Abstract
Placental development and imprinting co-evolved with parental conflict over resource distribution to mammalian offspring. The imprinted genes, IGF2 and IGF2R, code for the growth promoter insulin-like growth factor 2 and its binding inhibitor, mannose 6-phosphate/IGF2 receptor, respectively. M6P/IGF2R of birds and fish do not recognize IGF2. In monotremes that lack imprinting, IGF2 specifically bound M6P/IGF2R via a hydrophobic CD loop. We show that the DNA coding the CD loop in monotremes functions as an exon splice enhancer (ESE) and that structural evolution of binding site loops (AB, HI, FG) improved therian IGF2 affinity. We propose that evolution of this ESE led to the fortuitous acquisition of M6P/IGF2R IGF2 binding that drew IGF2R into parental conflict prior to imprinting, that may have accelerated subsequent affinity maturation.
Keywords: Genomic imprinting, parental conflict, protein evolution, Insulin-like growth factor 2, mannose 6 phosphate receptor, surface plasmon resonance, NMR structure, ligand-receptor binding kinetics, RNA splicing, exon splice enhancer, mammalian evolution, monotremes
The sequence of molecular evolutionary events that established placental viviparity, genomic imprinting and parental conflict in mammals remain poorly understood (1) . Genomic imprinting occurs when expression of one allele of a diploid gene is silenced depending on the parent-of-origin, e.g. either from the father or the mother. Parental conflict over the distribution of resources to offspring has been supported by the observation of reciprocal imprinting of genes coding for the growth promoter Insulin-like growth factor 2 (IGF2), and the cation-independent mannose 6-phosphate/ IGF2 receptor (M6P/IGF2R or IGF2R) (2) . IGF2 and IGF2R are two of the approximately 80 genes imprinted in mammals, and two of the five genes (with INS, MEST/PEG1 and PEG10) imprinted in marsupials. So far, no evidence supports the existence of imprinting in monotremes despite the presence of a chorio-vitelline placenta (3, 4). On the basis of functional data, IGF2R transports M6P modified acid hydrolases to the pre-lysosomes (5). Of the 15 extra-cellular domains of IGF2R, domain 11 binds IGF2 in therians, and internalizes the ligand for degradation, whereas M6P bind to domains 3, 5 and 9 (5). Igf2 rescues placental dependent embryonic lethality associated with laboratory created murine parthenogenesis, implicating IGF2 supply as a regulator of placental development (6). Disruption of the maternal Igf2r allele results in Igf2 dependent overgrowth and fatality, supporting that IGF2R antagonizes the function of IGF2 (7, 8). The structure of the unbound human domain 11 shows that the IGF2 binding site composed of defined loops (AB, CD, FG and HI, Fig. 1A and Fig. S1) but how this domain 11 evolved to bind IGF2, and the relationship to imprinting co-evolution, remain unknown (9-12).
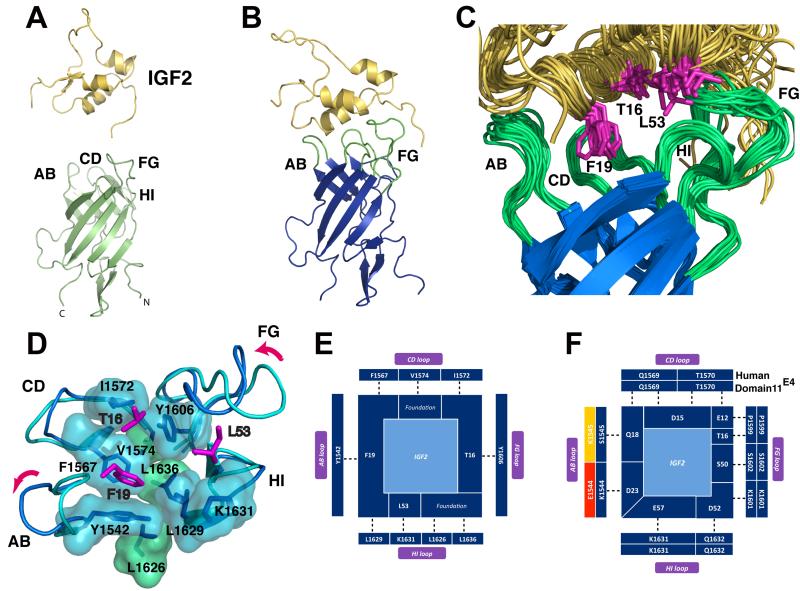
Figure 1.
