Abstract
Thymidylate synthase (TYMS; EC 2.1.1.15) catalyzes the reductive methylation of 2ʹ-deoxyuridine-5ʹ-monophosphate (dUMP) by N5,N10-methyhlenetetrahydrofolate, forming dTMP for the maintenance of DNA replication and repair. Inhibitors of TYMS have been widely used in the treatment of neoplastic disease. A number of fluoropyrimidine and folate analogs have been developed that lead to inhibition of the enzyme, resulting in dTMP deficiency and cell death. In the current study, we have examined the role of oxidative stress in response to TYMS inhibitors. We observed that intracellular reactive oxygen species (ROS) concentrations are induced by these inhibitors and promote apoptosis. Activation of the enzyme NADPH oxidase (NOX), which catalyzes one-electron reduction of O2 to generate superoxide (O2●−), is a significant source of increased ROS levels in drug-treated cells. However, gene expression profiling revealed a number of other redox-related genes that may contribute to ROS generation. TYMS inhibitors also induce a protective response, including activation of the transcription factor nuclear factor E2-related factor 2 (NRF2), a critical mediator of defense against oxidative and electrophilic stress. Our results show that exposure to TYMS inhibitors induces oxidative stress that leads to cell death, while simultaneously generating a protective response that may underlie resistance against such death.
Introduction
Thymidylate synthase (TYMS, EC 2.1.1.15) catalyzes the reductive methylation of 2ʹ-deoxyuridine-5ʹ-monophosphate (dUMP) by N5N10-methylenetetrahydrofolate (CH2H4PteGlu), to form dTMP and dihydrofolate (Carreras and Santi, 1995; Berger and Berger, 2006). The enzyme is indispensable to DNA synthesis and is therefore critical for replication and repair of the cellular genome in growing cells. As a consequence, it has been an attractive target at which chemotherapeutic drugs are directed (Chu et al., 2003; Longley et al., 2003; Wilson et al., 2014). Inhibitors of TYMS, including both fluoropyrimidine and folate analogs, reduce de novo synthesis of dTMP, which leads to impaired DNA replication, genome damage, and programmed cell death (Carreras and Santi, 1995; Berger and Berger, 2006; Barbour and Berger, 2008). Fluoropyrimidines such as 5-fluorouracil (FUra) and 5-fluoro-2ʹ-deoxyuridine (FdUrd) are metabolized within cells to the potent TYMS inhibitor 5-fluoro-2ʹ-deoxyuridylic acid and have been important in the therapy of a variety of cancers, including those of the ovary, breast, and gastrointestinal tract (Chu et al., 2003; Longley et al., 2003). Folate-based inhibitors of TYMS have been designed on the basis of information from the structures of the natural substrates and the enzyme’s active site cleft (Carreras and Santi, 1995; Bertino, 1997; Takemura and Jackman, 1997). Indeed, these inhibitors were among the first to be developed using such rationally based criteria. Several, including raltitrexed (RTX), LY231514 [(2R)-2-[[4-[2-(2-amino-4-oxo-1,7-dihydropyrrolo[2,3-d]pyrimidin-5-yl)ethyl]benzoyl]amino]pentanedioic acid], ZD9331 [(2S)-2-[[4-[(2,7-dimethyl-4-oxo-1H-quinazolin-6-yl)methyl-prop-2-ynylamino]-2-fluorobenzoyl]amino]-4-(2H-tetrazol-5-yl)butanoic acid], and GW1843U89 [(S)-2-(5-(((3-methyl-1-oxo-1,2-dihydrobenzo[f]quinazolin-9-yl)methyl)amino)-1-oxoisoindolin-2-yl)pentanedioic acid], have advanced to clinical trials, and have demonstrated significant activity against a variety of neoplasms (Bertino, 1997; Takemura and Jackman, 1997).
The mechanism by which dTMP deficiencies lead to the death of cells exposed to TYMS inhibitors is of central interest. Over a decade ago, Hwang et al. (Hwang et al., 2001) demonstrated that induction of reactive oxygen species (ROS) in FUra-treated cells results from increased expression of the mitochondrial enzyme ferredoxin reductase (FDXR), and is required for p53-dependent apoptotic response to drug. Studies by Liu and Chen (2002) similarly suggested that FDXR promotes ROS-mediated apoptosis in response to FUra in a p53-dependent fashion. Despite these interesting studies as well as others showing increased ROS levels after exposure to FUra (Akhdar et al., 2009; Matsunaga et al., 2010; Lamberti et al., 2012), we still lack a detailed mechanistic understanding of how exposure to TYMS inhibitors alters the intracellular redox environment, and how cells respond to such alterations. Currently, we have examined ROS-mediated oxidative stress and its function in apoptotic cell death induced by TYMS inhibitors. We have identified the superoxide-generating enzyme NADPH oxidase (NOX) as an important source of increased ROS levels during drug exposure. In addition, we have found that cells mount a protective response that includes activation of the transcription factor nuclear factor E2-related factor 2 (NRF2) and its downstream target genes, which may diminish the cytotoxic impact of TYMS inhibitors.
Materials and Methods
Reagents.
FUra, FdUrd, deoxythymidine (dThd), folinic acid, and N-acetylcysteine (NAC) were purchased from Sigma-Aldrich (St. Louis, MO). RTX was obtained from AstraZeneca (Macclesfield, Cheshire, UK). Diphenyleneiodonium (DPI) and NADPH were purchased from Calbiochem/EMD Biosciences (San Diego, CA). Apocyanin (APO) was obtained from Acros Organics (Geel, Belgium). Lucigenin and VAS-2870 (VAS [3-benzyl-7-(2-benzoxazolyl)thio-1,2,3-triazolo(4,5-d)pyrimidine]) were from Enzo Life Sciences (Plymouth Meeting, PA). Paraformaldehyde was from Alfa Aesar (Ward Hill, MA). Dihydroethidium (DHE) and 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA) were purchased from Invitrogen/Molecular Probes (Eugene, OR). Oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA).
Cell Culture.
Human colon tumor cell line HCT116 (originally obtained from Dr. Michael G. Brattain), FdUrd-resistant HCT116/200 (Berger et al., 1988), HCT-15 (American Tissue Culture Collection, Manassas, VA), and SW480 (American Tissue Culture Collection, Manassas, VA) were maintained in RPMI-1640 medium (Mediatech, Manassas, VA) containing 4.5 g/l glucose and supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Norcross, GA), 100 U/ml penicillin, 100 mg/ml streptomycin, and 250 ng/ml amphotericin B (Mediatech) at 37°C in a humidified 5% CO2 atmosphere. Cells were exposed to the indicated concentrations of TYMS inhibitors (FUra, FdUrd, RTX) based upon previous studies in our laboratory (Barbour and Berger, 2008); 10 µM folinic acid was included with FdUrd. Where indicated, NAC (10 mM), dThd (10 µM), DPI (10 µM), APO (1 mM), or VAS (10 µM) were included in the culture medium; all were added along with TYMS inhibitors except for DPI, which was added 3 hours before addition of TYMS inhibitors.
Detection of ROS.
Intracellular ROS production was measured by oxidation of cell permeable dyes 2ʹ,7ʹ-dichlorodihydrofluorescein diacetate (DCFDA) to detect H2O2 and DHE to detect superoxide (O2●−). Cells were grown to 60% confluence in chamber slides, fixed in 4% paraformaldehyde, and incubated at 37°C for 30 minutes in 5 µM DCFDA or DHE. After incubation, cells were washed twice with phosphate-buffered saline and imaged by fluorescence microscopy (Carl Zeiss, Thornwood, NY) using a charge-coupled camera (AxioCam HRm) linked to AxioVision software (version 4.7; Carl Zeiss).
For measurement of ROS by flow cytometry, cells were grown to 80% confluence in 60-mm plates and incubated in fluorescence-activated cell-sorting staining buffer (phosphate-buffered saline with 1% bovine serum albumin) at 37°C for 15 minutes in 50 nM DCFDA or DHE. After incubation, cells were washed twice with fluorescence-activated cell-sorting buffer and analyzed by flow cytometry (FC 500; Beckman Coulter, Fort Collins, CO). A shift to the right indicates increased ROS levels.
Measurement of Apoptotic Cell Death.
Cells were treated with TYMS inhibitors and inhibitory agents at the indicated concentrations for 48 hours. Apoptotic indices were determined by TUNEL assays using the In Situ Cell Death Detection Kit, POD (Roche Applied Science, Indianapolis, IN). Cells were stained according to the manufacturer’s instructions, counterstained with hematoxylin, and viewed under a light microscope at 400× magnification. Apoptotic nuclei were counted manually, based on staining and morphology, and the apoptotic index was calculated as the ratio of apoptotic/total cells. At least 1000 cells from several microscopic fields were counted in each determination.
NADPH Oxidase Activity Assay.
For each reaction, 105 cells were suspended in 500 µl reaction buffer [50 mM phosphate buffer (pH 7.0), 1 mM EGTA, 150 mM sucrose]. NADPH oxidase activity was detected by lucigenin-derived chemiluminescence with 100 µM NADPH as substrate and 5 µM lucigenin. Cells were incubated at 37°C for 10 minutes. Chemiluminescence was measured using a luminometer (Promega, Madison WI) and expressed as arbitrary light units per 105 cells.
RNA Extraction and Quantitative Reverse-Transcription Polymerase Chain Reaction.
