quadruplicate to confluent MARC-145 cells in a 96-well tissue culture plate (BD Falcon). Following a 4-day incubation at 37°C under 5% CO2, wells were examined for PRRSV-induced cytopathic effects, and the 50% tissue culture infectious dose (TCID50) per milliliter was calculated using the method of Reed and Muench (18).
The quantity of PCV2 was determined by titration on swine testicle (ST) cells. Briefly, serial 10-fold dilutions of the PCV2 challenge stock were plated in quadruplicate into rapidly dividing ST cells in a 96-well tissue culture plate (BD Falcon). Dilutions were prepared in Eagle's minimal essential medium (EMEM; Sigma-Aldrich) supplemented with 7% FBS (Sigma-Aldrich) and 50 μg/ml of gentamicin (Lonza). Following a 3-day incubation at 37°C under 5% CO2, the cells were fixed, permeabilized with 80% acetone, and then stained with fluorescein isothiocyanate (FITC)-labeled porcine anti-PCV (Veterinary Medical Research and Development, Inc.). Infected cells were visualized using an inverted fluorescence microscope, and the TCID50 per milliliter was calculated using the method of Reed and Muench (18).
The challenge viruses were recombined to yield a 2-ml dose consisting of 103.6 TCID50 PCV2 and 105 TCID50 PRRSV in MEM. The 2-ml dose was split, with 1 ml administered intranasally and the remaining 1 ml administered intramuscularly.
Clinical evaluation.
Pigs were evaluated daily for the presence of clinical signs associated with PCVAD, including dyspnea, aural cyanosis, coughing, nasal discharge, open-mouth breathing, poor body condition, muscle wasting, pallor or jaundice, lameness, joint effusion, depression, and lethargy. Each pig was visually examined by a veterinarian or veterinary assistant each day during the study period. Appropriate treatments were initiated for pigs that presented with moderate to severe clinical disease. Examples of clinical presentations where treatment was administered included (i) difficult respiration, (ii) mucoid nasal discharge, (iii) lameness with associated joint effusion, (iv) pallor or jaundice associated with muscle wasting, and (v) lethargy or depression, with a rectal temperature of ≥104°F. For clinically affected pigs, antibiotic therapy was administered, including ceftiofur hydrochloride for respiratory or systemic disease, oxytetracycline for infectious arthritis, and enrofloxacin for cases unresponsive to the previous two antibiotics. All pigs with overt clinical disease and rectal temperatures of ≥104°F were administered flunixin meglumine, a nonsteroidal anti-inflammatory drug (NSAID). Pigs with intractable fevers of >4 days' duration were given a 2-day washout period and were then administered oral meloxicam. All treatments were administered as directed by a veterinarian. Clinical signs and systemic treatments unrelated to PRRSV or PCVAD (e.g., lacerations, dermatitis, hoof wounds, congenital hernias) were documented but were not included in the data analysis related to clinical outcomes. Animals were humanely euthanized with pentobarbital sodium. Pigs that died or were humanely euthanized due to circumstances unrelated to the effects of coinfection were excluded from the mortality analysis. Average daily gain (ADG) was calculated as the change in weight divided by the number of days and was reported in kilograms per day.
Gross pathology and histopathology.
Lungs were removed in toto immediately after euthanasia. Gross lung lesions were scored using two techniques. First, the percentage of the lung affected by pneumonia was estimated for both the dorsal and ventral aspects of each lung lobe during gross necropsy. The results were reported as the percentage of the whole lung affected by pneumonia (ranging from 0 to 100%) (19). Second, the dorsal and ventral aspects of the whole lung were photographed (with an Olympus Stylus 7010 camera), and digital images were evaluated after gross necropsy using a photo scoring system. Gross anatomical photo scores were determined on a scale of 0 to 4, as follows: 0, no macroscopic lesions; 1, pneumonia affecting <25% of gross lung; 2, pneumonia affecting 25 to 50% of gross lung; 3, pneumonia affecting 50 to 75% of gross lung; 4, pneumonia affecting >75% of gross lung. The evaluator was blinded to the sources of the lung pictures.
For histopathology, tissues collected from the lung, tracheobronchial lymph node, and inguinal lymph node were immediately placed in 10% neutral buffered formalin and were allowed to fix for at least 7 days. Fixed tissues were processed in an automated tissue processor and were embedded in paraffin. Slide-mounted tissue sections were stained with hematoxylin and eosin (H&E) and were evaluated by a blinded board-certified pathologist. Microscopic lung lesions were estimated based on the following scoring system: 0, no significant microscopic lesions; 1, mild interstitial pneumonia with <50% lung lobe involvement; 2, mild to moderate multifocal interstitial pneumonia with 50 to 75% lung lobe involvement; 3, moderate to severe multifocal interstitial pneumonia with 50 to 75% lung lobe involvement; 4, severe diffuse interstitial pneumonia with >75% lung lobe involvement. The final score assigned to each pig was an average from two separate evaluations by the same pathologist, who remained blinded to the source of the lung tissue.
PCV2 immunohistochemical (IHC) staining.
PCV2 antigen staining in paraffin-embedded thin sections of tissue was performed by personnel in the Kansas State Veterinary Diagnostic Laboratory. Briefly, deparaffinized slide-mounted thin sections were first treated with proteinase K (1.2 mg/ml diluted in Bond enzyme diluent with 0.35% ProClin 950) for 10 min at room temperature (Bond enzyme pretreatment kit; Leica Biosystems). A rabbit anti-PCV2 antibody (Iowa State University) was diluted 1:500 in Bond primary antibody diluent (Leica Biosystems) and was applied to the tissue section for 15 min at room temperature. Bound antibody was detected by incubation with 25 μg/ml Poly-AP anti-rabbit IgG (Leica Biosystems) in antibody diluent for 25 min at room temperature. The complex was visualized using Fast Red chromogen (Bond Polymer Refine Red Detection kit; Leica Biosystems) and was counterstained with hematoxylin.
Measurement of PRRSV and PCV2 viremia.
Viral DNA and RNA were extracted simultaneously from 50 μl of serum by using Ambion's MagMAX-96 viral isolation kit (Applied Biosystems) in accordance with the manufacturer's instructions. PRRS viral RNA was quantified by using EZ-PRRSV MPX 4.0 real-time reverse transcription-PCR (RT-PCR) target-specific reagents (Tetracore) according to the manufacturer's instructions. For consistency, each plate contained Tetracore quantification standards and control sets for use with EZ-PRRSV MPX 4.0 RT-PCR reagents. All PCRs were carried out on a CFX96 Touch real-time PCR detection system (Bio-Rad) in a 96-well format using the recommended cycling parameters. PCV2 DNA was quantified using SsoAdvanced Universal SYBR green supermix (Bio-Rad). The forward and reverse PCR primers were 5′-AATGCAGAGGCGTGATTGGA-3′ and 5′-CCAGTATGTGGTTTCCGGGT-3′, respectively. The primers were used at a final concentration of 300 μM. Nuclease-free water was used to bring the master mix volume to 18 μl per reaction. The addition of 2 μl of template nucleic acid brought the final reaction volume for each sample to 20 μl. Standard curves and positive and negative controls were included on each plate. Plasmid DNA was used for the PCV2 standard curve and positive-control template. DNA inserted into the plasmid was obtained from a field strain of PCV2 (PCV2b 321/393). Plasmid DNA was isolated by using the PureYield Plasmid Miniprep system (Promega) according to the manufacturer's instructions. The DNA for the standard curve was quantified using a NanoDrop 8000 spectrophotometer. The standard curve was produced by diluting the purified plasmid DNA 1:1,000 in nuclease-free water, followed by five serial 1:10 dilutions in nuclease-free water. The final standard curve contained 6 points ranging from 107 to 102 copies of template DNA, which produced threshold crossing values between 15 and 33 cycles. Standard curves were run in duplicate with nuclease-free water as a negative control. The PCV2 PCR was carried out on a CFX96 Touch real-time PCR detection system using the following settings: activation at 98°C for 2 min, followed by 40 cycles of denaturation at 98°C for 5 s and annealing/extension at 60°C for 10 s. The melting curve was performed between 65 and 95°C using 0.5°C increments. The PCR assay results were reported as log10 PRRSV RNA starting quantity (copy number) per 50-μl reaction volume or log10 PCV2 DNA starting quantity per 20-μl reaction volume.
Microsphere immunoassay for detection of anti-PCV2 antibodies.