A. Solution structure of free IGF2 (yellow) and the free domain11E4 (light green) and characteristic domain 11 β-barrel and four loops involved in IGF2 binding (AB, CD, FG, HI). B. The 24.2 kDa complex of domain11E4 (blue) bound to IGF2 (yellow) solved by NMR. C. super-imposition of an ensemble of twenty domain 11E4 low energy NMR structures showing the IGF2 binding pocket. The AB, CD, FG and HI loops are shown in green. D. backbone and surface representation of the IGF2 binding pocket highlighting a group of nine hydrophobes on domain11E4, including the three foundation residues (L1626, L1636 and V1574) that support the binding pocket (green) and, in light blue, the hydrophobes that form the IGF2 binding pocket. The flexible AB and FG loops change conformation upon complex formation (purple arrows). E. hydrophobic binding residues on IGF2 (centre) and binding partners (dark blue, indicating favorable hydrophobic interactions) on domain11E4 over the AB, CD, FG and HI loops (clockwise). F. the non-hydrophobic groups (charged, polar and hydrogen bond interactions) of IGF2 that interact with domain11E4 are shown with matching complementarity in dark blue. Incorrect charge/ polar complementarity are shown in red. Yellow represents where either the acquisition of a charge or change in steric bulk of a residue cannot be assessed in the absence of a high-resolution structure.
We established a high resolution structure of the human IGF2R:IGF2 complex and then compared this to other phylogenetically informative vertebrates. We adopted an NMR approach as the side chain amino acid interactions across the binding interface were not resolved in our 4.1Å resolution co-crystal structures (9). Wild-type human domain 11 and IGF2 failed to form a stable association in initial NMR studies. However, we identified an AB loop mutant (domain 11E1544K, K1545S, L1547V or clone E4) with an increased affinity for IGF2 that formed a tight complex (KD 15nM vs 46-64nM for WT domain 11, Table 1 and 1H-15N correlation spectra Fig.S2 and S3) (12). We solved the solution structure of this 24.2 kDa complex (IGF2: domain 11E4) with NMR structural and quality statistics in Table S1. When free IGF2 (Fig. 1A) binds to the single domain11E4 (Fig. 1B), ~640-760Å2 of solvent accessible surface area is buried on domain 11E4 and ~600-820Å2 on IGF2 (Fig. 1C), similar to the crystal structure of IGF2 in complex (710Å2-750Å2) (9). Domain 11E4 retains a relatively fixed conformation of the CD loop upon complex formation (Fig. 1C and D). The mutated IGF2R AB loop also moves to accommodate IGF2 helix 1 and the packing of IGF2 residues T16 and F19, whilst the IGF2R FG loop is repositioned between helices 2 and 3 of IGF2 and accommodates burial of IGF2 residue L53 in the domain 11E4 binding site. All three of these IGF2 residues are critical for IGF2R binding (13). Both conformational changes allow the formation of complementary hydrophobic surfaces and correctly support a range of H-bonding and salt bridging interactions (Fig. 1D, E, F, Fig. S4, movie S1) (14). Importantly, three IGF2R domain 11 residues (V1574, L1626, L1636) form a foundation for the three pronged interactions of IGF2 with key domain 11 residues; namely hydrophobic residues IGF2 F19 with domain 11 F1567, L1629, Y1542 and IGF2 L53 with domain 11 K1631, with a third interaction where IGF2 specificity is conferred by T16 interactions with domain 11 Y1606 and I1572 (Fig.1).
Table 1.
Species specific real time binding kinetic analysis of recombinant IGF2R domain 11 interactions with IGF2
| Major binding kinetic variables (two state conformational change model) | Steady state binding kinetics | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| ka1 (x105 m-1 s-1) mean | kd1 (x10-2 s-1) mean | KD (x10-9 M) ± S.D. | Χ2 (RU2) mean | KD (x10-9 M) ± S.D. | Χ2 (RU2) mean | |
| Human domain 11E4 kinetics to IGF2 † | ||||||