Total RNA was isolated using RNeasy Mini Kit (Qiagen, Germantown, MD) with the addition of RNase Free DNase (Qiagen) to eliminate contaminating genomic DNA. The RNA concentration was determined by measuring the absorbance at 260 nm and 280 nm (NanoDrop ND-1000 Spectrophotometer; Thermo Fisher Scientific, Waltham, MA). For each reaction, 1 µg RNA was reverse transcribed using an IScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer’s instructions. For quantitative reverse-transcription polymerase chain reaction (qRT-PCR), cDNA (1μl) prepared as described earlier was amplified using Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. The PCR thermal profile was one cycle at 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds, 50°C for 15 seconds, and 72°C for 40 seconds using an Applied Biosystems 7300 Real Time PCR System. Relative mRNA levels were normalized to GAPDH, and calculated by the 2−ΔΔCt method. Relative changes in expression of each gene in response to TYMS inhibitors were expressed as fold-induction compared with the basal level of expression in nontreated cells. Gene-specific primer sets (Integrated DNA Technologies) used for qRT-PCR are listed in Table 1. Controls with no reverse transcriptase and no template RNA were used to monitor contamination.
TABLE 1.
Sequences of the primers for qRT-PCR
| Genes | Primers (5ʹ to 3ʹ) |
|---|---|
| AKR1B10 | Sense: 5ʹ-ACCTGTTCATCGTCAGCAAG-3ʹ |
| Antisense: 5ʹ-CATCCCCAGACTTGAATCCC-3ʹ | |
| ALDH3A1 | Sense: 5′-TGCTACGTGGACAAGAACTG-3′ |
| Antisense: 5ʹ-CACAATTTGGTTCTGGATCGAG-3ʹ | |
| GAPDH | Sense: 5ʹ-TCCCTGAGCTGAACGGGAAG-3ʹ |
| Antisense: 5ʹ-GGAGGAGTGGGTGTCGCTGT-3ʹ | |
| HSPB8 | Sense: 5ʹ-AAAGATGGATACGTGGAGGTG-3ʹ |
| Antisense: 5ʹ-GGGAAAGTGAGGCAAATACTG-3ʹ | |
| NOX2 | Sense: 5ʹ-GCCCAAAGGTGTCCAAGCT-3ʹ |
| Antisense: 5ʹ-TCCCCAACGATGCGGATAT-3ʹ | |
| p67phox | Sense: 5ʹ-ACCAGAAGCATTAACCGAGAC-3ʹ |
| Antisense: 5ʹ-TTCCCTCGAAGCTGAATCAAG-3ʹ | |
| SERPINE1 | Sense: 5ʹ-GTGGACTTTTCAGAGGTGGAG-3ʹ |
| Antisense: 5ʹ-GAAGTAGAGGGCATTCACCAG-3ʹ |
Transfection of Small-Interfering RNAs.
Small-interfering RNAs (siRNAs) were introduced into cells by transient transfection using the DharmaFECT 4 reagent (GE Dharmacon, Lafayette, CO) in accordance with the manufacturer’s instructions. HCT116 cells were transfected with ON-TARGETplus siRNA for NOX2 (Dharmafect J-011021-05-0005), NRF2 (Dharmafect J-003755-09-0005), or scrambled siRNA (Dharmafect D-001810-01-05) as a negative control. Twenty-four hours after transfection, the cells were treated with various drug combinations and harvested at appropriate times for assays.
Microarray Analysis.
Whole-genome cDNA microarray analyses were performed using Agilent’s platform (Agilent, Santa Clara, CA). Total RNA samples isolated from HCT116 were treated with FUra for 24 hours, then amplified and labeled using Agilent’s Low Input Quick Amp Labeling Kit (cat. no. 5190-2306) according to the manufacturer’s recommendations. Briefly, mRNA contained in 200 ng of total RNA was converted into cDNA using a poly-dT primer that also contained the T7 RNA polymerase promoter sequence. Subsequently, T7 RNA polymerase was added to cDNA samples to amplify original mRNA molecules and to simultaneously incorporate cyanine 3- or cyanine 5-labeled CTP (cRNA) into the amplification product. In addition, Agilent RNA spike-in controls (cat. no. 5188-5279) were added to samples before cDNA synthesis and were used as experimental quality control. In the next step, labeled RNA molecules were purified using Qiagen’s RNeasy Mini Kit (cat. no. 74104). After spectrophotometric assessment of dye incorporation and cRNA yield, samples were stored at −80°C until hybridization. Labeled cRNA samples were hybridized to Agilent Human GE 4x44K v2 Microarrays (cat. no. G4845A) at 65°C for 17 hours using Agilent’s Gene Expression Hybridization Kit (cat. no. 5188-5242) according to the manufacturer’s recommendations. Four (4) control sample replicates were hybridized against four (4) FUra-treated sample replicates in a dye swap design. After washes, arrays were scanned using an Agilent DNA Microarray Scanner System (cat. no. G2565CA).
Data were extracted from images with Feature Extractor Software version 10.7.3.1 (Agilent). In this process, background correction using additive and multiplicative detrending algorithms was performed. In addition, linear and LOWESS methods were used for dye normalization. Subsequently, data were uploaded into GeneSpring GX version 11.5.1 (Agilent) for analysis. In this process, data were log2 transformed, quantile normalized, and base line transformed using the median of all samples. Then, data were filtered by flags such that 100% of the samples in at least one of the two treatment groups have a “detected” flag.
Differentially expressed genes were determined by analysis of the data using an unpaired t test statistics that was corrected for multiple testing using the Benjamini-Hochberg algorithm. A cutoff of 1.5-fold change with a P value of 0.005 was used. To identify genes involved in redox metabolism, the GO (Gene Ontogeny) and KEGG (Kyoto Encyclopedia of Genes and Genomes) databases were queried with the terms “dehydro, dehydrog, hydro, hydrog, oxid, oxidase, oxidation, redox, reduc, reductase”; identified genes were assessed for their presence in the microarray.
Preparation of Extracts and Western Immunoblotting.
Cytoplasmic and nuclear protein extracts were prepared using NE-Per Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Pierce, Rockford, IL). Cells were harvested by trypsinization, and separation of cytoplasmic and nuclear fractions was performed according to the manufacturer’s instructions, with the following modifications: 1X SDS loading buffer was added directly to the nuclear pellet at 125 µl/1 × 107 cells, sonicated 3 times at 10-second pulses, and loaded on the gel. Immunoblotting was performed by standard techniques. After SDS/PAGE, the fractionated proteins were transferred to nitrocellulose membranes, probed with an antibody to NRF2 (Cell Signaling Technology, Beverly, MA) followed by anti-rabbit IgG-HRP secondary antibody (Bio-Rad Laboratories). The antigen–antibody complexes were visualized using an enhanced chemiluminescence kit (GE Healthcare Biosciences, Piscataway, NJ). To monitor the separation of cytoplasmic and nuclear fractions, and to control for equal loading, blots were reprobed with anti-lactate dehydrogenase (Rockland Immunochemicals, Limerick, PA) followed by anti-goat IgG-HRP secondary antibody (Bio-Rad Laboratories), and anti-lamin (Santa Cruz Biotechnology, Dallas, TX) followed by anti-rabbit IgG-HRP secondary antibody.
Statistical Analysis.
Results are reported as the mean ± S.E.M., based on at least two to five independent experiments. Significance was assessed by calculation of two-tailed P values by an unpaired Student’s t test.
Results
ROS Levels Are Induced during Exposure to TYMS-Directed Antimetabolites.
Several studies have shown that intracellular ROS levels increase in response to FUra (Hwang et al., 2001; Liu and Chen, 2002; Akhdar et al., 2009; Matsunaga et al., 2010; Lamberti et al., 2012). However, it has not been determined whether such increases are due to inhibition of TYMS as opposed to other effects of the antimetabolite. Indeed, FUra is well known to exert an impact on cellular processes other than thymidylate biosynthesis, such as RNA processing and glycoprotein synthesis (Longley et al., 2003).
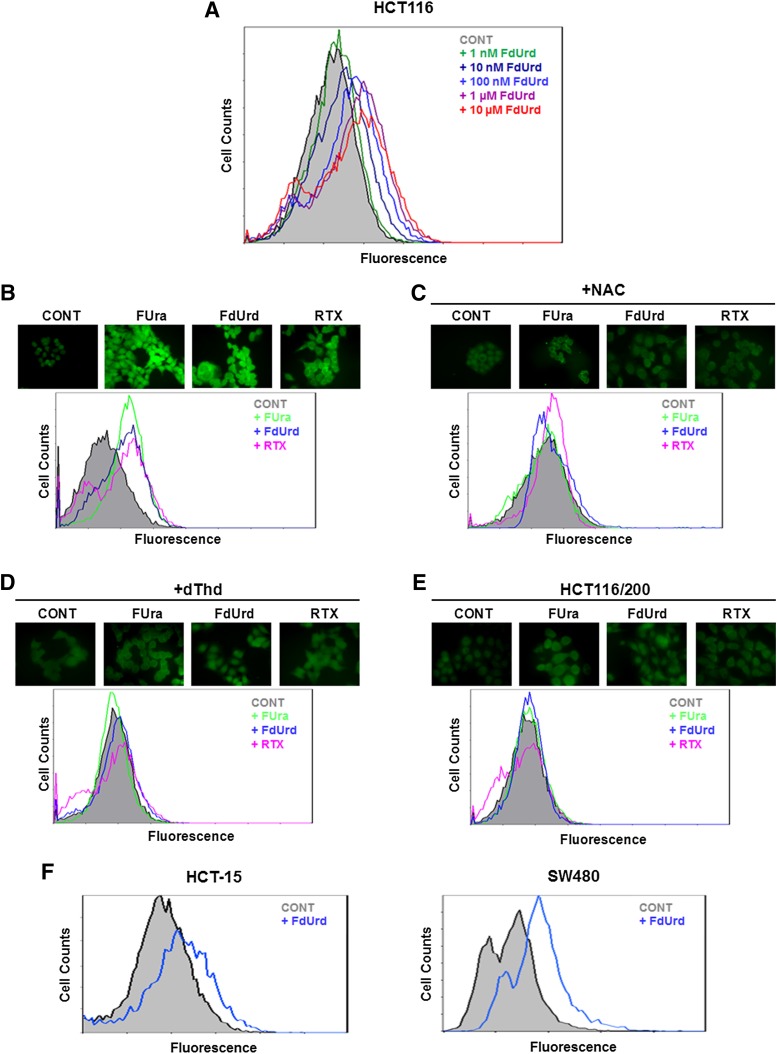
We measured the effects of TYMS inhibition on ROS levels in HCT116 cells treated with various concentrations of the TYMS-directed fluoropyrimidine FdUrd. Cells were analyzed by flow cytometry after the addition of the ROS-detecting fluorescent dye DCFDA to the media. As shown in Fig. 1A, a progressive increase in ROS levels over a drug concentration range of 1 nM to 10 µM was observed, with the maximum level being reached at ∼1 µM.
