Abstract
Background
Natural rubber, an important industrial raw material, is specifically synthesized in laticifers located inside the rubber tree (Hevea brasiliensis Muell. Arg.) trunk. Due to the absence of plasmodesmata, the laticifer water balance is mediated by aquaporins (AQPs). However, to date, the characterization of H. brasiliensis AQPs (HbAQPs) is still in its infancy.
Results
In this study, 51 full-length AQP genes were identified from the rubber tree genome. The phylogenetic analysis assigned these AQPs to five subfamilies, including 15 plasma membrane intrinsic proteins (PIPs), 17 tonoplast intrinsic proteins (TIPs), 9 NOD26-like intrinsic proteins (NIPs), 4 small basic intrinsic proteins (SIPs) and 6 X intrinsic proteins (XIPs). Functional prediction based on the analysis of the aromatic/arginine (ar/R) selectivity filter, Froger’s positions and specificity-determining positions (SDPs) showed a remarkable difference in substrate specificity among subfamilies. Homology analysis supported the expression of 44 HbAQP genes in at least one of the examined tissues. Furthermore, deep sequencing of the laticifer transcriptome in the form of latex revealed a key role of several PIP subfamily members in the laticifer water balance, and qRT-PCR analysis showed diverse expression patterns of laticifer-expressed HbAQP genes upon ethephon treatment, a widely-used practice for the stimulation of latex yield.
Conclusions
This study provides an important genetic resource of HbAQP genes, which will be useful to improve the water use efficiency and latex yield of Hevea.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-2152-6) contains supplementary material, which is available to authorized users.
Keywords: Rubber tree (Hevea brasiliensis Muell. Arg.), Laticifer, Transcriptome, Aquaporin, Plasma membrane intrinsic protein, Water balance
Background
Aquaporins (AQPs), also known as major intrinsic proteins (MIPs), are a class of integral membrane proteins that facilitate the passive transport of water and other small solutes across biological membranes [1]. Since their first discovery in 1990s, AQPs have been found in almost all living organisms [2]. Compared with animals and microbes, AQPs are particularly abundant and diverse in land plants and more than 20 homologs have been identified from Arabidopsis thaliana, Zea mays, Oryza sativa, Vitis vinifera, Populus trichocarpa, Gossypium hirsutum, Solanum tuberosum, Solanum lycopersicum and Glycine max [3–11]. According to the sequence similarity, the AQPs in model plant Arabidopsis are divided into four subfamilies, i.e. the plasma membrane intrinsic protein (PIP) subfamily that contains two subgroups, the tonoplast intrinsic protein (TIP) that contains five subgroups, the NOD26-like intrinsic protein (NIP) that contains seven subgroups and small basic intrinsic protein (SIP) that contains two subgroups. In moss and some dicots including P. trichocarpa, one more subfamily named X intrinsic protein (XIP) subfamily that contains three subgroups is also found [7, 12].
AQPs assemble in tetramers in the cell membrane, although each monomer can act as a water channel [13]. Even though the overall pairwise sequence similarity can be low, AQPs share some structural features such as harboring six transmembrane helices (TM1–TM6) connected by five loops (LA–LE). LB and LE from opposite sides dip into the membrane and form two half helices (HB and HE), at the N-termini of which, two highly conserved NPA (Asn-Pro-Ala) motifs form one selectivity region. And another region that determines the substrate specificity is known as the aromatic/arginine (ar/R) selectivity filter (H2 in TM2, H5 in TM5, LE1 and LE2 in LE) [13]. The NPA motifs create an electrostatic repulsion of protons and act as a size barrier, where the ar/R filter renders the pore constriction site diverse in both size and hydrophobicity [13]. In addition to water-conducting AQPs, certain AQPs called aquaglyceroporins (GLPs) transport glycerol instead. Statistical analysis indicated that GLPs feather five highly conserved amino acid residues (named Froger’s positions: P1–5): an aromatic residue at P1, an acidic residue at P2, a basic residue at P3, a proline followed by a nonaromatic residue at P4 and P5, as Y108-D207-K211-P236-L237 observed in the Escherichia coli glycerol facilitator GlpF in contrast to A103-S190-A194-F208-W209 in the pure water channel AqpZ [14]. Very recently, nine specificity-determining positions (SDPs) for non-aqua substrates, i.e. urea, boric acid, silicic acid, ammonia (NH3), carbon dioxide (CO2) and hydrogen peroxide (H2O2) were also proposed for each group via a comprehensive analysis of functionally characterized AQPs [15].