| Human | 27.6 | 4.4 | 15.3 ± 2.0 | 0.41 | 15.5± 3.3 | 0.77 |
| Human domain 11 kinetics to IGF2 † | ||||||
| Human IGF2 | 50 | 23 | 46 ± 1.8 | 0.33 | 64 ± 0.1 | 0.24 |
| Echidna IGF2‡ | 20 | 10.6 | 53 ± 2.7 | 0.25 | 77 ± 0.1 | 0.1 |
| Chicken IGF2∥ | 40 | 56 | 46 ± 0.4 | 0.15 | 63 ± 1.7 | 1.0 |
| Opossum domain 11 kinetics to IGF2 †§ | ||||||
| Human IGF2 | 6.9 | 10.8 | 130 ± 0.9 | 0.59 | 164 ± 0.7 | 0.86 |
| Echidna IGF2‡ | 6.5 | 10.6 | 135 ± 0.1 | 0.56 | 157 ± 2.5 | 0.70 |
| Chicken IGF2 | 17.6 | 35.0 | 171 ± 0.2 | 0.10 | 219 ± 6.6 | 0.08 |
| Echidna domain 11 kinetics to IGF2 † | ||||||
| Human IGF2 | 3.8 | 10.6 | 244 ± 2.0 | 0.08 | 329 ± 10 | 0.23 |
| Echidna IGF2‡ | 4.5 | 13.8 | 256 ± 2.0 | 0.54 | 385 ± 13 | 0.14 |
| Chicken IGF2 | 9.2 | 25.9 | 244 ± 2.0 | 0.08 | 412 ± 10 | 0.03 |
| Chicken and Zebrafish domain 11 kinetics to IGF2 † | ||||||
| Human IGF2 | - | - | - | - | - | - |
| Echidna IGF2‡ | - | - | - | - | - | - |
| Chicken IGF2 | - | - | - | - | - | - |
| Chicken with Echidna CD loop kinetics to IGF2 *§ | ||||||
| Human IGF2 | 1.1 | 4.7 | 427 | 0.74 | 325 ± 20 | 0.26 |
| Echidna IGF2‡ | 328 ± 28 | 0.18 | ||||
BIAcore 3000,
BIAcore T200 using purified biotinylated IGF2 on a streptavidin biosensor chip,
Echidna (peak 2) purified recombinant IGF2 (see Fig.S8). Recombinant proteins expressed in E. coli and
P. pastoris. All experiments repeated 4 times on at least two chips except
repeated twice on two different chips.‘-’ means no binding.
We expressed and performed binding studies of recombinant domain 11-His6 and NusA-IGF2 fusion proteins from different species in the yeast Pitchia pastoris and bacteria Escherichia coli, respectively (Fig. S5, S6, S7). IGF2 from each species bound human IGFBP-3 with similar nanomolar affinity (Fig.S7, S8). Initial analysis of species specific domain 11 binding to NusA-IGF2 fusion protein revealed an affinity (KD) of human and opossum between (100-130nM) and of platypus and echidna, revealed a lower affinity interaction, with KD range between 300-900nM (Fig. S6). We confirmed the absence of binding of purified IGF2 to IGF2R domain 11 in chicken and zebrafish, and the lower affinity binding in opossum (Table 1) (15, 16). Purified echidna IGF2 (7866.8 Da) bound human IGF1R, IGFBP-2, IGFBP-3 and purified echidna domain 11 with affinities similar to human IGF2 (14) (Fig. S8), and confirmed that purified echidna IGF2 binds echidna domain 11 with a steady state KD of 385±13 nM (Table 1). These data contradict a previous report where non-recombinant platypus IGF2R failed to bind human IGF2 at a fixed 2nM concentration, a concentration that our data suggests may be too low for binding detection (3).
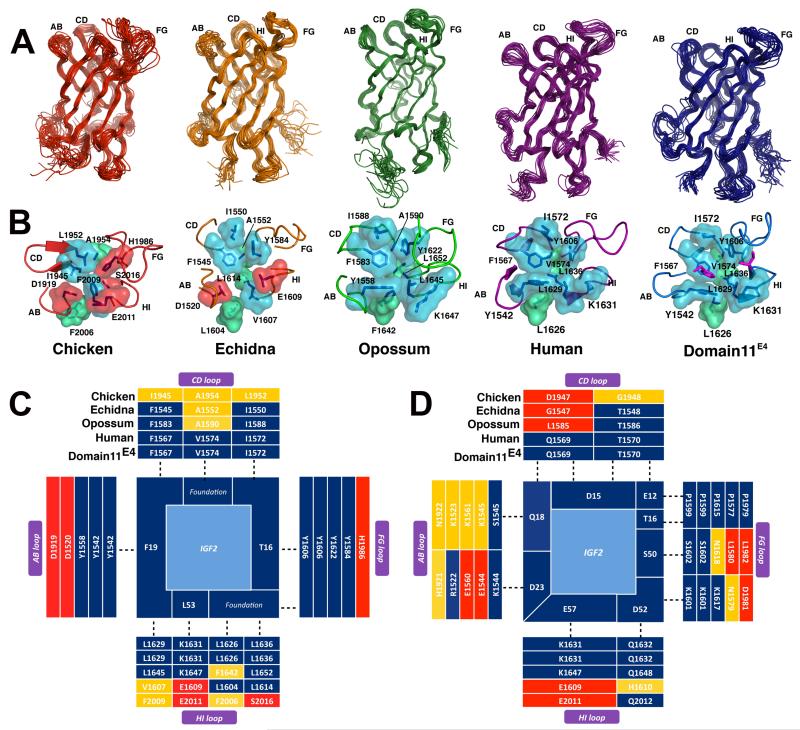
IGF2 is highly conserved across all vertebrate species including IGF2R interacting residues T16, F19 and L53 (Fig. S1). Evolution of the IGF2:IGF2R interaction was likely to be dependent on the changes to the binding pocket of domain 11. We solved the solution NMR structures of domain 11 from opossum, echidna and chicken (Table S2). These structures all adopt the same topology as human domain 11, with a flattened β-barrel (9) (Fig. 2A). Differences between the structures were observed in the flexible loops and foundation regions, in particular those comprising the IGF2 binding site (Fig. 2B). As domain 11 evolved from a non-IGF2 binding domain (chicken) to acquire IGF2 binding (echidna) and then to higher affinity binding (opossum, human), the surface surrounding the IGF2 binding region display a loss of charged residues, increased volume and hydrophobicity and assembly of the correct shape complementarity (Fig. 2C, D) (14). The charge distribution, primarily located in the AB and CD loops, changed from an overall negative potential to positive potential to complement the negative electrostatic surface of IGF2 (Fig. 2D, Fig. S10). The maps show the gradual accumulation of optimal