Fig. 1.
Exposure to TYMS inhibitors leads to increased ROS levels. (A) HCT116 cells were treated for 24 hours in the indicated concentrations of FdUrd and incubated with DCFDA; ROS levels were assessed by flow cytometry. (B–D) Cells were treated for 24 hours with TYMS inhibitors (10 µM FUra, 10 µM FdUrd, or 1 µM RTX), either alone (B), or in the presence of 10 mM NAC (C) or 10 µM dThd (D). After addition of DCFDA, ROS levels were measured by fluorescence microscopy (upper panel) or flow cytometry (lower panel). (E) TYMS-overproducing, FdUrd-resistant HCT116/200 cells were treated for 24 hours with drugs (10 µM FUra, 10 µM FdUrd, or 1 µM RTX), and ROS levels were analyzed by fluorescence microscopy (upper panel) or flow cytometry (lower panel). (F) ROS levels in cell lines HCT-15 (left panel) and SW480 (right panel) were analyzed by flow cytometry after treatment with 10 µM FdUrd for 24 hours. CONT, control, untreated cells.
To verify and extend these initial findings, we examined the impact of three TYMS-directed antimetabolites, including two fluoropyrimidines (FUra, FdUrd) and a folate analog (RTX), on intracellular levels of ROS. The concentration of each was based upon earlier studies in our laboratory (Barbour and Berger, 2008). Using both fluorescence microscopy and flow cytometry, we observed marked increases in ROS concentrations in response to each inhibitor (Fig. 1B). Similar results were obtained with a different fluorescent probe, DHE (data not shown). Further analyses showed that addition of NAC to the culture media abrogated drug-induced fluorescence (Fig. 1C), indicating that the observed increases in fluorescence indeed reflect augmentation of ROS levels after drug exposure. Furthermore, supplementation with dThd caused a loss of drug-mediated ROS induction (Fig. 1D), demonstrating that dTMP deprivation due to TYMS inhibition is occurring. Finally, ROS levels were profoundly diminished in the TYMS-overproducing cell line HCT116/200 (Fig. 1E), which is resistant to fluoropyrimidines as a consequence of TYMS overproduction derived from amplification of the TYMS gene (Berger et al., 1988). In all, these observations show that ROS induction in response to TYMS-directed antimetabolites arises from inhibition of TYMS and reduction in dTMP pools.
FdUrd elicited increased ROS levels in other human colon tumor cell lines, such as HCT-15 and SW480 (Fig. 1F). These lines also responded to FUra and RTX (data not shown). Thus, the observed induction of ROS after exposure to TYMS inhibitors is not a specific property of HCT116 cells but rather is broadly observed among distinct colon tumor cell lines.
ROS Induction by TYMS Inhibitors Promotes Apoptosis.
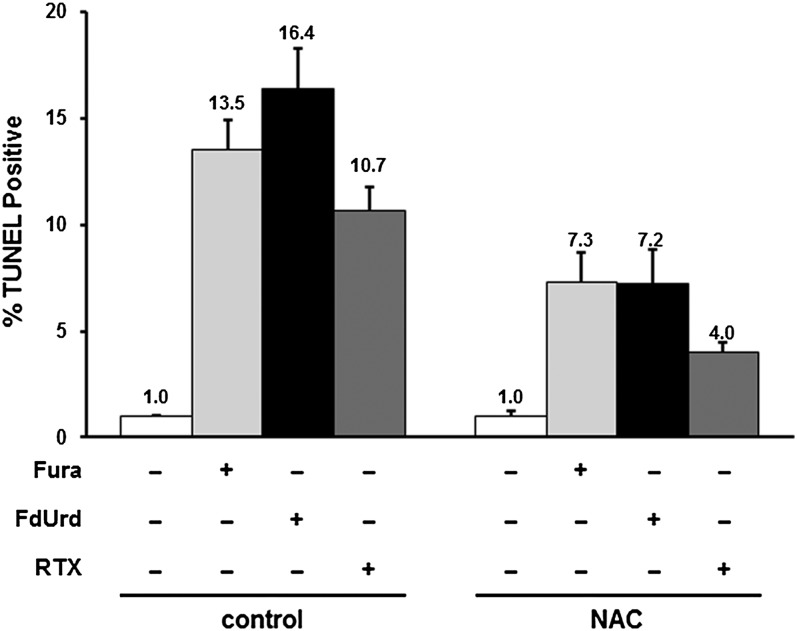
The observed augmentation of ROS levels in cells treated with TYMS inhibitors may promote oxidative stress that contributes to apoptotic cell death. In previous studies, we used TUNEL assays to show that apoptosis is increased by 10- to 20-fold in cells exposed to TYMS inhibitors, and was completely inhibited by the broad-spectrum caspase inhibitor Z-VAD-FMK [N-benzyloxycarbonyl-Val-Ala-Asp(O-Me) fluoromethyl ketone] (Barbour and Berger, 2008). We assessed the role of ROS in apoptosis induced by TYMS inhibitors. Figure 2 shows that the apoptotic index rose ∼11- to 17-fold in response to FUra (P = 3.7 × 10−6), FdUrd (P = 2.5 × 10−6), and RTX (P = 1.6 × 10−5). Furthermore, addition of NAC reduced the index by ∼50% (P = 0.017 for FUra, 4.0 × 10−4 for FdUrd, and 7.0 × 10−4 for RTX), indicating that abrogation of ROS levels reduces apoptotic response to TYMS inhibitors. Thus, oxidative stress is important to the cytotoxicity of these inhibitors.
Fig. 2.
Induction of ROS by TYMS inhibitors promotes apoptosis. Apoptotic indices in HCT116 cells in response to 48 hours of treatment with TYMS inhibitors (10 µM FUra, 100 µM FdUrd, or 1 µM RTX), either alone or in the presence of 10 mM NAC, were measured by TUNEL assays. Apoptotic indices were expressed relative to untreated cells.
NOX Contributes to ROS Induction after Exposure to TYMS Inhibitors.
Previous studies had indicated that the mitochondrial enzyme FDXR is an important source of expanded ROS pools after exposure to FUra (Hwang et al., 2001; Liu and Chen, 2002). Indeed, gene expression profiling conducted in our laboratory (described herein) identified FDXR mRNA as among the transcripts induced by FUra. However, given the abundance of genes that function in redox pathways, it is highly likely that there exist additional enzymes that generate ROS in response to TYMS inhibitors. We tested the role of one such enzyme, NOX, which catalyzes the formation of O2●− via one-electron reduction of O2. The enzyme, which exists as a membrane-bound, multisubunit complex whose sole function appears to be generation of O2●− (Sumimoto, 2008; Jiang et al., 2011; Brandes et al., 2014), is activated by a wide variety of chemical, physical, and inflammatory stress stimuli (Jiang et al., 2011). As such, it is of interest to test the contribution of NOX to expanded ROS pools in response to TYMS inhibitors.
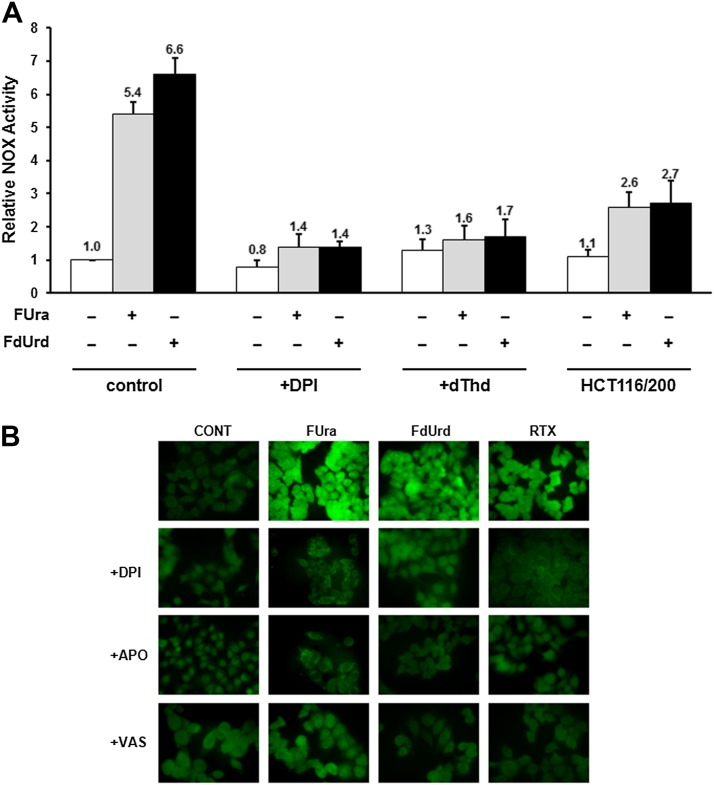
We used a whole-cell chemiluminescence assay to measure total NOX enzyme activity and the effect of TYMS inhibitors. As shown in Fig. 3A, both FUra and FdUrd elicit a 5- to 7-fold activation of enzyme activity (P = 2.7 × 10−3 for FUra and 1.6 × 10−3 for FdUrd). To be assured that NOX is being measured in these assays, we added the NOX inhibitor DPI (Wind et al., 2010) to the culture medium and observed complete inhibition of the drug-induced activity (P = 1.6 × 10−3 for FUra and 1.1 × 10−3 for FdUrd) (Fig. 3A). Attenuation of the increase in enzyme activity was also observed in the presence of dThd (P = 0.024 for FUra and 0.014 for FdUrd), and was significantly reduced in TYMS-overproducing HCT116/200 cells (P = 0.021 for FUra and 8.6 × 10−3 for FdUrd) (Fig. 3A). Thus, NOX activation in response to TYMS inhibitors is primarily due to inhibition of TYMS.