PCV2b capsid protein (CP) polypeptide fragments CP(43–233) and CP(160–233) were cloned and expressed in the Escherichia coli vector pHUE as described previously (14). For protein expression, bacteria were grown in Luria-Bertani (LB) broth plus ampicillin (0.01 mg/ml) and were incubated at 37°C with shaking. When the optical density at 600 nm (OD600) reached 0.4 to 0.6, protein expression was induced with isopropyl β-d-1-thiogalactopyranoside (IPTG; final concentration, 1 mM), and bacteria were harvested 4 h later. Protein was purified by using the USB PrepEase histidine-tagged protein purification kit (Affymetrix) under nondenaturing conditions, according to the manufacturer's directions. Purity was assessed by SDS-PAGE, and total protein was measured using the Bio-Rad protein assay.
Proteins were coupled to carboxylated Luminex MagPlex polystyrene microspheres according to the manufacturer's directions. For the assay, approximately 2,500 antigen-coated beads, suspended in 50 μl phosphate-buffered saline with 10% goat serum (PBS-GS), were placed in each well of a 96-well round-bottom polystyrene plate (Costar). Sera were diluted 1:400 in PBS-GS, and 50 μl was added to each well. The plate was wrapped in foil and was incubated for 30 min at room temperature with gentle shaking. The plate was placed on a magnet, and beads were washed three times with 190 μl of PBS-GS. For the detection of IgG, 50 μl of a Biotin-SP-conjugated, affinity-purified goat anti-swine secondary antibody (IgG; Jackson ImmunoResearch) was diluted to 2 μg/ml in PBS-GS, and 100 μl was added to each well. The plate was incubated at room temperature for 30 min and was washed three times, followed by the addition of 50 μl of streptavidin-conjugated phycoerythrin (SAPE) (2 μg/ml in PBS-GS). After 30 min, the plate was washed, and the microspheres were resuspended in 100 μl of PBS-GS. The microspheres were analyzed using a Magpix instrument (Luminex) and Luminex xPONENT software, version 4.2. A minimum of 100 microspheres were used for the calculation of mean fluorescence intensity (MFI). The sample-to-positive (S/P) ratio was calculated as (MFI of sample − MFI of negative control)/(MFI of standard positive control − MFI of negative control).
Statistical analyses.
A logistic mixed model was used to evaluate the effect of vaccination on the binary traits of blue ear, veterinary treatment for PCVAD, and mortality. The model included the design effects of vaccination, WUR, and the interaction between vaccination and WUR, with initial body weight as a covariate. Random effects included sire, dam, and pen. Odds ratios for the effect of vaccination were estimated as the vaccinated group over the nonvaccinated group (treatment and mortality) and the nonvaccinated group over the vaccinated group (blue ear). Odds ratios included Wald's confidence intervals (CI).
Data on the percentage of lung lobe involvement were analyzed as a normal quantitative response variable, whereas gross anatomical photo scores and microscopic lung lesion scores were analyzed as ordinal categorical variables in a mixed multinomial regression. The model included the fixed effects of PRRS vaccination, WUR, their interaction, and clinical signs (presence/absence). Sire and dam were included as random effects. All analyses were performed using the GLIMMIX procedure of SAS, version 9.3 (Statistical Analysis System Institute, Inc.). Comparisons of average daily gain, viremia, and antibody response between groups were performed using GraphPad Prism software, version 5.00, using the unpaired t test.
RESULTS
PRRSV viremia is decreased after PRRS vaccination, but PCV2 viremia is increased.
PRRS MLV replication in the vaccine group prior to challenge was assessed by RT-PCR on serum samples collected at 11 dpv. Of the 84 pig sera tested, 78 (93%) had detectable levels of the vaccine virus, confirming that the pigs supported active MLV replication (Fig. 1). The mean level of viremia at 11 dpv was 2.7 ± 1.7 log10 templates per PCR. Prior to challenge, the nonvaccine group was negative for PRRSV nucleic acid in serum (data not shown).
FIG 1.
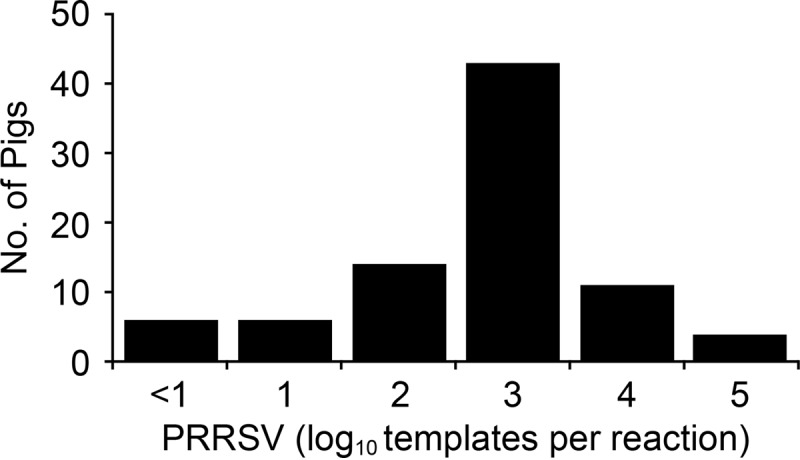
Distribution of viremia at 11 days after vaccination with PRRS MLV. Shown are PRRSV RT-PCR results for 84 pigs in the vaccine group.
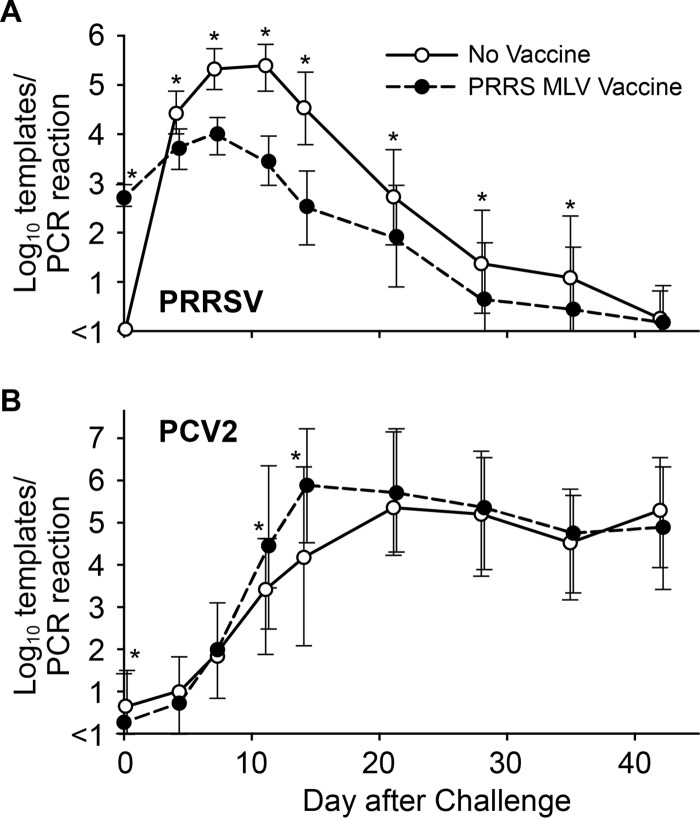
The results for PRRSV and PCV2 viremia after challenge are shown in Fig. 2. PRRSV infection in the nonvaccine group followed the typical course of viremia, peaking between 7 and 11 dpi, followed by decay and the eventual disappearance of virus from the blood by 42 dpi. In the vaccine group, 90% of pigs (102/113) had detectable levels of virus nucleic acid in serum at 28 days after vaccination or at the time of challenge. The PRRSV viremia in the vaccine group peaked at about 7 days after challenge and then decayed. Except for the day of challenge and day 42, the mean PRRSV level was significantly lower on all days in the vaccine group (Fig. 2A). Peak viremia for the vaccine group at days 7 and 11 was reduced by >1 log unit from that for nonvaccinated pigs. The results demonstrated that vaccination was effective in reducing PRRS viremia in a heterologous challenge model.
FIG 2.
PRRSV and PCV2 viremia in vaccinated and nonvaccinated pigs. Values are means ± 1 standard deviation. Asterisks indicate statistically significant differences between groups (P < 0.015 by Student's t test).
Mean PCV2 viremia levels for the vaccine and nonvaccine groups are presented in Fig. 2B. In the nonvaccine group, mean PCV2 viremia peaked at about 21 days after challenge and remained elevated for the remainder of the study. In contrast, mean PCV2 viremia for the vaccine group peaked at 14 dpi, when the mean virus level was approximately 1.5 log units greater for the vaccine group than for the nonvaccine group (P < 0.0001). Thus, PCV2 viremia in the vaccine group peaked much earlier.