residue interactions to IGF2 in all the loops, including the foundation residues, suggesting evolutionary maturation of the binding site topology and affinity (Fig. 2C and D). We then replaced the longer non-binding chicken CD loop (amino acids 1144–1154) with the shorter echidna CD loop, and rescued similar binding kinetics to the intact echidna domain, demonstrating the central importance of the CD loop in IGF2 binding (Table1, Fig. S11). The echidna CD loop driven binding site acquisition in chicken was augmented by the addition of echidna AB and FG loops that stabilized the interaction (Table S3). As expected, mutations of hydrophobic residues of the shortened echidna CD loop in chicken abolished binding, as did other amino acids (E1568D, G1571K, T1570P), underlining the importance of a rigid structural topology of CD loop for IGF2 binding (Table S3).
Figure 2.
A. high resolution NMR structures of domain 11 from chicken (red), echidna (orange), opossum (green) and human (magenta, PDB:2CNJ) and domain11E4 (blue). (Table S2 provides a summary of structural statistics). Ensembles of the lowest twenty energy models are shown for each species. B. surface representations of the binding pocket of IGF2R-domain 11 and the acquisition of an increased hydrophobicity surrounding the IGF2 binding pocket (Movie S1). C. hydrophobic binding residues on IGF2 (centre) and binding partners (dark blue) on domain11E4, human, opossum, echidna and chicken over the AB, CD, FG and HI loops. D. evolution of favorable charged, polar and hydrogen bond interactions between IGF2 and domain 11 species. (colors as in Fig. 1E, F).
A domain 11 hydrophobic pocket, although critical for IGF2 binding, is not the only factor necessary for a high affinity interaction between these proteins (9). A number of residues in the AB loop of echidna domain 11 are more similar to non-mammalian sequences than to the therian domain 11, and may result in a lower binding affinity (14). Human IGF2R domain 13 acts through a fibronectin type II domain-like insert to stabilize the AB loop of domain 11, an effect that enhances affinity that we also detected for echidna IGF2R domains 11-13 compared to the single domain (9) (Fig. S11).
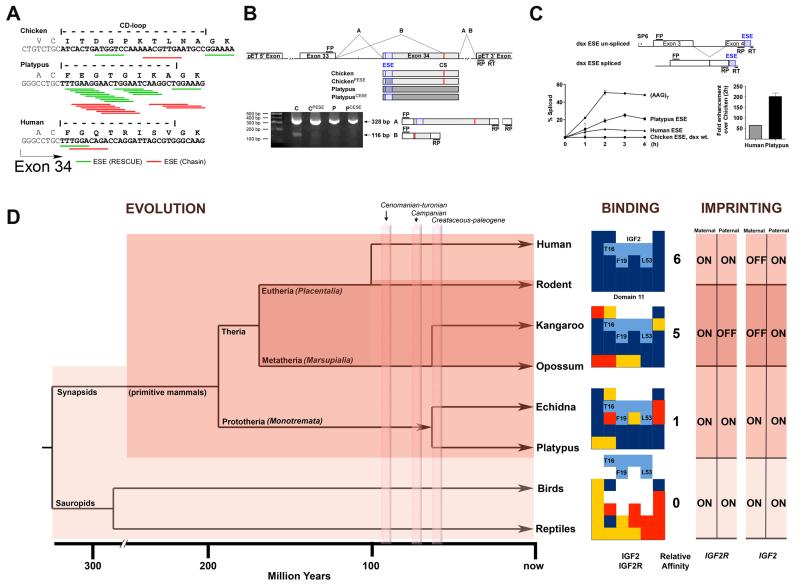
Evolutionary analysis of exon 33 and 34 that code for domain 11 predicted a dense exon splicing enhancer (ESE) cluster within exon 34 in platypus, a region that only codes for the CD loop of domain 11 (Fig. 3A, Fig. S12) (14). A shift in the dependency from intron to exon based gene splicing occurred during evolution to multi-cellular eukaryotes (17), presumably in order to improve efficient generation of mRNA without extended introns. In monotremes, ESE’s may have been important because of intronic expansion due to insertions of multiple repeat elements (18). We tested whether the ESE in the 5′ region of exon 34 was functional using in vivo splicing assays of mini-genes from the chicken and platypus CD loop sequences (Fig. 3B, C). Splicing of intron 33 containing mini-genes in chicken DF-1 cells identified a cryptic splice (CS) acceptor site that caused a frame shift and premature stop codon in chicken exon 34 (Fig. 3B, Fig. S13). In this system, replacing the chicken exon 34 ESE sequence with that of the same region of platypus, suppressed the appearance of the CS product, suggesting that the platypus CD loop sequence contained a functional ESE (Fig. 3B). We next tested the CD loop sequences in vitro with an ESE dependent dsx mini-gene splicing system compared to a positive control (AAG)7 (14) (Fig. 3C, Fig. S14). In agreement with ESE predictions, platypus and human ESE’s promoted rapid accumulation of spliced product quantified using RT-PCR and capillary electrophoresis.
Figure 3.