Fig. 3.
Role of NOX in ROS induction by TYMS inhibitors. (A) Total NOX activities were measured in HCT116 cells after 24 hours of exposure to TYMS inhibitors (10 µM FUra or 10 µM FdUrd), either alone or in the presence of 10 µM DPI or 10 µM dThd. HCT116/200 cells were analyzed after exposure to TYMS inhibitors alone. Activities were normalized to those in control, untreated cells. (B) HCT116 cells were exposed to TYMS inhibitors (10 µM FUra, 10 µM FdUrd, or 1 µM RTX) either alone or in the presence of 10 µM DPI, 1 µM APO, or 10 µM VAS. Cells were stained with DCFDA and observed by fluorescence microscopy.
We determined whether NOX functions in drug-mediated accumulation of ROS. DPI inhibited ROS induction in response to FUra, FdUrd, and RTX, as measured by fluorescence assays with DCFDA (Fig. 3B). Furthermore, another NOX inhibitor, APO, which has a mechanism of action distinct from DPI (Aldieri et al., 2008), also prevented ROS induction by the three TYMS-targeted drugs (Fig. 3B). Because both DPI and APO are nonspecific and exert NOX-independent effects (Aldieri et al., 2008), we tested the impact of VAS, a triazolo pyrimidine that exhibits a high degree of specificity for NOX (Wind et al., 2010). VAS, similar to DPI and APO, reduced drug-mediated ROS accumulation (Fig. 3B). Similar findings as these were made using DHE as the fluorescent probe for ROS detection (data not shown). These results implicate NOX activation as functioning in augmentation of ROS levels after exposure to TYMS inhibitors.
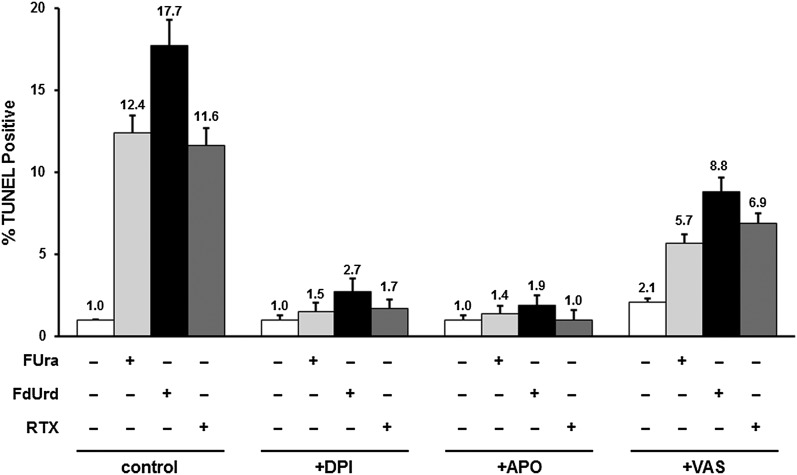
We measured the impact of drug-mediated NOX activation on apoptosis. As shown in Fig. 4, increases in the apoptotic index after exposure to FUra, FdUrd, or RTX were abolished by both DPI (P = 9.7 × 10−6 for FUra, 1.0 × 10−5 for FdUrd, and 3.4 × 10−5 for RTX) and APO (P = 4.1 × 10−5 for FUra, 1.1 × 10−5 for FdUrd, and 1.6 × 10−5 for RTX), indicating an important role for NOX in drug response. VAS also abrogated drug-induced increases in apoptosis, but only by ∼50% (P = 0.013 for FUra, 0.047 for FdUrd, and 0.063 for RTX) (Fig. 4). The effect of VAS was consistently less than that of either DPI or APO, which may reflect the fact that the latter two exhibit a broader range of specificity than the former (Aldieri et al., 2008; Wind et al., 2010). This implies that factors other than NOX may contribute to the apoptotic response to TYMS-targeted agents.
Fig. 4.
Role of NOX in apoptotic response to TYMS inhibitors. HCT116 cells were exposed for 48 hours to TYMS inhibitors (10 µM FUra, 100 µM FdUrd, or 1 µM RTX), either alone or in the presence of 10 µM DPI, 1 µM APO, or 10 µM VAS. Apoptotic indices were measured by TUNEL assays.
Expression of NOX Regulatory Subunit p67phox Is Stimulated by TYMS Inhibitors.
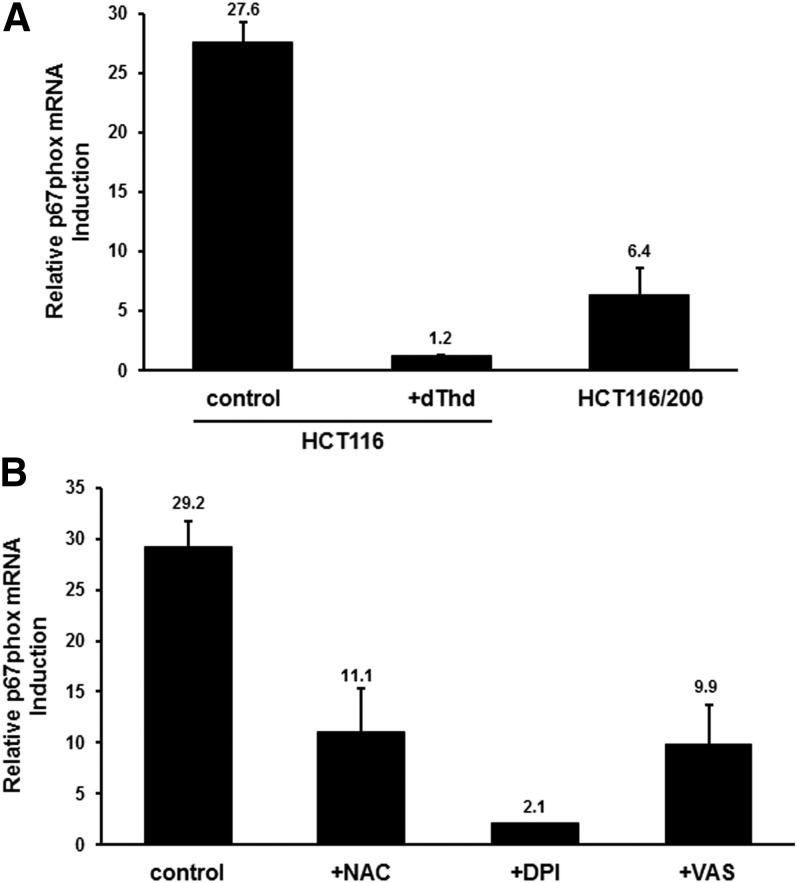
The NOX family is organized into a number of distinct multisubunit complexes, each member of which is comprised of one of seven known core catalytic subunits along with several regulatory subunits ( Sumimoto, 2008; Block and Gorin, 2012; Brandes et al., 2014). To gain insight into the mechanism by which NOX is activated in cells treated with TYMS-directed drugs, we examined expression of mRNAs encoding several NOX subunits, including catalytic (NOX1-4 and p22phox) as well as regulatory (p47phox, p67phox, p40phox, NOX01, NOXA1, and Rac1). For most mRNAs, there were no detectable changes in levels of mRNA expression after exposure to FdUrd, as measured by qRT-PCR (data not shown). The exception was the NOX2 regulatory subunit p67phox, which is required for maximal activity of the NOX2 isoform (Italiano et al., 2012). Expression of p67phox mRNA was up-regulated ∼20- to 25-fold by FdUrd (P = 5.1 × 10−5) (Fig. 5A). This up-regulation was prevented by addition of dThd (P = 5.0 × 10−6), and was significantly attenuated in TYMS-overproducing HCT116/200 cells (P = 2.0 × 10−5) (Fig. 5A). Thus, TYMS inhibition and subsequent reduction of dTMP pool levels are crucial to the up-regulation of p67phox.
Fig. 5.
Induction of p67phox mRNA by TYMS inhibitors. (A) HCT116 cells were treated for 24 hours with 10 µM FdUrd either alone or in the presence of 10 µM dThd. RNA was extracted and analyzed for p67phox mRNA levels by qRT-PCR. HCT116/200 cells were analyzed after exposure to FdUrd alone. The induction of p67phox mRNA concentrations relative to that in untreated cells is shown. (B) HCT116 cells were treated for 24 hours with 10 µM FdUrd either alone or in the presence of 10 µM NAC, 10 µM DPI, or 10 µM VAS, and p67phox mRNA levels were analyzed by qRT-PCR. Induction of p67phox mRNA concentrations relative to that in untreated cells is shown.
Interestingly, the antioxidant NAC as well as NOX inhibitors DPI and VAS reduced the drug-mediated increases in p67phox mRNA (P = 1.3 × 10−3 for NAC, 8.0 × 10−5 for DPI, and 9.0 × 10−4 for VAS) (Fig. 5B). This suggests a “feed-forward” mechanism whereby ROS generated by NOX activation leads to induction of p67phox and NOX activity.