PCV2 immunization of dams provides temporary protection of piglets from PCV2 infection. However, by 35 days after weaning, passive immunity decays to the point that pigs become susceptible to PCV2, a virus that is normally present in the environment. At the time of challenge, low but detectable levels of PCV2 nucleic acid were present in 7 of 115 vaccinated pigs (6%) and 25 of 111 nonvaccinated pigs (22%). Mean PCV2 viremia levels prior to challenge were 2.3 and 2.9 log10 templates per PCR for PCR-positive vaccinated and nonvaccinated pigs, respectively. The increased proportion of nonvaccinated pigs with evidence of PCV2 exposure prior to challenge may account for the differences in the outcomes between the two groups. However, exclusion of these 32 pigs did not alter the conclusions of the study.
PRRSV vaccination results in reduced clinical signs and pathology during the first 21 days after coinfection.
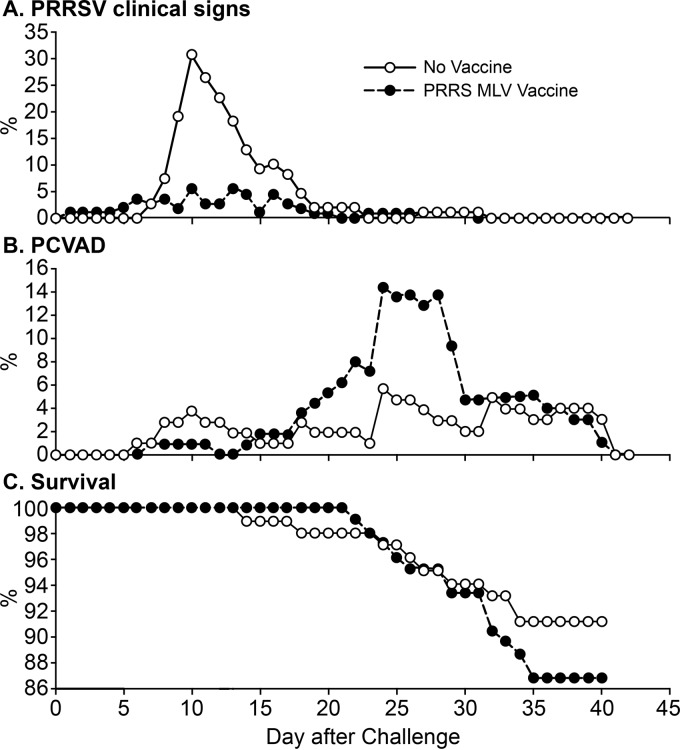
Prior to virus challenge, no clinical signs were apparent in either the vaccinated or the nonvaccinated group. After infection, two clinical syndromes emerged. The first was a PRRSV-associated syndrome, aural cyanosis, commonly known as “blue ear” (20, 21), which was easily identified in pigs by the presence of red, cyanotic, or blue discoloration of the ear tissue. Even though blue ear is not pathognomonic for PRRS, it often coincides with acute infection. A representative example of a pig with blue ear is shown in Fig. 3. No blue ear was observed among either the vaccinated or the nonvaccinated pigs prior to challenge. However, during the postchallenge period, 64 of all 226 pigs (28.3%) were documented as having blue ear on one or more days. As shown in Fig. 4A, the percentage of pigs with blue ear peaked between 8 and 17 dpi, which corresponded to the peak in PRRS viremia (compare Fig. 2A). Overall, 19% of vaccinated pigs (22/115) and 38% of nonvaccinated pigs (42/111) were documented with blue ear. A nonvaccinated pig was 3.02 times (95% CI, 1.7, 5.9) more likely to develop blue ear than a vaccinated pig (P = 0.001). The total numbers of days with blue ear were 65 and 201 for the vaccine and nonvaccine groups, respectively. The reduction in the number of pigs with blue ear in the vaccine group is consistent with a beneficial effect of the PRRS MLV.
FIG 3.
Aural cyanosis, or blue ear. (A) Photograph representative of the ear discoloration associated with aural cyanosis during PRRSV infection. The photograph was taken at 11 days after virus challenge. (B) A normal ear is shown for comparison.
FIG 4.
Clinical outcomes for the vaccinated and nonvaccinated groups after a dual challenge with PRRSV and PCV2. Panel A shows the percentage of pigs with aural cyanosis, a PRRSV-associated syndrome. Clinical signs were assessed as described in Materials and Methods.
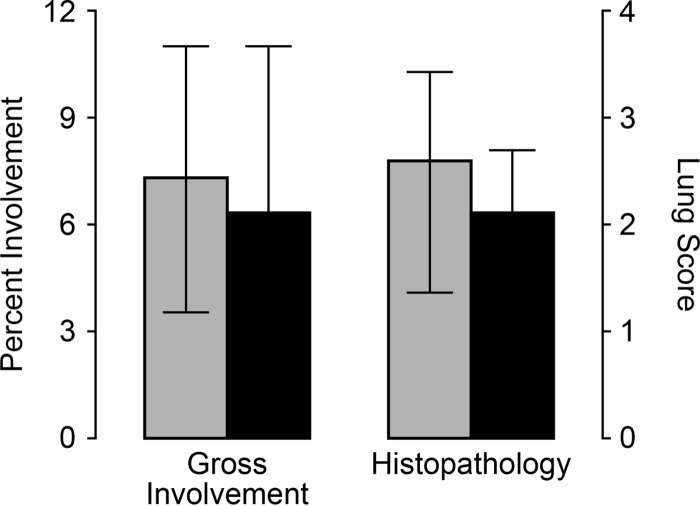
The primary clinical sign associated with acute PRRSV infection is respiratory disease resulting from interstitial pneumonia. Lungs were removed from 20 euthanized pigs (10 pigs from each group), which were randomly selected at 11 dpi. Figure 5 shows a summary of the gross and microscopic lung scores. Mean scores for the percentage of gross lung lobe involvement and microscopic scoring for interstitial pneumonia were higher in the nonvaccine group; however, the differences were not statistically significant (P = 0.62 for the percentage of lung involvement and 0.30 for the histopathology score). Together, the results showed that vaccination with the PRRS MLV had an overall protective effect by reducing PRRSV viremia and decreasing PRRS-associated clinical signs.
FIG 5.
Assessment of lung pathology at 11 days after PRRSV/PCV2 challenge. Ten vaccinated pigs (filled bars) and 10 nonvaccinated pigs (shaded bars) were randomly removed from the study at 11 days and were assessed for pneumonia. Results (means ± standard deviations) are presented as the percentage of gross lung involvement and the histopathology score. The differences between the vaccine and nonvaccine groups were not significant (P > 0.05).
PRRS vaccination results in increased clinical signs and pathology at 22 to 42 days after coinfection.
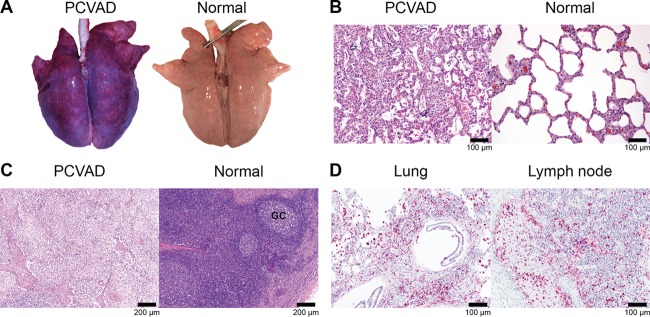
Beyond the acute period of infection, a second clinical syndrome appeared, which first became apparent by an increase in the number of pigs receiving systemic veterinary treatment due to clinical signs associated with PCVAD, such as tachypnea, dyspnea, pyrexia, loss of condition, muscle wasting, mucoid nasal discharge, lethargy, and pallor or jaundice (Fig. 4B). Lesions typical of PCVAD were found by gross anatomical and microscopic examinations of lungs and lymph nodes from pigs that died or were euthanized. Representative pictures and photomicrographs showing the lesions associated with clinically affected pigs are presented in Fig. 6. Lungs showed multifocal to diffuse interstitial pneumonia with mottling of lung tissue, hemorrhage, and consolidation (Fig. 6A). At the microscopic level, multifocal to diffuse interstitial pneumonia with lymphohistiocytic infiltration into the alveolar septa and peribronchiolar areas was easily visible (Fig. 6B). The lymph nodes of affected pigs showed depletion of lymphocytes (Fig. 6C). Positive staining for PCV2 antigen was observed in the lymph nodes and lungs of affected pigs (Fig. 6D). Analysis of gross and microscopic lesions combined with the accumulation of PCV2 antigen in target organs confirmed the presence of PCVAD.