A. Exonic splicing enhancer (ESE) densities at the CD-loop coding region of exon 34. The positions of predicted hexamer (Rescue-ESE) and Octamer (Chasin) ESEs are illustrated (Fig. S12). B. in vivo splicing of chicken, platypus, or hybrid exons 34 in chicken DF-1 cells (sequences of splice products provided in Fig. S13). Two complete splice products A and B (cryptic splice site, CS) are shown, with RT-PCR gel products showing expression of A product and suppression of B product by ESEs. FP, RP, and RT are forward, reverse and reverse transcriptase primers, respectively. C. mini-gene constructs and comparative enhancement of dsx mini-gene splicing in HeLa cell nuclear extracts by control (AAG7 repeat) and ESEs (Fig. S14). D. phylogenetic context implies that IGF2-IGF2R binding site acquisition (light shade) occurred prior to the appearance of imprinting (dark shade), but was present within prototheria. Relative affinity increased in methatheria compared to prototheria, in keeping with a transition in binding site structure (CD loop). IGF2R is bi-allelically expressed in human, presumably because less selection pressure exists to maintain mono-allelic expression, and limits IGF2R imprinting to non-primate eutherians and metatheria. In terms of binding, favourable protein interactions are shown in blue, with incorrect charge and unpredictable complementarity shown in red and yellow, respectively. Silenced (imprinted) IGF2R allele is shown as ‘OFF’ compared to the expressed allele as ’ON’. For IGF2, the reciprocal imprinted alleles are present in both methatheria and eutheria, but absent in prototheria.
Comparison of ESE densities of all 6144 possible combinations of codons for the platypus CD loop, we found 6124 of the 6144 sequences, i.e. >99.67%, to have a lower ESE density than the sequence present in platypus, with only 10 codons containing more ESE’s (Fig. S12). Compared to platypus, the human CD loop DNA sequence exhibited relatively reduced ESE density, but showed a higher degree of codon adaptation for the coded amino acids (14) . While possible that the ESE arose after the CD loop dependent binding site was established, these data suggest that a functional ESE in exon 34 changed the amino acid sequence of the CD loop and thereby lead to the fortuitous creation of the IGF2 binding site in a M6P receptor. The evolution of splicing is underpinned by a complicated multi-factorial network of molecular interactions, which cannot be expected to reduce to a single factor (19). We speculate that the observed monotreme ESE clustering may have occurred in a common mammalian ancestor, perhaps as a response to intron expansion, in agreement with genome wide correlations (20). The subsequent reduction of ESE density observed in human, may have been due to further selective pressure to improve IGF2 binding.
Our data suggest that the IGF2 binding site on IGF2R was induced through evolution of an ESE specifically coding for the CD loop, and the CD loop alone. As the structure and function of the CD loop is conserved in therians, we propose that binding initially occurred in a common ancestor of monotremes, marsupials and eutherian mammals. The absence of IGF2R imprinting in in monotremes (3, 21) is supported by a therian specific mechanism of silencing of transposons (LTR) by methylation (22) and by lack of germ-line restriction of BORIS expression (23) . These data suggest that the acquisition of the IGF2 binding site occurred prior to IGF2R imprinting. We speculate that once the binding interaction with IGF2R was established, the conserved transportation function of IGF2R as a M6P receptor mediated a reduction of bio-available IGF2. The subsequent increase in IGF2 affinity was selected for with respect to non-CD, AB, FG and HI loop changes, coincident with the onset of imprinting, and consistent with parental conflict (24). Affinity maturation through purifying selection for improved regulation of IGF2 supply may have been accelerated by imprinting induced mono-allelic IGF2R expression (25).