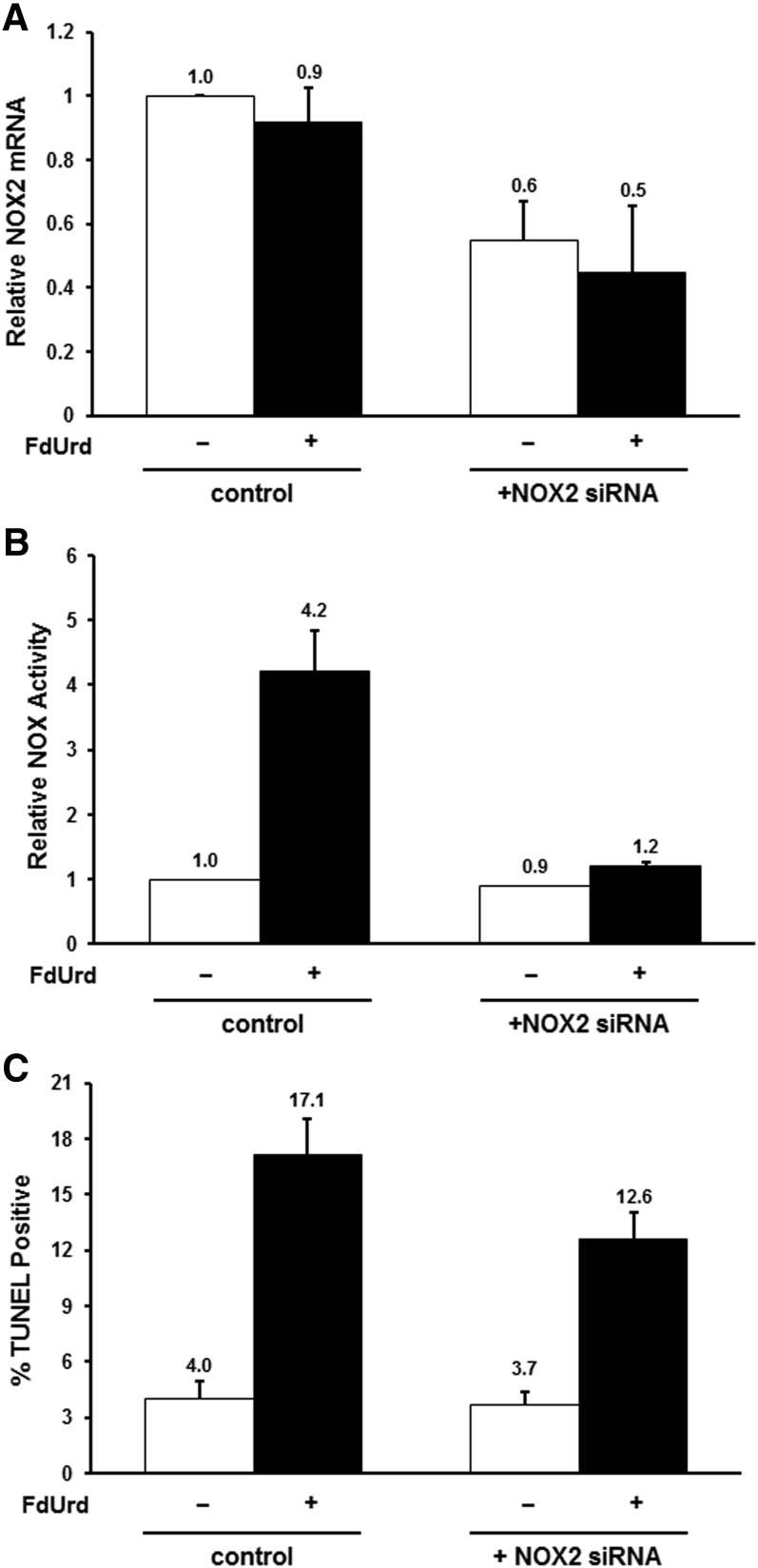
The observation of induced p67phox expression in response to TYMS inhibitors, coupled with the fact that p67phox specifically regulates the NOX2 isoform (Sumimoto, 2008; Jiang et al., 2011; Block and Gorin, 2012), implies the latter as significant in activation of NOX activity and subsequent apoptosis accompanying drug exposure. Therefore, we assessed the impact of NOX2 down-regulation using siRNA transfection. Introduction of NOX2-specific siRNAs led to ∼50% reduction in NOX2 mRNA in both control and FdUrd-treated cells (P = 1.5 × 10−3 and 2 × 10−2, respectively), while a control nonspecific “scrambled” siRNA had no effect (Fig. 6A). Reduction of NOX2 expression had essentially no impact on basal NOX activity levels before drug exposure (Fig. 6B), indicating that the isoform contributes little to NOX activity in normal cells. In contrast, the NOX2-specific siRNA completely inhibited the FdUrd-mediated increase in enzyme activity (P = 0.011) (Fig. 6B), signifying that this increase is largely due to NOX2. A modest, yet significant, ∼30% reduction in the apoptotic index was observed in FdUrd-treated cells transfected with the NOX2-specific siRNA (P = 0.013), while there was measurable effect of the siRNA in untreated cells (Fig. 6C). Thus, associated with down-regulation of NOX2 mRNA expression was attenuation of drug-mediated induction of both NOX activity and apoptotic cell death, implying that the NOX2 isoform plays a significant role in the apoptotic response to TYMS inhibitors.
Fig. 6.
Down-regulation of the NOX2 isoform attenuates response to TYMS inhibitors. (A) HCT116 cells were transfected for 24 hours with a scrambled, nonspecific control siRNA or a NOX2-specific siRNA, and exposed to 10 µM FdUrd for an additional 24 hours. NOX2 mRNA levels were analyzed by qRT-PCR. (B) Cells transfected for 24 hours with a scrambled, nonspecific control siRNA or a NOX2-specific siRNA were exposed to 10 µM FdUrd for an additional 24 hours, and assayed for total NOX enzyme activities. (C) Cells transfected for 24 hours with a scrambled, nonspecific control siRNA or a NOX2-specific siRNA were exposed to 100 µM FdUrd for an additional 48 hours, and the apoptotic index was measured by TUNEL assays.
Alterations in the Redox Transcriptome by TYMS Inhibitors.
In light of the results reported in the previous sections, we were interested in determining the impact of TYMS-directed antimetabolites on the cellular transcriptome, particularly with regard to mRNAs involved in redox metabolism. We therefore performed a whole-genome cDNA microarray analysis of HCT116 cells both before and after 24 hours of exposure to 10 µM FUra. Data were filtered to include transcripts exhibiting at least a 1.5-fold change with P < 0.005. We found 1975 genes whose levels of expression were altered by FUra; among these, 1119 (56.6%) were induced in response to drug, while 856 (43.4%) were repressed (Supplemental Table 1). For the induced genes, 452 (41%) increased by 2-fold or more, 63 (5.6%) by 4-fold or more, and 16 (1.4%) by 6-fold or more; the largest induction was observed for CEACAM1, which increased by ∼14-fold. Among the repressed genes, 243 (28%) decreased by 2-fold or more, 12 (1.4%) by 4-fold or more, and 4 (0.5%) by 6-fold or more; the largest change was exhibited by HIST1H2BF, which decreased by ∼8-fold.
To identify redox-related transcripts (the redox transcriptome), we queried both the GO (Gene Ontogeny) and KEGG (Kyoto Encyclopedia of Genes and Genomes) databases using the terms “dehydro, dehydrog, hydro, hydrog, oxid, oxidase, oxidation, redox, reduc, reductase.” Those present in the microarray were considered FUra-regulated redox genes. This strategy led to identification of 54 genes, 27 of which were up-regulated after drug exposure (e.g., AKR1B10, ALDH3A1, FDXR, GGT1, GPX1, GLRX2, SAT1, SOD2, HMOX1, LOXL4, CBR3), and 27 that were down-regulated (e.g., DHRS2, CYB5RL, HSD17B8, ACADSB, CRYL1, SORD) (Supplemental Table 2). The presence of FDXR among the up-regulated genes corroborates earlier findings by others (Hwang et al., 2001; Liu and Chen, 2002). We found that p67phox did not show up in the microarray, likely due to its low levels of expression. Our findings, overall, indicate that exposure to TYMS inhibitors modifies the redox transcriptome within the cell through altered expression of a variety of redox-related genes.
NRF2-Regulated Genes Are among Those Induced by TYMS Inhibitors.
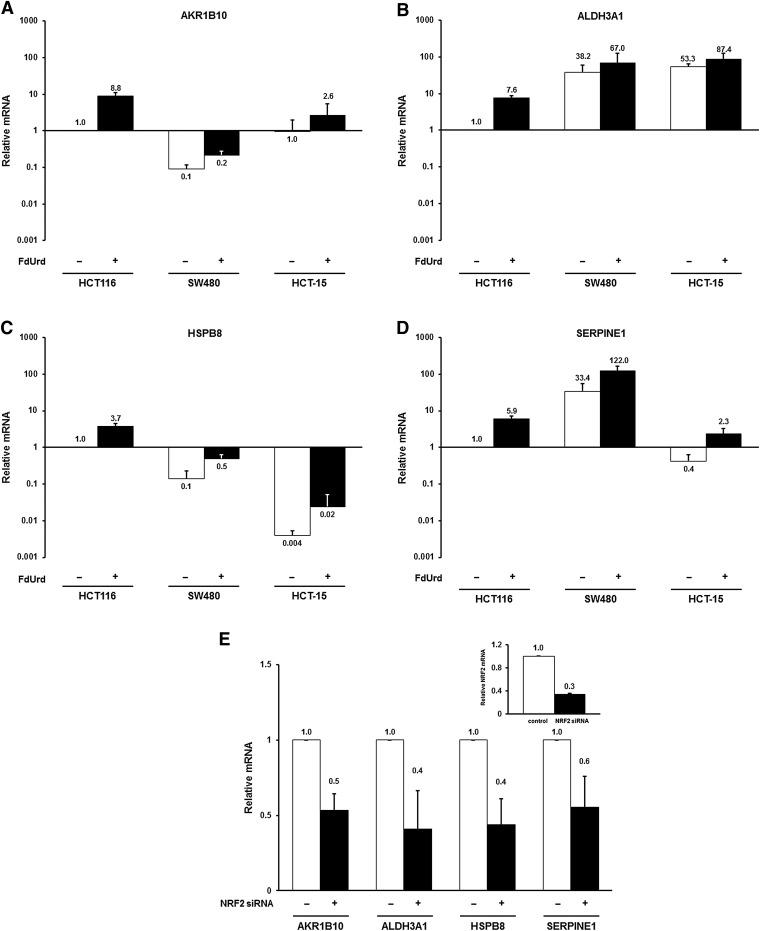
Notably, several of the redox transcripts whose expression is modulated by FUra (e.g., AKR1B10, ALDH3A1, GGT1, HMOX1) are known to be regulated by transcription factor NRF2. NRF2 is a member of the cap ’n collar family of transcription factors that controls expression of genes conferring protection against oxidative and electrophilic stress (Baird and Dinkova-Kostova, 2011; Jaramillo and Zhang, 2013). Table 2 lists 27 transcripts from the microarray whose expression levels have been identified in the literature as regulated by NRF2 (Agyeman et al., 2012; Suzuki et al., 2013; Choi et al., 2014; Hayes and Dinkova-Kostova, 2014). We tested 15 of these by qRT-PCR and confirmed all as inducible by FdUrd (Table 2). An additional six transcripts, representing known NRF2-regulated genes that did not appear in the microarray, were examined by qRT-PCR and shown to be induced by FdUrd (Table 3). In all, we identified 33 NRF2-modulated genes whose levels of expression change in response to TYMS inhibitors.