FIG 6.
Gross and microscopic lesions associated with PCVAD. The images shown are representative of the lesions of PCVAD-affected pigs necropsied between 32 and 42 days after combined PRRSV and PCV2 challenge. (A) Set of lungs from a challenged pig with pneumonia, mottling, and consolidation. A normal set of lungs from an age-matched pig is shown for comparison. (B) H&E-stained lung from a challenged pig showing moderate to severe multifocal interstitial pneumonia with lymphohistiocytic infiltration of alveolar septa. A normal lung is shown for comparison. (C) Lymphoid depletion in a lymph node from a challenged pig. A normal lymph node with prominent germinal centers (GC) is shown for comparison. (D) Immunohistochemical staining showing the accumulation of PCV2 antigen in a lung and a lymph node from a challenged pig.
The number of pigs undergoing treatment as a result of PCVAD-associated clinical signs peaked between 22 and 35 dpi (Fig. 4B). During this time, 39 pigs received at least 1 day of veterinary treatment, including 12 nonvaccinated pigs (12/101 [12%]) and 27 pigs in the vaccine group (27/105 [26%]). A vaccinated pig was 2.67 times (95% CI, 1.23, 5.80) more likely to receive veterinary treatment during peak PCVAD than a nonvaccinated pig (P = 0.01). Large amounts of PCV2 in serum were associated with the 39 pigs that went on to develop PCVAD. At 14 dpi, significantly higher levels of circulating PCV2 were present in the 39 PCVAD-affected pigs (mean, 5.8 log10 templates/PCR) than in the 163 pigs without clinical signs (mean, 4.8 log10 templates/PCR) (P = 0.004). The different treatments administered to the 39 pigs with clinical signs included a single antibiotic and an NSAID (16/39 [41%]), multiple antibiotics and an NSAID (7/39 [18%]), a single antibiotic (6/39 [15%]), multiple antibiotics (3/39 [8%]), and an NSAID alone (3/39 [8%]). Four of the 39 pigs (10%) were humanely euthanized after the initial treatment due to the severity of the clinical presentations. The decline in the percentage of pigs with PCVAD clinical signs was largely the result of increased mortality or the euthanization of pigs that were moribund or nonresponsive to treatment (compare Fig. 4B and C). Over the entire study period, 49 pigs received at least 1 day of systemic veterinary treatment: 16% in the nonvaccine group (18/111) and 27% in the vaccine group (31/115). A vaccinated pig was 1.79 times (90% CI, 0.99, 3.25) more likely to receive veterinary treatment than a nonvaccinated pig (P = 0.11) over the entire study period.
Macroscopic and microscopic changes in organs and tissues were evaluated between 32 and 42 dpi for 11 clinically affected pigs, which were humanely euthanized as a result of failure to respond to treatment. For comparison, 7 pigs without clinical signs were also necropsied. As summarized in Table 1, all 11 pigs with clinical signs showed some form of macroscopic lung involvement, as determined by the photographic score. The macroscopic scores for the group without clinical signs were significantly lower (P = 0.04). A similar trend for clinically affected versus nonaffected pigs appeared at the microscopic level; however, the difference was not statistically significant (P = 0.16). Mild to severe lymphoid depletion was observed in 8 of the 11 pigs with clinical signs (73%), compared to only 3 of the 7 (43%) pigs without clinical signs. Even though the pigs without clinical signs appeared normal, almost all showed some form of pathology related to PCVAD, such as mild to moderate pneumonia and/or mild lymphoid depletion.
TABLE 1.
Lung and lymph node lesions of pigs with clinical signs associated with PCVAD and apparently healthy pigs between 32 and 42 dpia
| Score or severity | No. (%) of pigs: |
|
|---|---|---|
| With clinical signs | Apparently healthy | |
| Macroscopic lung lesion scoreb | ||
| 0 | 0 (0) | 1 (14) |
| 1 | 4 (36) | 4 (57) |
| 2 | 2 (18) | 1 (14) |
| 3 | 1 (9) | 1 (14) |
| 4 | 4 (36) | 0 (0) |
| Microscopic lung lesion scorec | ||
| 0 | 0 (0) | 0 (0) |
| 1 | 1 (9) | 1 (14) |
| 2 | 3 (27) | 4 (57) |
| 3 | 7 (64) | 2 (29) |
| 4 | 0 (0) | 0 (0) |
| Level of lymphoid depletiond | ||
| None | 3 (27) | 4 (57) |
| Mild | 4 (36) | 3 (43) |
| Moderate | 2 (18) | 0 (0) |
| Severe | 2 (18) | 0 (0) |
A total of 18 pigs, of which 11 showed clinical signs associated with PCVAD and 7 were apparently healthy, were necropsied.
Determined by evaluation of ventral and dorsal photographs of lungs. Scores were assigned as follows: 0, no macroscopic lesions; 1, pneumonia affecting <25% of gross lung; 2, pneumonia affecting 25 to 50% of gross lung; 3, pneumonia affecting 50 to 75% of gross lung; 4, pneumonia affecting >75% of gross lung. The data for pigs with clinical signs were statistically different (P = 0.0426) from those for apparently healthy pigs, based on ordinal logistic regression.
Determined by evaluation of tissue sections stained with hematoxylin and eosin. Scores were assigned as follows: 0, no significant microscopic lesions; 1, mild interstitial pneumonia with <50% lung lobe involvement; 2, mild to moderate multifocal interstitial pneumonia with 50 to 75% lung lobe involvement; 3, moderate to severe multifocal interstitial pneumonia with 50 to 75% lung lobe involvement; 4, severe diffuse interstitial pneumonia with >75% lung lobe involvement.
Determined by evaluation of lymph node and tonsil tissue sections stained with hematoxylin and eosin. Characterization as mild, moderate, or severe indicates a small, intermediate, or large extent of lymphocyte depletion, respectively, with replacement by histiocytes.
Effect of PRRS MLV vaccination on mortality.
As shown in Fig. 4C, of the 101 nonvaccinated pigs, 9 died, resulting in an overall survival rate of 91.1%. Of the 105 vaccinated pigs, 14 died, for an overall survival rate of 86.7% (Fig. 4C). A vaccinated pig was 1.7 times (95% CI, 0.89, 3.72) more likely to die during the overall study period than a nonvaccinated pig (P = 0.35). Increased mortality became apparent after 20 days and was associated with the appearance of PCVAD. Of the 39 pigs that developed clinical signs of PCVAD, 21 died prior to the end of the study (14 vaccinated and 7 nonvaccinated), resulting in a mortality rate of approximately 54% in pigs exhibiting clinical signs. Between 22 and 35 dpi, a vaccinated pig was 2.1 times (90% CI, 1.03, 5.87) more likely to die than a nonvaccinated pig (P = 0.09). Even though mortality was higher in the vaccine group, the differences between the vaccine and nonvaccine groups were not significantly different.
Vaccination increases the appearance of antibodies against a PCV2 decoy epitope.
In previous work, we identified the presence of antibodies against a decoy epitope in the capsid protein of PCV2, CP(160–180), correlated with PCVAD (13–15). The presence of an anti-CP(160–180) response is associated with the absence of PCV2 neutralizing activity in serum. Pigs vaccinated for PCV2 and protected from disease produce little anti-CP(160–180) activity and preferentially recognize a larger epitope, CP(43–233). Pigs that are naturally infected with PCV2 show a mixture of the two antibody responses. As a means to standardize results across plates, the anti-PCV2 antibody responses for the vaccine and nonvaccine groups were presented as CP(160–233)/CP(43–233) ratios. The higher the ratio, the more the immune response is skewed toward the recognition of the decoy epitope. The results, presented in Fig. 7, showed a significant (P = 0.0006) difference between the CP(160–233)/CP(43–233) mean ratios: 0.85 for nonvaccinated pigs versus 0.97 for vaccinated pigs. Exclusion of the 32 pigs that showed the presence of PCV2 nucleic acid at the time of challenge changed the ratios but did not affect the conclusion; CP(160–233)/CP(43–233) mean ratios were 0.92 for nonvaccinated pigs (n = 61) and 0.98 for vaccinated pigs (n = 72) (P = 0.03).
FIG 7.