The para rubber tree (Hevea brasiliensis Muell. Arg.) is a perennial tropical plant species that belongs to the Euphorbiaceae family. Although it is native to the Amazon basin, the economic importance and increasing demand of natural rubber has prompted its wide-domestication to Southeast Asia. To date, the rubber tree is still the only commercial source of natural rubber for its high production and quality of rubber. Natural rubber (cis-1,4-polyisoprene) is specifically synthesized in the highly differentiated vessels termed laticifers, which are located in the secondary phloem of the tree trunk and are periodically differentiated from the vascular cambium [16]. The rubber-containing latex which represents the cytoplasmic content of laticifers is harvested by tapping the bark every 2–5 days, and the latex yield is determined by the initial flow rate, duration of latex flow and latex regeneration between two tappings [17].
Ethylene, a gas phytohormone, plays crucial roles in numerous aspects of growth, development, and response to biotic and abiotic stresses in plants [18]. In Arabidopsis, the ethylene signaling pathway has been well established and its involvement in certain agronomically important processes such as seed dormancy, fruit ripening, abscission and senescence has made ethylene a target for manipulation by chemical and biotechnological methodologies [19]. Ethephon (also known as Ethrel), an ethylene releaser, was initiatively tested for rubber yield promotion as early as the 1970s and now is widely used in rubber production all over the world. Although the molecular mechanism on ethephon stimulation of latex yield is still unclear, researches showed that the treatment of rubber tree barks with ethephon could significantly decrease latex dry rubber content (DRC) or total solid content (TSC), extend the bark drainage area, and prolong latex flow duration [20–23]. These effects are mainly benefited from water influx toward laticifers and the resultant latex dilution.
Since water accounts for 60–70 % of total latex upon each tapping, sufficient water supply is essential for both latex flow and latex regeneration [24]. Nevertheless, the mature laticiferous vessel rings are devoid of functional plasmodesmata connections [25], and thus the water inflow into laticifers is mediated largely by AQPs. However, the molecular characterization of rubber tree AQPs (HbAQPs) is still in its infancy. As of June 2015, only eight full-length HbAQP cDNAs have been reported [21–23, 26–29]. Among them, the water transport activity of HbPIP1, HbPIP2, HbPIP1;1, HbPIP2;1 HbPIP2;3 and HbTIP1;1 was characterized by Tungngoen et al. and our group using Xenopus laevis oocytes: HbTIP1;1, HbPIP2;1 and HbPIP2;3 were shown to be highly efficient, whereas HbPIP1;1; HbPIP1;4 and HbPIP2;7 were less efficient [21–23, 26].
Lately, the rubber tree genome was sequenced by Rahman et al. [30] and Rubber Research Institute, Chinese Academy of Tropical Agricultural Sciences. In addition, more than 50,000 ESTs (expressed sequence tags) and a high number of RNA sequencing reads derived from several tissues such as shoot apex, leaf, laticifer, bark, root and somatic embryogenesis are also available in NCBI SRA [30–35]. These datasets provide a good chance to analyze the rubber tree AQP gene family from a global view. In the present study, a genome-wide search was carried out to identify AQP genes encoded in the rubber tree genome. Further, functional prediction was performed based on the analysis of the ar/R filter, Froger’s positions and SPDs, and deep transcriptome sequencing and qRT-PCR expression analysis were also adopted to identify the most important AQP members expressed in the laticifer.
Results
Identification and classification of rubber tree aquaporin genes
Via a comprehensive homology analysis, 57 or 54 loci putatively encoding AQP-like genes were identified from the rubber tree genome of clone RY7-33-97 or RRIM600, respectively (data not shown). Since all AQP-encoding loci identified in the RRIM600 genome were found in the genome of RY7-33-97 and some genes from RRIM600 are incomplete and/or the sequences have a high number of “N”s, the AQP genes identified from the RY7-33-97 genome were selected for further analyses. After discarding loci encoding partial AQP-like sequences which are truncated and lacking any of the NPA motifs, 51 full-length AQP genes were retained and the gene models are available in Additional file 1.