TABLE 2.
Identification of FUra-inducible NRF2 target genes by gene expression profiling
NRF2 target genes identified in the whole genome microarray analysis (Supplemental Table 2) are listed. Classification of each gene as an NRF2 target is based upon the literature (Agyeman et al., 2012; Suzuki et al., 2013; Choi et al., 2014; Hayes and Dinkova-Kostova, 2014). The fold change represents the extent of induction by exposure to 10 µM FUra for 24 hours (see text). The P value is corrected for multiple testing using the Benjamini-Hochberg algorithm (see Materials and Methods). Targets in boldface were tested by qRT-PCR, and confirmed as inducible by FdUrd.
| Gene | Description | Fold Change | Corrected P Value |
|---|---|---|---|
| ABCC2 | ATP-binding cassette, subfamily C, member 2 | 1.5861826 | 0.000741887 |
| AGPAT9 | Acylglycerol-3-phosphate O-acyltransferase 9 | 1.6375676 | 0.000784155 |
| AHR | Aryl hydrocarbon receptor | 1.742765 | 0.001298002 |
| AKR1B10 | Aldo-keto reductase family 1, member B10 | 6.701502 | 2.20317E-05 |
| AKR1B15 | Aldo-keto reductase family 1, member B15 | 6.0408144 | 0.000258688 |
| AKR1C1 | Aldo-keto reductase family 1, member C1 | 2.3066108 | 0.001858692 |
| AKR1C3 | Aldo-keto reductase family 1, member C3 | 1.9071698 | 0.001267411 |
| ALDH3A1 | Aldehyde dehydrogenase family 3, member A1 | 2.0209148 | 0.001248846 |
| ALDH4A1 | Aldehyde dehydrogenase family 4, member A1 | 2.560456 | 0.000557924 |
| ATF3 | Activating transcription factor 3 | 3.492217 | 6.41362E-05 |
| BLVRB | Biliverdin reductase B | 2.0149345 | 0.001494992 |
| GADD45A | Growth arrest and DNA-damage-inducible, alpha | 2.0611608 | 0.000269189 |
| GBE1 | Glucan (1,4-alpha-), branching enzyme 1 | 1.7280452 | 6.94566E-05 |
| GGT1 | Gamma-glutamyltransferase 1 | 2.475309 | 0.000289259 |
| GLRX2 | Glutaredoxin 2 | 1.6844928 | 0.000622618 |
| GLS | Glutaminase | 2.4446886 | 0.000183834 |
| HMOX1 | Heme oxygenase 1 | 2.0041764 | 1.4295E-05 |
| HSPB8 | Heat shock 22-kDa protein 8 | 2.7588468 | 0.00043559 |
| KIFAP3 | Kinesin-associated protein 3 | 1.807134 | 0.00020369 |
| LIPH | Lipase, member H | 1.9393841 | 0.000126275 |
| MAFF | v-Mafavian musculoaponeurotic fibrosarcoma oncogene homolog F | 2.1331222 | 8.87782E-05 |
| MGST3 | Microsomal glutathione S-transferase 3 | 1.5016723 | 0.0031992 |
| MT1B | Metallothionein 1B | 1.5310463 | 0.000482686 |
| MT2A | Metallothionein 2A | 1.7345982 | 0.002288793 |
| SAT1 | Spermidine/spermine N1-acetyltransferase 1 | 2.4873772 | 6.04632E-05 |
| SERPINE1 | Serpin peptidase inhibitor, clade E, member 1 | 5.988837 | 7.4388E-05 |
| SPP1 | Secreted phosphoprotein 1 | 3.0035641 | 5.6968E-05 |
TABLE 3.
Additional NRF2 target genes induced by TYMS inhibitors, identified by qRT-PCR
Genes that did not show up in the microarray analysis but are known from the literature to be NRF2 targets (Agyeman et al., 2012; Suzuki et al., 2013; Choi et al., 2014; Hayes and Dinkova-Kostova, 2014) were tested for response to TYMS inhibitors by qRT-PCR. The fold-change represents the extent of induction by 10 µM FdUrd (with 10 µM folinic acid) for 24 hours before harvesting.
| Gene | Description | Fold Change |
|---|---|---|
| ABCC5 | ATP-binding cassette, subfamily C, member 5 | 2.1 ± 0.1 |
| AOX1 | Aldehyde oxidase 1 | 2.2 ± 0.5 |
| CES1 | Carboxylesterase 1 | 4.9 ± 2.4 |
| GSTA1 | Glutathione S-transferase α 1 | 7.6 ± 2.4 |
| GSTM1 | Glutathione S-transferase μ 1 | 5.2 ± 0.2 |
| GSTM4 | Glutathione S-transferase μ 4 | 2.8 ± 0.5 |
We determined the extent to which these findings apply to additional colon tumor cell lines. Figure 7, A–D, shows the results of qRT-PCR analysis of mRNA levels for four genes (AKR1B10, ALDH3A1, HSPB8, and SERPINE1) in cell lines SW480 and HCT-15, as compared with HCT116. Though considerable heterogeneity in expression of these genes was observed among the three cell lines, induction of expression by FdUrd was observed in all three, though to variable extents. It is thus apparent that increased expression of NRF2-regulated genes in the presence of TYMS inhibitors is not specific to cell line HCT116, but occurs among distinct colon tumor cell lines.
Fig. 7.
NRF2-regulated genes are induced by TYMS inhibitors. HCT116, SW480, and HCT-15 cells were treated for 24 hours with 10 µM FdUrd, and expression of mRNAs corresponding to several NRF2 target genes were analyzed by qRT-PCR, including (A) AKR1B10, (B) ALDH3A1, (C) HSPB8, and (D) SERPINE1. For each mRNA, values were expressed relative to those in untreated HCT116 cells. (E) HCT116 cells were transfected for 24 hours with a scrambled, nonspecific control siRNA or the NRF2-specific siRNAs. Expression of the indicated mRNAs, as well as NRF2 (inset), was analyzed by qRT-PCR.
To verify that these genes are indeed NRF2-regulated, we examined their expression after siRNA-mediated down-regulation of the transcription factor in HCT116 cells. As shown in Fig. 7E, we observed a 40%–60% reduction in mRNA levels for AKR1B10 (P = 0.017), ALDH3A1 (P = 0.016), HSPB8 (P = 0.043), and SERPINE1 (P = 0.019) after introduction of an NRF2-specific siRNA.
Nuclear Accumulation of NRF2 and Its Role in Apoptotic Response to TYMS Inhibitors.
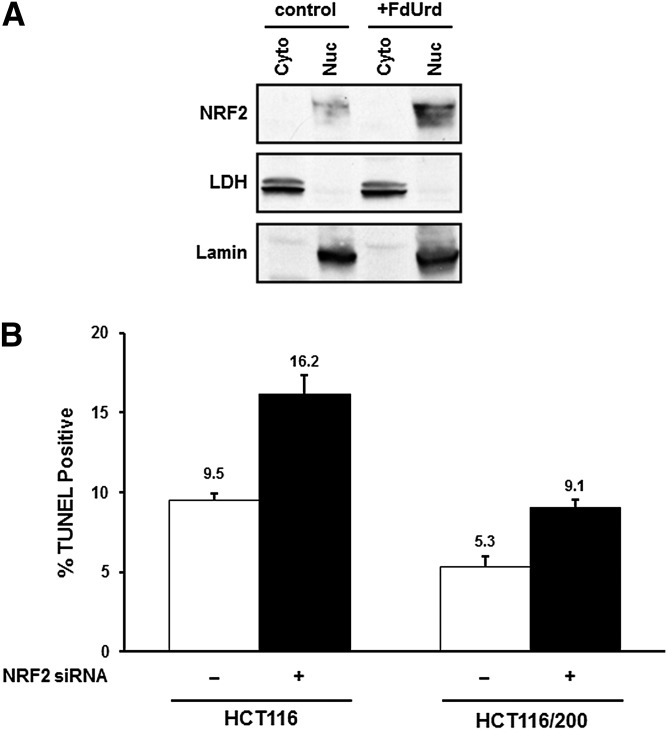
Under normal circumstances, NRF2 is maintained at low levels within the cell, a status maintained by the transcription factor’s intrinsically short half-life. NRF2 is degraded by ubiquitin-mediated proteasomal degradation facilitated by the adaptor molecule Kelch-like ECH-associated protein 1 (KEAP1) (Baird and Dinkova-Kostova, 2011; Ma and He, 2012; Jaramillo and Zhang, 2013; Kansanen et al., 2013; Geismann et al., 2014). Under conditions of oxidative or electrophilic stress, KEAP1 becomes disabled, allowing NRF2 to escape degradation and accumulate in the nucleus, where it binds antioxidant response elements within promoter regions and induces transcription (Baird and Dinkova-Kostova, 2011; Jaramillo and Zhang, 2013). To determine the concentration and intracellular locale of NRF2 both before and after exposure to TYMS inhibitors, we performed Western immunoblotting of cytoplasmic and nuclear fractions of HCT116 cells. Figure 8A shows that NRF2 was present in the nucleus, with little, if any, in the cytoplasm. Furthermore, its concentration in the nucleus increases after treatment with FdUrd.