Supplementary Material
Acknowledgements
We thank C Graham, J Crosby, N Barclay, C Ponting, S Murphy and N Proudfoot for discussion. Work supported by Cancer Research UK (ABH, MPC and EYJ, C429, C375), Wellcome Trust (MPC and HWB NMR facility), Science Foundation Ireland (CMN), Algerian Government (DR), NHMRC and ARC (Australia, BF, FG is an ARC Research Fellow). The research described in this manuscript is the subject of a worldwide patent application WO2007020402A, granted in Europe as EP1945663B and pending in the US as US20090304651A. The ensembles of NMR structures and associated NMR chemical shifts have been deposited with the protein database and BioMagResBank with the following accession codes: Chicken 2L21/17110; Echidna 2L5W/17287; Opossum 2L2G/17134; Domain11E4 2L2A/17128; Domain11E4 /IGF2 complex 2L29/17127.
Footnotes
There are no competing financial interests.
Supporting Material Online (www.sciencemag.)
Supplementary Information (Material and Methods, Supplementary Text, References, Supplementary Figures 1-14, Tables)
Supplementary Movie S1
References and Notes
- 1.Renfree MB, Hore TA, Shaw G, Graves JA, Pask AJ. Evolution of genomic imprinting: insights from marsupials and monotremes. Annu Rev Genomics Hum Genet. 2009;10:241. doi: 10.1146/annurev-genom-082908-150026. [DOI] [PubMed] [Google Scholar]
- 2.Haig D, Graham C. Genomic imprinting and the strange case of the insulin-like growth factor II receptor. Cell. 1991 Mar 22;64:1045. doi: 10.1016/0092-8674(91)90256-x. [DOI] [PubMed] [Google Scholar]
- 3.Killian JK, et al. M6P/IGF2R imprinting evolution in mammals. Mol Cell. 2000 Apr;5:707. doi: 10.1016/s1097-2765(00)80249-x. [DOI] [PubMed] [Google Scholar]
- 4.Killian JK, et al. Monotreme IGF2 expression and ancestral origin of genomic imprinting. J Exp Zool. 2001 Aug 15;291:205. doi: 10.1002/jez.1070. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: new twists in the tale. Nat Rev Mol Cell Biol. 2003;4:202. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 6.Kono T, et al. Birth of parthenogenetic mice that can develop to adulthood. Nature. 2004 Apr 22;428:860. doi: 10.1038/nature02402. [DOI] [PubMed] [Google Scholar]
- 7.Lau MM, et al. Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes Dev. 1994 Dec 15;8:2953. doi: 10.1101/gad.8.24.2953. [DOI] [PubMed] [Google Scholar]
- 8.Wang ZQ, Fung MR, Barlow DP, Wagner EF. Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene. Nature. 1994 Dec 1;372:464. doi: 10.1038/372464a0. [DOI] [PubMed] [Google Scholar]
- 9.Brown J, et al. Structure and functional analysis of the IGF-II/IGF2R interaction. Embo J. 2008 Jan 9;27:265. doi: 10.1038/sj.emboj.7601938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown J, et al. Structure of a functional IGF2R fragment determined from the anomalous scattering of sulfur. Embo J. 2002 Mar 1;21:1054. doi: 10.1093/emboj/21.5.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams C, et al. Structural insights into the interaction of insulin-like growth factor 2 with IGF2R domain 11. Structure. 2007 Sep;15:1065. doi: 10.1016/j.str.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Zaccheo OJ, et al. Kinetics of insulin-like growth factor II (IGF-II) interaction with domain 11 of the human IGF-II/mannose 6-phosphate receptor: function of CD and AB loop solvent-exposed residues. J Mol Biol. 2006 Jun 2;359:403. doi: 10.1016/j.jmb.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 13.Delaine C, et al. A novel binding site for the human insulin-like growth factor-II (IGF-II)/mannose 6-phosphate receptor on IGF-II. J Biol Chem. 2007 Jun 29;282:18886. doi: 10.1074/jbc.M700531200. [DOI] [PubMed] [Google Scholar]
- 14.I. o. m. a. m. i. a. o. S. Online.