To analyze the evolutionary relationship and their putative function, an unrooted phylogenetic tree was constructed from the deduced amino acid sequences of HbAQPs together with that from Arabidopsis (AtAQPs) and poplar (PtAQPs) (the Phytozome accession numbers are available in Additional file 2) . The reasons for choosing these two species are mainly as follows: the complete set of AQP genes in Arabidopsis was firstly identified and then well characterized; the well-studied wood plant poplar harbors one more subfamily (XIP) that is not found in Arabidopsis. According to the phylogenetic analysis, 51 HbAQPs were grouped into five subfamilies, i.e. PIP (15), TIP (17), NIP (9), SIP (4) and XIP (6) (Table 1; Fig. 1). Following the nomenclature of Arabidopsis and poplar [3, 36], the HbPIP subfamily was further divided into two phylogenetic subgroups (5 HbPIP1s and 10 HbPIP2s), the HbTIP subfamily into five subgroups (8 HbTIP1s, 4 HbTIP2s, 2 HbTIP3s, 1 HbTIP4 and 2 HbTIP5s), the HbNIP subfamily into seven subgroups (2 HbNIP1s, 1 HbNIP2, 1 HbNIP3, 2 HbNIP4s, 1 HbNIP5, 1 HbNIP6 and 1 HbNIP7), the HbSIP subfamily into two subgroups (3 HbSIP1s and 1 HbSIP2) and the HbXIP subfamily into three subgroups (4 HbXIP1s, 1 HbXIP2 and 1 HbXIP3) (Fig. 1). Although the closest homolog of HbNIP2;1 and HbNIP3;1 is not AtNIP2;1 or AtNIP3;1, their counterparts in poplar were identified and thus the names were assigned. As shown in Fig. 1, many HbAQPs were grouped in pairs, i.e. HbPIP1;1/HbPIP1;2, HbPIP1;3/HbPIP1;4, HbPIP2;1/HbPIP2;2, HbPIP2;3/HbPIP2;4, HbPIP2;5/HbPIP2;6, HbPIP2;7/HbPIP2;8, HbTIP1;1/HbTIP1;2, HbTIP1;3/HbTIP1;4, HbTIP1;5/HbTIP1;6, HbTIP1;7/HbTIP1;8, HbTIP2;1/HbTIP2;2, HbTIP2;3/HbTIP2;4, HbTIP3;1/HbTIP3;2, HbTIP5;1/HbTIP5;2, HbNIP1;1/HbNIP1;2, HbSIP1;2/HbSIP1;3, HbXIP1;3/HbXIP1;4, which exhibit sequence identities of 78.9–96.3 % and 71.9–98.3 % at the nucleotide or amino acid level, respectively (Additional file 3).
Table 1.
List of 51 HbAQP genes identified in this study
| Name | Nucleotide length (bp, from start to stop codons) | Intron NO. | EST hits in GenBank | Shoot apexa | Leafb | laticiferc | Barkd | Roote | Somatic embryogenesisf | Cloning strategy | Phytozome ID of AtAQP ortholog | Phytozome ID of PtAQP ortholog | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | CDS | |||||||||||||
| PIP | HbPIP1;1 | 1161 | 864 | 3 | 10 | Yes | Yes | Yes | Yes | Yes | Yes | RT-PCR | AT4G00430 | Pt_0010s19930 |
| HbPIP1;2 | 1155 | 864 | 3 | 6 | Yes | Yes | Yes | Yes | Yes | Yes | RT-PCR | AT4G00430 | Pt_0010s19930 | |