Fig. 8.
(A) Cytoplasmic (Cyto) and nuclear (Nuc) extracts were prepared from control HCT116 cells and from those treated for 24 hours with 10 μM FdUrd. NRF2 levels were assessed by Western blotting. Lactate dehydrogenase and lamin were measured to ensure separation of cytoplasmic and nuclear fractions, respectively. (B) Cells (HCT116 and TYMS-overproducing HCT116/200) were transfected for 24 hours with a scrambled, nonspecific control siRNA or NRF2-specific siRNAs, and treated for an additional 48 hours with 10 µM FdUrd. Apoptotic indices were measured by TUNEL assays.
The finding that induction of NRF2-regulated genes by TYMS inhibitors is associated with an increase in the nuclear NRF2 concentration prompted us to ask whether such induction impacts apoptosis in FdUrd-treated cells. Therefore, we analyzed apoptotic indices in cells in which NRF2 levels were down-regulated by siRNAs (see Fig. 7E, above). For this analysis, we reduced the FdUrd concentrations to 10 µM to maximize the chances of observing an increase in apoptosis after reduction of NRF2 concentrations; this resulted in slightly lower extents of apoptosis as compared with experiments in Figs. 2, 4, and 6. As shown in Fig. 8B, the apoptotic index increased by ∼80% in FdUrd-treated HCT116 cells transfected with NRF2-specific siRNAs, as compared with cells with a control siRNA (P = 0.017).
We also examined TYMS-overproducing HCT116/200 cells, which are relatively resistant to TYMS inhibitors (see Fig. 1) (Berger et al., 1988). These cells, as expected, exhibited a reduced apoptotic index relative to parental HCT116 cells; similar to the parental cells, there was a 70% increase in apoptosis after exposure to FdUrd (P = 0.024). Thus, down-regulation of NRF2 levels results in sensitization to drug exposure.
Discussion
ROS are well-known mediators of both tumorigenesis as well as response to cytotoxic agents (Ozben, 2007; Trachootham et al., 2009; Gupta et al., 2012; Nogueira and Hay, 2013). They are generally maintained at higher concentrations in tumor cells relative to normal cells, and can function as protumorigenic signaling molecules that promote cell growth and proliferation. However, due to their highly reactive nature, ROS levels that are excessive can lead to intracellular molecular damage, which includes chemical modification of macromolecules and small metabolites, oxidative stress, and cell death. Thus, cancer cells must maintain levels of ROS that are high enough to sustain robust cell growth yet below the threshold that initiates intracellular oxidative damage. This requires an intricate balance between pro-oxidants and antioxidants. Augmented ROS levels in tumor cells puts them closer to this threshold, making them vulnerable to excessive oxidative stress. This creates an opportunity to augment the efficacy of therapeutic agents used in the control of cancer (Trachootham et al., 2009). Indeed, it is likely that modification of the intracellular redox environment in the direction of excess ROS will enhance the sensitivity of tumors to cytotoxic drugs used in cancer chemotherapy.
In the current study, we examined the role of oxidative stress in the cytotoxic effects of TYMS-targeted antimetabolites. We have shown that ROS levels are increased after exposure to TYMS inhibitors such as FUra, FdUrd, and RTX (Fig. 1). Such increases are attenuated by the antioxidant NAC, by dThd, and by overproduction of TYMS. Thus, it is TYMS inhibition, as opposed to some other drug effect, that underlies stimulation of ROS levels. Drug-mediated apoptotic cell death, like the ROS levels, is reduced by NAC, indicating that ROS induction is critical to promotion of apoptosis (Fig. 2). These observations, in total, strongly suggest that oxidative stress plays a central role in cell death mediated by TYMS-targeted agents. It is evident that TYMS inhibitors stimulate increases in ROS concentrations that exceed the threshold required for initiation of oxidative stress and eventual cell death.
We have identified NOX as a source of increased ROS levels in cells responding to TYMS inhibitors. NOX occurs as a family of at least seven multisubunit complexes that catalyze transfer of an electron from NADPH to O2, generating O2●− (Sumimoto, 2008; Jiang et al., 2011; Block and Gorin, 2012; Brandes et al., 2014). Activities of NOX complexes respond to a wide range of hormonal, chemical, viral, and bacterial stimuli, and have been implicated in a variety of physiologic functions, including host defense, cell signaling, and the regulation of gene expression (Sumimoto, 2008; Jiang et al., 2011; Block and Gorin, 2012).
Several findings described in our report support the notion that NOX contributes to oxidative stress and apoptotic cell death brought on by exposure to TYMS inhibitors. First, total NOX enzyme activity was significantly increased in the presence of drug (Fig. 3A). Second, three NOX inhibitors (DPI, APO, and VAS) markedly attenuated increases in both ROS levels (Fig. 3B) and apoptotic indices (Fig. 4) in response to TYMS inhibitors. Third, expression of p67phox, a regulatory subunit for the NOX2 isoform, was up-regulated in cells exposed to TYMS inhibitors (Fig. 5). Finally, introduction of a NOX2-specific siRNA into cells completely abrogated increases in NOX activity and resulted in attenuation of apoptosis after drug treatment (Fig. 6). These findings, collectively, indicate that NOX, in particular the NOX2 isoform, is a source of increased ROS in response to TYMS-directed agents.
Interestingly, drug-induced NOX enzyme activity was completely inhibited by the NOX2-specific siRNA (Fig. 6B) whereas apoptosis was reduced by only ∼30% (Fig. 6C). This suggests that increased NOX2 activity does not fully account for the apoptotic response to drug exposure. Other NOX isoforms may be involved, a notion that will require further investigation. Other enzymes of redox metabolism that are induced or activated by TYMS inhibitors (e.g., FDXR) may also be important. This possibility is consistent with analysis of the effects of FUra on the cellular transcriptome, in which mRNAs corresponding to a number of dehydrogenases, reductases, and oxidases were impacted (Supplemental Table 2). Thus, TYMS-targeted antimetabolites elicit marked modifications in the redox transcriptome, which may determine the oxidative environment within the cell, and as a consequence, the cytotoxic impact of drug exposure.
Of note is the observation that several transcripts induced by TYMS inhibitors are known target genes of transcription factor NRF2, which plays a central role in ameliorating oxidative and electrophilic stress within cells. Genes regulated by NRF2 exert their protective effects in a variety of ways, including export of xenobiotic compounds, reduction in ROS levels, and removal or metabolism of oxidative by-products (Baird and Dinkova-Kostova, 2011; Jaramillo and Zhang, 2013; Hayes and Dinkova-Kostova, 2014). NRF2 is normally maintained at low levels within the cell due to its high rate of proteasomal degradation, which is mediated by the factor’s binding to Kelch-like ECH-associated protein 1 (KEAP1) (Baird and Dinkova-Kostova, 2011; Ma and He, 2012; Jaramillo and Zhang, 2013; Kansanen et al., 2013; Geismann et al., 2014). During oxidative or electrophilic stress, KEAP1 is inactivated, resulting in stabilization and accumulation of NRF2, eventually leading to induction of antioxidant response element–containing genes (Baird and Dinkova-Kostova, 2011; Jaramillo and Zhang, 2013). Consistent with this model of NRF2 activation, we find that exposure to FdUrd leads to an accumulation of the transcription factor in the nucleus (Fig. 8), providing a mechanism by which TYMS-directed antimetabolites activate NRF2-regulated transcription. Furthermore, we found that siRNA-mediated down-regulation of NRF2 leads to increased apoptosis in FdUrd-treated cells, indicating that the transcription factor inhibits apoptotic response to TYMS inhibitors and generates a chemoresistant state.
Analysis of several colon tumor cell lines showed that while considerable heterogeneity in expression of the NRF2-regulated genes exists, induction of expression by FdUrd was observed in all three, though to variable extents (Fig. 7). This heterogeneity among cell lines is not unexpected. It should be recalled that the cDNA microarray experiment leading to the identification of these genes was performed in HCT116 cells; it is therefore feasible that an overlapping yet distinct set of drug-inducible genes exist in the other cells lines, and would have been recognized had the microarray analyses been performed in those lines.
In a number of circumstances, NRF2 has been shown to confer relative resistance to chemotherapeutic agents (Kensler et al., 2007; Wang et al., 2008; Ma and He, 2012). Such may be the case for TYMS inhibitors. Our observations invite speculation that expression of NRF2-regulated genes and their induction by TYMS inhibitors may constrain therapeutic response to this class of drugs. Further studies will be necessary to determine which of the many NRF2-modulated genes mediate reduced drug response. Furthermore, the cDNA microarray analysis presented in the current report makes it clear that many genes that are not regulated by NRF2 are induced by exposure to TYMS inhibitors and could be involved in drug response. Thus, it is likely that NRF2 is one of many molecular players that control cellular response to TYMS inhibitors.