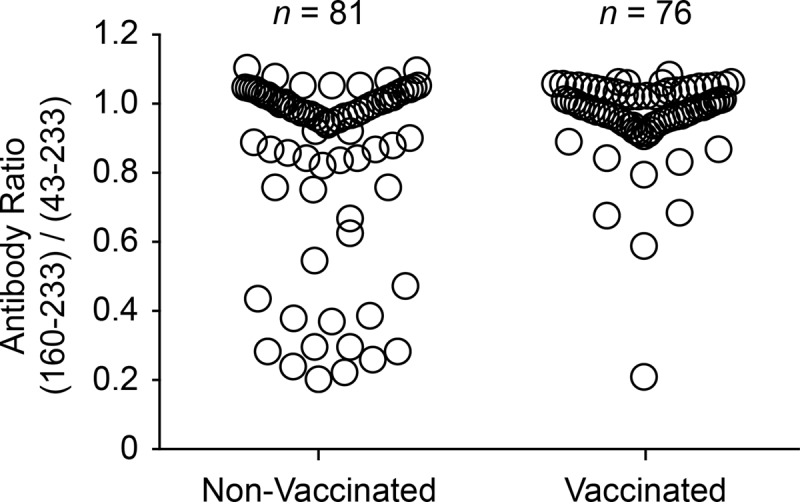
PCV2 CP(160–233)/CP(43–233) ratio at 42 days after coinfection. Sera were analyzed for anti-PCV2 CP antibodies. The serum antibody ratio was calculated as the MFI for reactivity with the CP(160–233) decoy epitope divided by the MFI for the CP(43–233) conformational antigen. The difference between the means for the PRRSV-vaccinated and nonvaccinated groups was significant (P = 0.0006 by Student's t test).
As discussed above, previous work showed that pigs with low CP(160–233)/CP(43–233) ratios are protected from disease. Therefore, nonvaccinated pigs with ratios of <0.5 (n = 13) were compared to nonvaccinated pigs with PCV2 antibody ratios of >0.5 (n = 68). Pigs with antibody ratios of <0.5 had significantly lower levels of circulating PCV2 in the serum at 21 (P = 0.0001), 28 (P = 0.0008), 35 (P = 0.007), and 42 (P = 0.03) dpi than pigs with higher ratios. The results confirm earlier findings describing the nonprotective effect of anti-CP(160–233) antibodies (13).
Effect of PRRS MLV vaccination on ADG.
Over the entire 70-day study period, the mean average daily gain (ADG) for the vaccine group (n = 91) was 0.65 ± 0.11 kg, compared to 0.68 ± 0.10 kg for the nonvaccine group (n = 92). The means differed significantly between the two groups (P = 0.029). Decreased mean ADG was also observed in the vaccine group during the 42-day post–virus challenge period. However, the difference between the vaccine group (ADG, 0.82 ± 0.14 kg) (n = 91) and the nonvaccine group (ADG, 0.86 ± 0.14 kg) (n = 92) was not statistically significant (P = 0.061). In addition, ADG differences between the two groups during the 70-day study period were no longer significant after the exclusion of the 32 pigs with PCV2 detected prior to challenge.
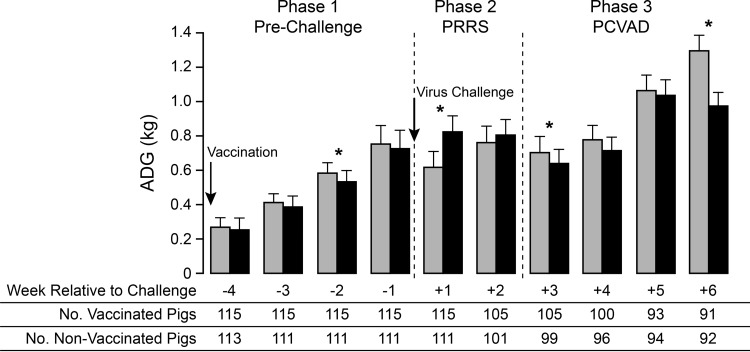
Therefore, a more-detailed analysis was conducted by calculating ADG on a weekly basis (Fig. 8). The results showed that ADG differences between the nonvaccine and vaccine groups could be divided into three distinct phases. In the first phase, covering the prechallenge period, mean ADG after vaccination was reduced, with a significant difference between the vaccinated and nonvaccinated groups appearing at 3 weeks after vaccination. The second phase covered the period of acute PRRSV infection. At 1 week after virus challenge, mean ADG was significantly increased for the vaccine group. ADG remained higher for the vaccine group in the second week postchallenge, but the difference was not significant. The improved ADG for the vaccinated pigs likely resulted from the positive effect of vaccination in reducing PRRS-associated clinical signs and virus load (Fig. 2A and 4A). The third phase covered the period from the onset of PCVAD, beginning at about 3 weeks after virus challenge, to the end of the study. During this phase, ADG was lower in the vaccinated group at every time point, with significantly lower mean ADG values on week 6 after virus challenge. A significant decrease in ADG was also initially detected for vaccinated pigs on week 3 postchallenge; however, this difference was no longer significant after the 32 PCV2-positive pigs had been excluded (P = 0.08). The lower mean ADG values are consistent with the effects of PCVAD, which include poor growth performance and muscle wasting.
FIG 8.
Weekly average daily gain (ADG) before and after virus challenge. Mean ADG values for the nonvaccinated (shaded bars) and vaccinated (filled bars) groups were calculated on a weekly basis. The data show means and standard deviations. The numbers of pigs used in the analysis are given below the bar graph. Asterisks indicate statistically significant differences in ADG between groups (P < 0.03 by Student's t test).
The negative effect of vaccination on ADG could have been the result of increased numbers of pigs with clinically apparent PCVAD. Therefore, a separate analysis was performed after the 26 pigs with clinical signs that survived the length of the study had been excluded from the vaccine and nonvaccine groups. The removal of pigs with clinical signs increased the ADGs of the two groups to 0.69 ± 0.09 kg for nonvaccinated pigs without clinical signs (n = 83) and 0.66 ± 0.10 kg for vaccinated pigs without clinical signs (n = 74) (P = 0.047). However, even in the absence of overt clinical signs, PRRSV vaccination had a negative effect on weight gain.
DISCUSSION
Enhanced PCV2 infection leading to PCVAD is typically associated with immune stimulation (22, 23). PCV2 replication is located in the nuclei of permissive cells and is dependent on cellular enzymes expressed during the S phase of the cell cycle (24). It is presumed that actively dividing lymphocytes, in response to an immune stimulus, provide a cellular environment ideal for supporting PCV2 replication. PRRSV, porcine parvovirus, and Mycoplasma hyopneumoniae are common copathogens linked with lymphoproliferation and increased PCV2 pathogenesis (10–13, 25–31). Examples of noninfectious immunostimulators include immunization with keyhole limpet hemocyanin in incomplete Freund's adjuvant (22, 32) and inactivated vaccines, such as Mycoplasma hyopneumoniae (32–34) and Actinobacillus pleuropneumoniae (33, 34). The results of this study showed that the PRRS MLV initially had a beneficial effect in reducing PRRS-associated clinical signs and PRRS viremia; however, PRRSV-vaccinated pigs showed increased PCV2 replication, reduced average daily gain, and increased clinical signs associated with PCVAD.
Previous experimental studies documenting interactions between the PRRS MLV and PCV2 infection have yielded conflicting results. Allan et al. (35) found that colostrum-deprived, specific-pathogen-free (SPF) pigs infected with PCV2 at the age of 5 weeks and administered the PRRS MLV 1 week later had greater amounts of PCV2 antigen in tissues and more-severe histologic lesions, characteristic of PMWS, than pigs infected with PCV2 alone. However, the pigs failed to exhibit clinical signs or gross lesions typical of PCVAD (35). In contrast, Opriessnig et al. (36) evaluated the effects of PCV2 infection on the efficacy of the PRRS MLV in groups of 10 early-weaned SPF pigs. Pigs were inoculated with PCV2 at the age of 6 weeks, vaccinated with the PRRS MLV 2 weeks later, and then challenged with PRRSV at the age of 12 weeks. The group with PCV2 and the MLV exhibited lower ADG and more-severe lung lesions after PRRSV challenge than the group that was vaccinated and received a PRRSV challenge without PCV2. Because PCV2 was not detected in affected lungs by IHC, the authors attributed the lesions to PRRSV infection and further concluded that the effect of PCV2 was to reduce the efficacy of the PRRS MLV (36). Park et al. (37) investigated the potential for the PRRS MLV to reduce PRRSV-associated amplification of PCV2 pathogenesis after coinfection with PRRSV and PCV2. Groups of 8 conventional pigs were subjected to a variety of treatments involving different combinations of the PRRS MLV, wild-type PRRSV, a PCV2 vaccine, and PCV2. Pigs were vaccinated with the PRRS MLV, a PCV2 subunit vaccine, or both and were then challenged, 4 weeks later, with PRRSV, PCV2, or both. The group that received the PRRS MLV followed by coinfection showed no differences in PCV2 viremia, PCV2-associated pathology, or the number of PCV2-positive cells in lymph nodes and lungs from the coinfected group that was not vaccinated with the PRRS MLV (37). In contrast to our study, in which vaccination enhanced PCVAD, these authors found that coinfected pigs, with or without previous PRRS MLV vaccination, had similar PCV2 replication and pathogenesis.