- 15.Yandell CA, Dunbar AJ, Wheldrake JF, Upton Z. The kangaroo cation-independent mannose 6-phosphate receptor binds insulin-like growth factor II with low affinity. J Biol Chem. 1999 Sep 17;274:27076. doi: 10.1074/jbc.274.38.27076. [DOI] [PubMed] [Google Scholar]
- 16.Canfield WM, Kornfeld S. The chicken liver cation-independent mannose 6-phosphate receptor lacks the high affinity binding site for insulin-like growth factor II. J Biol Chem. 1989 May 5;264:7100. [PubMed] [Google Scholar]
- 17.Warnecke T, Weber CC, Hurst LD. Why there is more to protein evolution than protein function: splicing, nucleosomes and dual-coding sequence. Biochem Soc Trans. 2009 Aug;37:756. doi: 10.1042/BST0370756. [DOI] [PubMed] [Google Scholar]
- 18.Warren WC, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008 May 8;453:175. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao X, Wang Z, Jang M, Burge CB. Coevolutionary networks of splicing cis-regulatory elements. Proc Natl Acad Sci U S A. 2007 Nov 20;104:18583. doi: 10.1073/pnas.0707349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dewey CN, Rogozin IB, Koonin EV. Compensatory relationship between splice sites and exonic splicing signals depending on the length of vertebrate introns. BMC Genomics. 2006;7:311. doi: 10.1186/1471-2164-7-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Killian JK, et al. Divergent evolution in M6P/IGF2R imprinting from the Jurassic to the Quaternary. Hum Mol Genet. 2001 Aug 15;10:1721. doi: 10.1093/hmg/10.17.1721. [DOI] [PubMed] [Google Scholar]
- 22.Pask AJ, et al. Analysis of the platypus genome suggests a transposon origin for mammalian imprinting. Genome Biol. 2009;10:R1. doi: 10.1186/gb-2009-10-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hore TA, Deakin JE, Graves J. A.. Marshall. The evolution of epigenetic regulators CTCF and BORIS/CTCFL in amniotes. PLoS Genet. 2008 Aug;4:e1000169. doi: 10.1371/journal.pgen.1000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trivers RL. Parent-offspring conflict. Integrative and Comparative Biology. 1974;14:249. [Google Scholar]
- 25.Chess A. Mechanisms and consequences of widespread random monoallelic expression. Nat Rev Genet. 2012 Jun;13:421. doi: 10.1038/nrg3239. [DOI] [PubMed] [Google Scholar]
- 26.Denley A, et al. Structural determinants for high-affinity binding of insulin-like growth factor II to insulin receptor (IR)-A, the exon 11 minus isoform of the IR. Mol Endocrinol. 2004 Oct;18:2502. doi: 10.1210/me.2004-0183. [DOI] [PubMed] [Google Scholar]
- 27.Carrick FE, et al. Interaction of insulin-like growth factor (IGF)-I and -II with IGF binding protein-2: mapping the binding surfaces by nuclear magnetic resonance. J Mol Endocrinol. 2005 Jun;34:685. doi: 10.1677/jme.1.01756. [DOI] [PubMed] [Google Scholar]
- 28.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995 Nov;6:277. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 29.Vranken WF, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005 Jun 1;59:687. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 30.Brunger AT, Adams PD, Rice LM. Recent developments for the efficient crystallographic refinement of macromolecular structures. Curr Opin Struct Biol. 1998 Oct;8:606. doi: 10.1016/s0959-440x(98)80152-8. [DOI] [PubMed] [Google Scholar]
- 31.Nederveen AJ, et al. RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins. 2005 Jun 1;59:662. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- 32.Rieping W, et al. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007 Feb 1;23:381. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 33.La D, et al. 3D-SURFER: software for high-throughput protein surface comparison and analysis. Bioinformatics. 2009 Nov 1;25:2843. doi: 10.1093/bioinformatics/btp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laskowski RA. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001 Jan 1;29:221. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipsitz RS, Tjandra N. Residual dipolar couplings in NMR structure analysis. Annu Rev Biophys Biomol Struct. 2004;33:387. doi: 10.1146/annurev.biophys.33.110502.140306. [DOI] [PubMed] [Google Scholar]
- 36.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006 Oct;62:1243. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 37.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 38.Abril JF, Castelo R, Guigo R. Comparison of splice sites in mammals and chicken. Genome Res. 2005 Jan;15:111. doi: 10.1101/gr.3108805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kol G, Lev-Maor G, Ast G. Human-mouse comparative analysis reveals that branch-site plasticity contributes to splicing regulation. Hum Mol Genet. 2005 Jun 1;14:1559. doi: 10.1093/hmg/ddi164. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz SH, et al. Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res. 2008 Jan;18:88. doi: 10.1101/gr.6818908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desmet FO, et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009 May;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puigbo P, Bravo IG, Garcia-Vallve S. CAIcal: a combined set of tools to assess codon usage adaptation. Biol Direct. 