In conclusion, our study shows that TYMS-targeted antimetabolites induce oxidative stress that promotes cell death; at the same time, these agents activate antioxidative mechanisms that inhibit or reverse such stress and constrain drug response. Thus, the relative balance between these two processes determines the cytotoxic impact of drug exposure. This has important implications for the clinical use of TYMS inhibitors in cancer treatment. By reducing NRF2 levels within the cell and preventing the nuclear accumulation of the transcription factor that occurs in the presence of TYMS inhibitors, it may be possible to increase the efficacy of these drugs, thereby enhancing their clinical impact. With this in mind, ongoing efforts are aimed at a deeper understanding of the role of NRF2 as a determinant of response to TYMS-directed antimetabolites.
Supplementary Material
Acknowledgments
The authors thank Dr. Diego Altomare of the South Carolina College of Pharmacy, DNA Microarray Core Facility, for assistance with microarrays and data analysis.
Abbreviations
- APO
apocyanin
- DCFDA
2ʹ,7ʹ-dichlorodihydrofluorescein diacetate
- DHE
dihydroethidium
- DPI
diphenyleneiodonium
- dThd
deoxythymidine
- dUMP
2ʹ-deoxyuridine-5ʹ-monophosphate
- FdUrd
5-fluoro-2ʹ-deoxyuridine
- FDXR
ferredoxin reductase
- FUra
5-fluorouracil
- GW1843U89
(S)-2-(5-(((3-methyl-1-oxo-1,2-dihydrobenzo[f]quinazolin-9-yl)methyl)amino)-1-oxoisoindolin-2-yl)pentanedioic acid
- LY231514
(2R)-2-[[4-[2-(2-amino-4-oxo-1,7-dihydropyrrolo[2,3-d]pyrimidin-5-yl)ethyl]benzoyl]amino]pentanedioic acid
- NAC
N-acetylcysteine
- NOX
NADPH oxidase
- NRF2
nuclear factor E2-related factor 2
- O2●−
superoxide
- qRT-PCR
quantitative reverse-transcription polymerase chain reaction
- ROS
reactive oxygen species
- RTX
raltitrexed
- siRNAs
small interfering RNAs
- TYMS
thymidylate synthase
- VAS
VAS-2870, VAS-2870, 3-benzyl-7-(2-benzoxazolyl)thio-1,2,3-triazolo(4,5-d)pyrimidine
- ZD9331
(2S)-2-[[4-[(2,7-dimethyl-4-oxo-1H-quinazolin-6-yl)methyl-prop-2-ynylamino]-2-fluorobenzoyl]amino]-4-(2H-tetrazol-5-yl)butanoic acid
Authorship Contributions
Participated in research design: Ozer, Barbour, Clinton, Berger.
Conducted experiments: Ozer, Barbour, Clinton.
Performed data analysis: Ozer, Barbour, Clinton, Berger.
Wrote or contributed to the writing of the manuscript: Ozer, Barbour, Berger.
Footnotes
This work was supported by the National Cancer Institute [Grant CA44013]; and the National Institutes of Health Centers of Biomedical Research Excellence program of the National Institute of General Medical Sciences [Grant GM103336]. The Microarray Core Facility receives partial support from South Carolina IDeA Networks of Biomedical Research Excellence [Grant P20GM103499].
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Agyeman AS, Chaerkady R, Shaw PG, Davidson NE, Visvanathan K, Pandey A, Kensler TW. (2012) Transcriptomic and proteomic profiling of KEAP1 disrupted and sulforaphane-treated human breast epithelial cells reveals common expression profiles. Breast Cancer Res Treat 132:175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhdar H, Loyer P, Rauch C, Corlu A, Guillouzo A, Morel F. (2009) Involvement of Nrf2 activation in resistance to 5-fluorouracil in human colon cancer HT-29 cells. Eur J Cancer 45:2219–2227. [DOI] [PubMed] [Google Scholar]
- Aldieri E, Riganti C, Polimeni M, Gazzano E, Lussiana C, Campia I, Ghigo D. (2008) Classical inhibitors of NOX NAD(P)H oxidases are not specific. Curr Drug Metab 9:686–696. [DOI] [PubMed] [Google Scholar]
- Baird L, Dinkova-Kostova AT. (2011) The cytoprotective role of the Keap1-Nrf2 pathway. Arch Toxicol 85:241–272. [DOI] [PubMed] [Google Scholar]
- Barbour KW, Berger FG. (2008) Cell death in response to antimetabolites directed at thymidylate synthase. Cancer Chemother Pharmacol 61:189–201. [DOI] [PubMed] [Google Scholar]
- Berger FG, Berger SH. (2006) Thymidylate synthase as a chemotherapeutic drug target: where are we after fifty years? Cancer Biol Ther 5:1238–1241. [DOI] [PubMed] [Google Scholar]
- Berger SH, Barbour KW, Berger FG. (1988) A naturally occurring variation in thymidylate synthase structure is associated with a reduced response to 5-fluoro-2′-deoxyuridine in a human colon tumor cell line. Mol Pharmacol 34:480–484. [PubMed] [Google Scholar]
- Bertino JR. (1997) Chemotherapy of colorectal cancer: history and new themes. Semin Oncol 24(Suppl 18):S18-13–S18-17. [PubMed] [Google Scholar]
- Block K, Gorin Y. (2012) Aiding and abetting roles of NOX oxidases in cellular transformation. Nat Rev Cancer 12:627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes RP, Weissmann N, Schröder K. (2014) Nox family NADPH oxidases: Molecular mechanisms of activation. Free Radic Biol Med 76:208–226. [DOI] [PubMed] [Google Scholar]
- Carreras CW, Santi DV. (1995) The catalytic mechanism and structure of thymidylate synthase. Annu Rev Biochem 64:721–762. [DOI] [PubMed] [Google Scholar]
- Choi BH, Kang KS, Kwak MK. (2014) Effect of redox modulating NRF2 activators on chronic kidney disease. Molecules 19:12727–12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu E, Callender MA, Farrell MP, Schmitz JC. (2003) Thymidylate synthase inhibitors as anticancer agents: from bench to bedside. Cancer Chemother Pharmacol 52 (Suppl 1):S80–S89. [DOI] [PubMed] [Google Scholar]
- Geismann C, Arlt A, Sebens S, Schäfer H. (2014) Cytoprotection “gone astray”: Nrf2 and its role in cancer. Onco Targets Ther 7:1497–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SC, Hevia D, Patchva S, Park B, Koh W, Aggarwal BB. (2012) Upsides and downsides of reactive oxygen species for cancer: the roles of reactive oxygen species in tumorigenesis, prevention, and therapy. Antioxid Redox Signal 16:1295–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Dinkova-Kostova AT. (2014) The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci 39:199–218. [DOI] [PubMed] [Google Scholar]
- Hwang PM, Bunz F, Yu J, Rago C, Chan TA, Murphy MP, Kelso GF, Smith RA, Kinzler KW, Vogelstein B. (2001) Ferredoxin reductase affects p53-dependent, 5-fluorouracil-induced apoptosis in colorectal cancer cells. Nat Med 7:1111–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano D, Lena AM, Melino G, Candi E. (2012) Identification of NCF2/p67phox as a novel p53 target gene. Cell Cycle 11:4589–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo MC, Zhang DD. (2013) The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev 27:2179–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhang Y, Dusting GJ. (2011) NADPH oxidase-mediated redox signaling: roles in cellular stress response, stress tolerance, and tissue repair. Pharmacol Rev 63:218–242. [DOI] [PubMed] [Google Scholar]
- Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL. (2013) The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol 1:45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S. (2007) Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47:89–116. [DOI] [PubMed] [Google Scholar]
- Lamberti M, Porto S, Marra M, Zappavigna S, Grimaldi A, Feola D, Pesce D, Naviglio S, Spina A, Sannolo N, et al. (2012) 5-Fluorouracil induces apoptosis in rat cardiocytes through intracellular oxidative stress. J Exp Clin Cancer Res 31:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Chen X. (2002) The ferredoxin reductase gene is regulated by the p53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene 21:7195–7204. [DOI] [PubMed] [Google Scholar]
- Longley DB, Harkin DP, Johnston PG. (2003) 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 3:330–338. [DOI] [PubMed] [Google Scholar]
- Ma Q, He X. (2012) Molecular basis of electrophilic and oxidative defense: promises and perils of Nrf2. Pharmacol Rev 64:1055–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga T, Tsuji Y, Kaai K, Kohno S, Hirayama R, Alpers DH, Komoda T, Hara A. (2010) Toxicity against gastric cancer cells by combined treatment with 5-fluorouracil and mitomycin c: implication in oxidative stress. Cancer Chemother Pharmacol 66:517–526. [DOI] [PubMed] [Google Scholar]
- Nogueira V, Hay N. (2013) Molecular pathways: reactive oxygen species homeostasis in cancer cells and implications for cancer therapy. Clin Cancer Res 19:4309–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozben T. (2007) Oxidative stress and apoptosis: impact on cancer therapy. J Pharm Sci 96:2181–2196. [DOI] [PubMed] [Google Scholar]
- Sumimoto H. (2008) Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J 275:3249–3277. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Motohashi H, Yamamoto M. (2013) Toward clinical application of the Keap1-Nrf2 pathway. Trends Pharmacol Sci 34:340–346. [DOI] [PubMed] [Google Scholar]
- Takemura Y, Jackman AL. (1997) Folate-based thymidylate synthase inhibitors in cancer chemotherapy. Anticancer Drugs 8:3–16. [DOI] [PubMed] [Google Scholar]
- Trachootham D, Alexandre J, Huang P. (2009) Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov 8:579–591. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, et al. (2008) Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 29:1235–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PM, Danenberg PV, Johnston PG, Lenz HJ, Ladner RD. (2014) Standing the test of time: targeting thymidylate biosynthesis in cancer therapy. Nat Rev Clin Oncol 11:282–298. [DOI] [PubMed] [Google Scholar]
- Wind S, Beuerlein K, Eucker T, Müller H, Scheurer P, Armitage ME, Ho H, Schmidt HH, Wingler K. (2010) Comparative pharmacology of chemically distinct NADPH oxidase inhibitors. Br J Pharmacol 161:885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.