Field studies have also yielded conflicting results. A survey conducted on 70 pig farms in the Netherlands included questions regarding PRRS MLV use on farms with and without PMWS or PCVAD. The results showed that the PRRS MLV is a significant risk factor for PMWS outbreaks (38). In contrast, an analysis of the effect of the PRRS MLV on farms affected by PCVAD in the United States found that farms incorporating the PRRS MLV have significantly lower levels of PCV2 viremia than nonvaccinating farms in the age group in which peak wasting disease occurred (39). In that study, quantitative PCR (qPCR) was used to measure PCV2 in serum samples collected at different time points from 6 herds vaccinated with the PRRS MLV and 12 nonvaccinated herds. These results suggest that the PRRS MLV can reduce PCV2 viremia (39).
The failure of experimental studies to find a consistent link between the PRRS MLV and PCVAD was likely due in part to the length of the observation period. For example, the studies described above were terminated on day 25 after infection with PCV2 (35) and 21 days after challenge with PRRSV and PCV2 (37). In the current study, peak PCVAD occurred between 22 and 35 days after challenge with PRRSV and PCV2. Another important difference is the sizes of the experimental groups. Typically, the prevalence of clinically ill pigs on farms affected by PCVAD is only 2 to 25% (40–42). In small groups of pigs, animals with clinical disease may not be apparent or present. The current study utilized more than 200 pigs, mimicking the environment found in the field and providing the depth of data required for the observation and quantification of low-percentage outcomes. PCVAD morbidity rates were 11.9% and 25.7% in nonvaccinated and vaccinated pigs, respectively. Therefore, PCVAD should be assessed by comparing mortality rates, clinical disease presentation, viremia, and weight gain in relatively large groups evaluated for several weeks after infection.
Because large groups of pigs were required to ensure clinical disease expression, some control groups, including pigs challenged with PCV2 or PRRSV alone, were not incorporated into the study design. Therefore, conclusions are based on the findings for vaccinated and nonvaccinated cochallenged pigs. Caution should be exercised in generalizing results to single infections (i.e., the effect of the PRRS MLV on PCV2-challenged pigs).
PCV2 is ubiquitous in swine populations, and elimination of the virus from the environment is extremely difficult. As demonstrated in this study, 32 of 226 pigs had detectable PCV2 in serum at the time of challenge. Although these pigs were in the minority (14%), and group titers were relatively low (2.3 and 2.9 log10 templates per PCR for vaccinated and nonvaccinated pigs, respectively), the presence of PCV2 prior to challenge had to be considered a possible factor in postchallenge response. This was especially true due to the difference between the proportions of PCV2-positive pigs in the two groups, despite randomized allocation of pigs and balanced genotypes. This difference was likely due to the failure of randomization to distribute PCV2-positive pigs equally across the two groups. Therefore, analyses were also completed after the 32 initially PCV2 positive pigs were excluded, and all conclusions of the study were confirmed. Regardless, this highlights the difficulty of eliminating PCV2 from the environment and also suggests that pigs should be balanced according to PCV2 status prior to challenge.
Significantly increased levels of PCV2 viremia in PRRS MLV-treated pigs were observed at 11 and 14 days after challenge, but not at later time points (Fig. 2B). This effect of the PRRS MLV on PCV2 infection is similar to that seen in a previous study by us, which showed a significant increase in the level of PCV2 viremia at 23 days after coinfection with PRRSV and PCV2 (13). In the current study, it is noteworthy that PCV2 viremia levels did not differ significantly between the vaccine and nonvaccine groups during peak PCVAD. However, the 39 clinically ill pigs did maintain significantly higher levels (P < 0.02) of circulating PCV2 during these later time points (21 and 35 dpi) (data not shown). The increased incidence of PCVAD in vaccinated pigs between 22 and 35 dpi may also be the result of greater levels of localized PCV2 in tissues. Further studies are needed to assess differences in the quantity and tissue distribution of PCV2 between vaccinated and nonvaccinated pigs.
Average daily gain (ADG) is used in swine production as an objective measure of overall health and performance. The negative effect of the PRRS MLV on growth performance is well documented. For example, Opriessnig et al. (43) reported that pigs vaccinated with the PRRS MLV at the age of 2 weeks exhibited significantly lower ADG than nonvaccinated pigs. Pretzer et al. (44) found that PRRS MLV-vaccinated weaned pigs had lower ADG between 0 and 14 dpv than nonvaccinated pigs. While this effect on ADG was no longer apparent between 21 and 42 dpv, the vaccinated pigs maintained lighter weights overall (44). We confirmed the negative effect of the PRRS MLV on ADG during the 28-day period prior to coinfection; significantly reduced ADG was observed during the third week after vaccination (Fig. 8). The benefit of PRRSV vaccination was documented during the first 2 weeks after PCV2 and PRRSV challenge, when the MLV had a positive effect on ADG. Although vaccinated pigs in this study had increased ADG in the first 2 weeks postchallenge, this effect was quickly outweighed when ADG was decreased in the presence of PCVAD. Vaccination decreased ADG in both clinically affected pigs and pigs without clinical signs during the study period, demonstrating that poor growth performance may be a subclinical manifestation of PCVAD in apparently healthy pigs.
At least three mechanisms may be involved in the enhancement of PCVAD following PRRSV vaccination. First, the PRRS MLV may function to stimulate the immune system and increase the number of PCV2-permissive cells. As with wild-type viruses, lymphocytes undergo mitosis in response to vaccination with the PRRS MLV, thereby increasing the population of cells with the ability to support PCV2 replication. In addition, the vaccine likely stimulates PRRSV-specific lymphocyte populations that are restimulated after challenge with a wild-type PRRSV. Second, like wild-type PRRS viruses, the PRRS MLV may suppress innate immunity, thereby blocking anti-PCV2 responses. For example, PRRSV nonstructural proteins, such as nsp1 and nsp2, block the induction of interferon and response of cells to interferon (5). Viral proteins such as nsp1α and nsp1β antagonize the type I interferon response by degrading key components needed for interferon gene expression and inhibiting interferon signaling pathways (45, 46). Finally, the third mechanism is based on the possibility that the PRRS MLV may skew the immune response toward the production of nonneutralizing PCV2-specific antibodies.
PCV2 has circulated in the swine population for at least 25 years. In 2005, the emergence of PCV2b in North America was attributed to outbreaks of PCVAD (47–49). Since then, the disease has been effectively managed through the use of PCV2 vaccines (50–52). Therefore, the negative effect of the MLV on PCV2 infection may not be relevant. However, there remain several countries in which PRRS MLV vaccination is in wide use, but in the absence of a comprehensive PCVAD vaccination program. Further, there is the potential for new and emerging PCV2 strains to escape current vaccine protection. Emerging PCV2 mutant strains have been documented in China (53) and more recently have been associated with PCVAD outbreaks in vaccinated herds in the United States and Korea (54, 55). Overall, this study supports the notion that maintaining a successful PCV2 control program and assessing the risk of virulent PRRSV exposure is critical to weighing the benefits of the PRRS MLV.
ACKNOWLEDGMENT
This work was supported by USDA NIFA award 2013-68004-20362.