2008;3:38. doi: 10.1186/1745-6150-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voelker RB, Berglund JA. A comprehensive computational characterization of conserved mammalian intronic sequences reveals conserved motifs associated with constitutive and alternative splicing. Genome Res. 2007 Jul;17:1023. doi: 10.1101/gr.6017807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002 Aug 9;297:1007. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 45.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004 Jun 1;18:1241. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auch D, Reth M. Exon trap cloning: using PCR to rapidly detect and clone exons from genomic DNA fragments. Nucleic Acids Res. 1990 Nov 25;18:6743. doi: 10.1093/nar/18.22.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka K, Watakabe A, Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol Cell Biol. 1994 Feb;14:1347. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edery I, Sonenberg N. Cap-dependent RNA splicing in a HeLa nuclear extract. Proc Natl Acad Sci U S A. 1985 Nov;82:7590. doi: 10.1073/pnas.82.22.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11:1475. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lam BJ, et al. Enhancer-dependent 5′-splice site control of fruitless pre-mRNA splicing. J Biol Chem. 2003 Jun 20;278:22740. doi: 10.1074/jbc.M301036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Girault I, et al. Altered expression pattern of alternatively spliced estrogen receptor beta transcripts in breast carcinoma. Cancer Lett. 2004 Nov 8;215:101. doi: 10.1016/j.canlet.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 52.Gabory A, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009 Oct;136:3413. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 53.Smits G, et al. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat Genet. 2008 Aug;40:971. doi: 10.1038/ng.168. [DOI] [PubMed] [Google Scholar]
- 54.Weidman JR, Dolinoy DC, Maloney KA, Cheng JF, Jirtle RL. Imprinting of opossum Igf2r in the absence of differential methylation and air. Epigenetics. 2006 Jan-Mar;1:49. doi: 10.4161/epi.1.1.2592. [DOI] [PubMed] [Google Scholar]
- 55.Yotova IY, et al. Identification of the human homolog of the imprinted mouse Air non-coding RNA. Genomics. 2008 Dec;92:464. doi: 10.1016/j.ygeno.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chandrashekaran IR, et al. The N-terminal subdomain of insulin-like growth factor (IGF) binding protein 6. Structure and interaction with IGFs. Biochemistry. 2007 Mar 20;46:3065. doi: 10.1021/bi0619876. [DOI] [PubMed] [Google Scholar]
- 57.Terasawa H, et al. Solution structure of human insulin-like growth factor II; recognition sites for receptors and binding proteins. Embo J. 1994 Dec 1;13:5590. doi: 10.1002/j.1460-2075.1994.tb06896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Torres AM, et al. Solution structure of human insulin-like growth factor II. Relationship to receptor and binding protein interactions. J Mol Biol. 1995 Apr 28;248:385. doi: 10.1016/s0022-2836(95)80058-1. [DOI] [PubMed] [Google Scholar]
- 59.Culler SJ, Hoff KG, Voelker RB, Berglund JA, Smolke CD. Functional selection and systematic analysis of intronic splicing elements identify active sequence motifs and associated splicing factors. Nucleic Acids Res. 2010 Aug;38:5152. doi: 10.1093/nar/gkq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang XH, Kangsamaksin T, Chao MS, Banerjee JK, Chasin LA. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol Cell Biol. 2005 Aug;25:7323. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chamary JV, Hurst LD. Biased codon usage near intron-exon junctions: selection on splicing enhancers, splice-site recognition or something else? Trends Genet. 2005 May;21:256. doi: 10.1016/j.tig.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 62.Parmley JL, Hurst LD. Exonic splicing regulatory elements skew synonymous codon usage near intron-exon boundaries in mammals. Mol Biol Evol. 2007 Aug;24:1600. doi: 10.1093/molbev/msm104. [DOI] [PubMed] [Google Scholar]
- 63.Parmley JL, Urrutia AO, Potrzebowski L, Kaessmann H, Hurst LD. Splicing and the evolution of proteins in mammals. PLoS Biol. 2007 Feb;5:e14. doi: 10.1371/journal.pbio.0050014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Warnecke T, Parmley JL, Hurst LD. Finding exonic islands in a sea of non-coding sequence: splicing related constraints on protein composition and evolution are common in intron-rich genomes. Genome Biol. 2008;9:R29. doi: 10.1186/gb-2008-9-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z, et al. Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay. BMC Biol. 2009;7:23. doi: 10.1186/1741-7007-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bryson K, et al. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005 Jul 1;33:W36. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krebs WG, Gerstein M. The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 2000 Apr 15;28:1665. doi: 10.1093/nar/28.8.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.