REFERENCES
- 1.Baekbo P, Kristensen CS, Larsen LE. 2012. Porcine circovirus diseases: a review of PMWS. Transbound Emerg Dis 59:60–67. doi: 10.1111/j.1865-1682.2011.01288.x. [DOI] [PubMed] [Google Scholar]
- 2.Ramamoorthy S, Meng XJ. 2009. Porcine circoviruses: a minuscule yet mammoth paradox. Anim Health Res Rev 10:1–20. doi: 10.1017/S1466252308001461. [DOI] [PubMed] [Google Scholar]
- 3.Benfield DA, Nelson E, Collins JE, Harris L, Goyal SM, Robison D, Christianson WT, Morrison RB, Gorcyca D, Chladek D. 1992. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J Vet Diagn Invest 4:127–133. doi: 10.1177/104063879200400202. [DOI] [PubMed] [Google Scholar]
- 4.Conzelmann KK, Visser N, Van Woensel P, Thiel HJ. 1993. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology 193:329–339. doi: 10.1006/viro.1993.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chand RJ, Trible BR, Rowland RR. 2012. Pathogenesis of porcine reproductive and respiratory syndrome virus. Curr Opin Virol 2:256–263. doi: 10.1016/j.coviro.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Gómez-Laguna J, Salguero FJ, Pallares FJ, Carrasco L. 2013. Immunopathogenesis of porcine reproductive and respiratory syndrome in the respiratory tract of pigs. Vet J 195:148–155. doi: 10.1016/j.tvjl.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Opriessnig T, Gimenez-Lirola LG, Halbur PG. 2011. Polymicrobial respiratory disease in pigs. Anim Health Res Rev 12:133–148. doi: 10.1017/S1466252311000120. [DOI] [PubMed] [Google Scholar]
- 8.Renukaradhya GJ, Alekseev K, Jung K, Fang Y, Saif LJ. 2010. Porcine reproductive and respiratory syndrome virus-induced immunosuppression exacerbates the inflammatory response to porcine respiratory coronavirus in pigs. Viral Immunol 23:457–466. doi: 10.1089/vim.2010.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pallarés FJ, Halbur PG, Opriessnig T, Sorden SD, Villar D, Janke BH, Yaeger MJ, Larson DJ, Schwartz KJ, Yoon KJ, Hoffman LJ. 2002. Porcine circovirus type 2 (PCV-2) coinfections in US field cases of postweaning multisystemic wasting syndrome (PMWS). J Vet Diagn Invest 14:515–519. doi: 10.1177/104063870201400614. [DOI] [PubMed] [Google Scholar]
- 10.Allan GM, McNeilly F, Ellis J, Krakowka S, Meehan B, McNair I, Walker I, Kennedy S. 2000. Experimental infection of colostrum deprived piglets with porcine circovirus 2 (PCV2) and porcine reproductive and respiratory syndrome virus (PRRSV) potentiates PCV2 replication. Arch Virol 145:2421–2429. doi: 10.1007/s007050070031. [DOI] [PubMed] [Google Scholar]
- 11.Harms PA, Sorden SD, Halbur PG, Bolin SR, Lager KM, Morozov I, Paul PS. 2001. Experimental reproduction of severe disease in CD/CD pigs concurrently infected with type 2 porcine circovirus and porcine reproductive and respiratory syndrome virus. Vet Pathol 38:528–539. doi: 10.1354/vp.38-5-528. [DOI] [PubMed] [Google Scholar]
- 12.Rovira A, Balasch M, Segales J, Garcia L, Plana-Duran J, Rosell C, Ellerbrok H, Mankertz A, Domingo M. 2002. Experimental inoculation of conventional pigs with porcine reproductive and respiratory syndrome virus and porcine circovirus 2. J Virol 76:3232–3239. doi: 10.1128/JVI.76.7.3232-3239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trible BR, Ramirez A, Suddith A, Fuller A, Kerrigan M, Hesse R, Nietfeld J, Guo B, Thacker E, Rowland RR. 2012. Antibody responses following vaccination versus infection in a porcine circovirus-type 2 (PCV2) disease model show distinct differences in virus neutralization and epitope recognition. Vaccine 30:4079–4085. doi: 10.1016/j.vaccine.2012.04.022. [DOI] [PubMed] [Google Scholar]
- 14.Trible BR, Kerrigan M, Crossland N, Potter M, Faaberg K, Hesse R, Rowland RR. 2011. Antibody recognition of porcine circovirus type 2 capsid protein epitopes after vaccination, infection, and disease. Clin Vaccine Immunol 18:749–757. doi: 10.1128/CVI.00418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trible BR, Suddith AW, Kerrigan MA, Cino-Ozuna AG, Hesse RA, Rowland RR. 2012. Recognition of the different structural forms of the capsid protein determines the outcome following infection with porcine circovirus type 2. J Virol 86:13508–13514. doi: 10.1128/JVI.01763-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Federation of Animal Science Societies. 2010. Guide for the care and use of agricultural animals in research and teaching. Federation of Animal Science Societies, Champaign, IL. [Google Scholar]
- 17.Boddicker N, Waide EH, Rowland RR, Lunney JK, Garrick DJ, Reecy JM, Dekkers JC. 2012. Evidence for a major QTL associated with host response to porcine reproductive and respiratory syndrome virus challenge. J Anim Sci 90:1733–1746. doi: 10.2527/jas.2011-4464. [DOI] [PubMed] [Google Scholar]
- 18.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg 27:493–497. [Google Scholar]
- 19.Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, Lum MA, Andrews JJ, Rathje JA. 1995. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol 32:648–660. doi: 10.1177/030098589503200606. [DOI] [PubMed] [Google Scholar]
- 20.Done SH, Paton DJ. 1995. Porcine reproductive and respiratory syndrome: clinical disease, pathology and immunosuppression. Vet Rec 136:32–35. doi: 10.1136/vr.136.2.32. [DOI] [PubMed] [Google Scholar]
- 21.Paton DJ, Brown IH, Edwards S, Wensvoort G. 1991. ‘Blue ear’ disease of pigs. Vet Rec 128:617. doi: 10.1136/vr.128.26.617. [DOI] [PubMed] [Google Scholar]
- 22.Krakowka S, Ellis JA, McNeilly F, Ringler S, Rings DM, Allan G. 2001. Activation of the immune system is the pivotal event in the production of wasting disease in pigs infected with porcine circovirus-2 (PCV-2). Vet Pathol 38:31–42. doi: 10.1354/vp.38-1-31. [DOI] [PubMed] [Google Scholar]
- 23.Opriessnig T, Halbur PG. 2012. Concurrent infections are important for expression of porcine circovirus associated disease. Virus Res 164:20–32. doi: 10.1016/j.virusres.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tischer I, Peters D, Rasch R, Pociuli S. 1987. Replication of porcine circovirus: induction by glucosamine and cell cycle dependence. Arch Virol 96:39–57. doi: 10.1007/BF01310989. [DOI] [PubMed] [Google Scholar]
- 25.Opriessnig T, Thacker EL, Yu S, Fenaux M, Meng XJ, Halbur PG. 2004. Experimental reproduction of postweaning multisystemic wasting syndrome in pigs by dual infection with Mycoplasma hyopneumoniae and porcine circovirus type 2. Vet Pathol 41:624–640. doi: 10.1354/vp.41-6-624. [DOI] [PubMed] [Google Scholar]
- 26.Allan GM, Kennedy S, McNeilly F, Foster JC, Ellis JA, Krakowka SJ, Meehan BM, Adair BM. 1999. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J Comp Pathol 121:1–11. doi: 10.1053/jcpa.1998.0295. [DOI] [PubMed] [Google Scholar]
- 27.Krakowka S, Ellis JA, Meehan B, Kennedy S, McNeilly F, Allan G. 2000. Viral wasting syndrome of swine: experimental reproduction of postweaning multisystemic wasting syndrome in gnotobiotic swine by coinfection with porcine circovirus 2 and porcine parvovirus. Vet Pathol 37:254–263. doi: 10.1354/vp.37-3-254. [DOI] [PubMed] [Google Scholar]
- 28.Ellis J, Krakowka S, Lairmore M, Haines D, Bratanich A, Clark E, Allan G, Konoby C, Hassard L, Meehan B, Martin K, Harding J, Kennedy S, McNeilly F. 1999. Reproduction of lesions of postweaning multisystemic wasting syndrome in gnotobiotic piglets. J Vet Diagn Invest 11:3–14. doi: 10.1177/104063879901100101. [DOI] [PubMed] [Google Scholar]
- 29.Kennedy S, Moffett D, McNeilly F, Meehan B, Ellis J, Krakowka S, Allan GM. 2000. Reproduction of lesions of postweaning multisystemic wasting syndrome by infection of conventional pigs with porcine circovirus type 2 alone or in combination with porcine parvovirus. J Comp Pathol 122:9–24. doi: 10.1053/jcpa.1999.0337. [DOI] [PubMed] [Google Scholar]
- 30.Fan P, Wei Y, Guo L, Wu H, Huang L, Liu J, Liu C. 2013. Synergistic effects of sequential infection with highly pathogenic porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. Virol J 10:265. doi: 10.1186/1743-422X-10-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinha A, Shen HG, Schalk S, Beach NM, Huang YW, Meng XJ, Halbur PG, Opriessnig T. 2011. Porcine reproductive and respiratory syndrome virus (PRRSV) influences infection dynamics of porcine circovirus type 2 (PCV2) subtypes PCV2a and PCV2b by prolonging PCV2 viremia and shedding. Vet Microbiol 152:235–246. doi: 10.1016/j.vetmic.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Krakowka S, Ellis J, McNeilly F, Waldner C, Rings DM, Allan G. 2007. Mycoplasma hyopneumoniae bacterins and porcine circovirus type 2 (PCV2) infection: induction of postweaning multisystemic wasting syndrome (PMWS) in the gnotobiotic swine model of PCV2-associated disease. Can Vet J 48:716–724. [PMC free article] [PubMed] [Google Scholar]
- 33.Allan GM, McNeilly F, Kennedy S, Meehan B, Ellis J, Krakowka S. 2000. Immunostimulation, PCV-2 and PMWS. Vet Rec 147:170–171. [PubMed] [Google Scholar]
- 34.Opriessnig T, Yu S, Gallup JM, Evans RB, Fenaux M, Pallares F, Thacker EL, Brockus CW, Ackermann MR, Thomas P, Meng XJ, Halbur PG. 2003. Effect of vaccination with selective bacterins on conventional pigs infected with type 2 porcine circovirus. Vet Pathol 40:521–529. doi: 10.1354/vp.40-5-521. [DOI] [PubMed] [Google Scholar]
- 35.Allan GM, Caprioli A, McNair I, Lagan-Tregaskis P, Ellis J, Krakowka S, McKillen J, Ostanello F, McNeilly F. 2007. Porcine circovirus 2 replication in colostrum-deprived piglets following experimental infection and immune stimulation using a modified live vaccine against porcine respiratory and reproductive syndrome virus. Zoonoses Public Health 54:214–222. doi: 10.1111/j.1863-2378.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- 36.Opriessnig T, McKeown NE, Harmon KL, Meng XJ, Halbur PG. 2006. Porcine circovirus type 2 infection decreases the efficacy of a modified live porcine reproductive and respiratory syndrome virus vaccine. Clin Vaccine Immunol 13:923–929. doi: 10.1128/CVI.00074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park C, Oh Y, Seo HW, Han K, Chae C. 2013. Comparative effects of vaccination against porcine circovirus type 2 (PCV2) and porcine reproductive and respiratory syndrome virus (PRRSV) in a PCV2-PRRSV challenge model. Clin Vaccine Immunol 20:369–376. doi: 10.1128/CVI.00497-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Jong MF, Elbers A, Wellenberg GJ. 2003. Factors associated with PMWS and PDNS: a case-control study, p 215 In Proceedings of the 4th International Symposium on Emerging and Re-emerging Pig Diseases—Rome, 29 June to 2 July 2003. [Google Scholar]
- 39.Genzow M, Schwartz K, Gonzalez G, Anderson G, Chittick W. 2009. The effect of vaccination against porcine reproductive and respiratory syndrome virus (PRRSV) on the porcine circovirus-2 (PCV-2) load in porcine circovirus associated disease (PCVAD) affected pigs. Can J Vet Res 73:87–90. [PMC free article] [PubMed] [Google Scholar]
- 40.Sorden SD, Harms PA, Sirinarumitr T, Morozov I, Halbur PG, Yoon K-J, Paul PS. 1998. Porcine circovirus and PRRS virus co-infection in pigs with chronic bronchointerstitial pneumonia and lymphoid depletion: an emerging syndrome in Midwestern swine, p 75 In Proceedings of the 41st Annual Meeting of the American Association of Veterinary Laboratory Diagnosticians. American Association of Veterinary Laboratory Diagnosticians, Visalia, CA. [Google Scholar]
- 41.Quintana J, Segales J, Rosell C, Calsamiglia M, Rodriguez-Arrioja GM, Chianini F, Folch JM, Maldonado J, Canal M, Plana-Duran J, Domingo M. 2001. Clinical and pathological observations on pigs with postweaning multisystemic wasting syndrome. Vet Rec 149:357–361. doi: 10.1136/vr.149.12.357. [DOI] [PubMed] [Google Scholar]
- 42.Kyriakis SC, Saoulidis K, Lekkas S, Miliotis CC, Papoutsis PA, Kennedy S. 2002. The effects of immuno-modulation on the clinical and pathological expression of postweaning multisystemic wasting syndrome. J Comp Pathol 126:38–46. doi: 10.1053/jcpa.2001.0520. [DOI] [PubMed] [Google Scholar]
- 43.Opriessnig T, Pallares FJ, Nilubol D, Vincent AL, Thacker EL, Vaughn EM, Roof M, Halbur PG. 2005. Genomic homology of ORF 5 gene sequence between modified live vaccine virus and porcine reproductive and respiratory syndrome virus challenge isolates is not predictive of vaccine efficacy. J Swine Health Production 13:246–253. [Google Scholar]
- 44.Pretzer SD, Claussen KM, Bergstrom JR, Henry SC, Phillips R, Tokach MD, Goodband RD, Nelssen JL, Dritz SS. 1996. The effects of porcine reproductive and respiratory syndrome (PRRS) vaccination on postweaning growth performance, p 83–86. In Swine Day, 1996. Kansas State University, Agricultural Experiment Station and Cooperative Extension Service, Manhattan, KS. [Google Scholar]
- 45.Chen Z, Lawson S, Sun Z, Zhou X, Guan X, Christopher-Hennings J, Nelson EA, Fang Y. 2010. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology 398:87–97. doi: 10.1016/j.virol.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han M, Yoo D. 2014. Modulation of innate immune signaling by nonstructural protein 1 (nsp1) in the family Arteriviridae. Virus Res 194:100–109. doi: 10.1016/j.virusres.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carman S, Cai HY, DeLay J, Youssef SA, McEwen BJ, Gagnon CA, Tremblay D, Hazlett M, Lusis P, Fairles J, Alexander HS, van Dreumel T. 2008. The emergence of a new strain of porcine circovirus-2 in Ontario and Quebec swine and its association with severe porcine circovirus associated disease—2004–2006. Can J Vet Res 72:259–268. [PMC free article] [PubMed] [Google Scholar]
- 48.Horlen KP, Schneider P, Anderson J, Nietfeld JC, Henry SC, Tokach LM, Rowland RRR. 2007. A cluster of farms experiencing severe porcine circovirus associated disease: clinical features and association with the PCV2b genotype. J Swine Health Production 15:270–278. [Google Scholar]
- 49.Cheung AK, Lager KM, Kohutyuk OI, Vincent AL, Henry SC, Baker RB, Rowland RR, Dunham AG. 2007. Detection of two porcine circovirus type 2 genotypic groups in United States swine herds. Arch Virol 152:1035–1044. doi: 10.1007/s00705-006-0909-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horlen KP, Dritz SS, Nietfeld JC, Henry SC, Hesse RA, Oberst R, Hays M, Anderson J, Rowland RR. 2008. A field evaluation of mortality rate and growth performance in pigs vaccinated against porcine circovirus type 2. J Am Vet Med Assoc 232:906–912. doi: 10.2460/javma.232.6.906. [DOI] [PubMed] [Google Scholar]
- 51.Velasova M, Alarcon P, Werling D, Nevel A, Wieland B. 2013. Effectiveness of porcine circovirus type 2 vaccination in reducing the severity of post-weaning multisystemic wasting syndrome in pigs. Vet J 197:842–847. doi: 10.1016/j.tvjl.2013.05.043. [DOI] [PubMed] [Google Scholar]
- 52.Kixmöller M, Ritzmann M, Eddicks M, Saalmuller A, Elbers K, Fachinger V. 2008. Reduction of PMWS-associated clinical signs and co-infections by vaccination against PCV2. Vaccine 26:3443–3451. doi: 10.1016/j.vaccine.2008.04.032. [DOI] [PubMed] [Google Scholar]
- 53.Guo LJ, Lu YH, Wei YW, Huang LP, Liu CM. 2010. Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virol J 7:273. doi: 10.1186/1743-422X-7-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seo HW, Park C, Kang I, Choi K, Jeong J, Park SJ, Chae C. 2014. Genetic and antigenic characterization of a newly emerging porcine circovirus type 2b mutant first isolated in cases of vaccine failure in Korea. Arch Virol 159:3107–3111. doi: 10.1007/s00705-014-2164-6. [DOI] [PubMed] [Google Scholar]
- 55.Opriessnig T, Xiao CT, Gerber PF, Halbur PG. 2013. Emergence of a novel mutant PCV2b variant associated with clinical PCVAD in two vaccinated pig farms in the U.S. concurrently infected with PPV2. Vet Microbiol 163:177–183. doi: 10.1016/j.vetmic.2012.12.019. [DOI] [PubMed] [Google Scholar]