| HbPIP1;3 | 2952 | 864 | 3 | 0 | Yes | Yes | Yes | Yes | Yes | Yes | RT-PCR | AT4G00430 | Pt_0003s12870 | |
| HbPIP1;4 | 2285 | 864 | 3 | 4 | Yes | Yes | Yes | Yes | Yes | Yes | RACE | AT4G00430 | Pt_0003s12870 | |
| HbPIP1;5 | 1370 | 810 | 3 | 2 | Yes | Yes | Yes | Yes | RT-PCR | AT4G00430 | Pt_0016s12070 | |||
| HbPIP2; 1 | 1150 | 867 | 3 | 4 | Yes | Yes | Yes | Yes | Yes | Yes | RT-PCR | AT3G53420 | Pt_0006s12980 | |
| HbPIP2;2 | 1174 | 867 | 3 | 2 | Yes | Yes | Yes | Yes | Yes | AT3G53420 | Pt_0006s12980 | |||
| HbPIP2;3 | 1801 | 861 | 3 | 2 | Yes | Yes | Yes | Yes | Yes | Yes | RT-PCR | AT3G53420 | Pt_0006s12980 | |
| HbPIP2;4 | 1590 | 861 | 3 | 0 | Yes | Yes | Yes | Yes | Yes | Yes | RT-PCR | AT3G53420 | Pt_0006s12980 | |
| HbPIP2;5 | 1673 | 858 | 3 | 0 | Yes | Yes | Yes | Yes | Yes | Yes | RT-PCR | AT3G53420 | Pt_0010s22950 | |
| HbPIP2;6 | 2927 | 861 | 3 | 0 | Yes | Yes | Yes | Yes | Yes | AT3G53420 | Pt_0010s22950 | |||
| HbPIP2; 7 | 1263 | 837 | 3 | 15 | Yes | Yes | Yes | Yes | Yes | Yes | RACE | AT3G53420 | Pt_0004s18240 | |
| HbPIP2;8 | 1263 | 843 | 3 | 1 | Yes | Yes | Yes | Yes | AT3G53420 | Pt_0004s18240 | ||||
| HbPP2;9 | 1120 | 843 | 3 | 0 | Yes | Yes | Yes | Yes | Yes | AT3G53420 | Pt_0004s18240 | |||
| HbPIP2; 1 | 1401 | 858 | 3 | 0 | Yes | AT3G53420 | Pt_0005s11110 | |||||||
| TIP | HbTIP1;1 | 853 | 759 | 1 | 1 | Yes | Yes | Yes | Yes | Yes | RT-PCR | AT2G36830 | Pt_0006s12350 | |
| HbTIP1;2 | 851 | 759 | 1 | 0 | Yes | Yes | Yes | Yes | Yes | Yes | RT-PCR | AT2G36830 | Pt_0006s12350 | |
| HbTP1;3 | 848 | 762 | 1 | 0 | Yes | Yes | Yes | AT2G36830 | Pt_0009s01070 | |||||
| HbTIP1;4 | 820 | 684 | 1 | 0 | AT2G36830 | Pt_0009s01070 | ||||||||
| HbTP1;5 | 931 | 759 | 2 | 2 | Yes | Yes | Yes | Yes | Yes | AT4G01470 | Pt_0008s05050 | |||
| HbTIP1;6 | 974 | 759 | 2 | 0 | Yes | Yes | Yes | Yes | Yes | Yes | RT-PCR | AT4G01470 | Pt_0008s05050 | |
| HbTIP1;7 | 1189 | 759 | 2 | 0 | Yes | Yes | Yes | AT4G01470 | Pt_0001s24200 | |||||
| HbTIP1;8 | 997 | 759 | 2 | 0 | Yes | Yes | Yes | Yes | Yes | AT4G01470 | Pt_0001s24200 | |||
| HbTP2;1 | 1574 | 747 | 2 | 0 | Yes | Yes | Yes | Yes | Yes | AT3G16240 | Pt_0001s18730 | |||
| HbTIP2;2 | 1165 | 747 | 2 | 0 | Yes | Yes | Yes | Yes | Yes | Yes | RT-PCR | AT3G16240 | Pt_0001s18730 | |
| HbTIP2;3 | 1010 | 753 | 2 | 0 | Yes | Yes | Yes | AT4G17340 | Pt_0003s07550 | |||||
| HbTIP2;4 | 1009 | 753 | 2 | 0 | Yes | AT4G17340 | Pt_0003s07550 | |||||||
| HbTIP3;1 | 967 | 774 | 2 | 0 | Yes | Yes | Yes | AT1G17810 | Pt_0017s03540 | |||||
| HbTIP3;2 | 915 | 742 | 2 | 0 | Yes | AT1G17810 | Pt_0017s03540 | |||||||
| HbTIP4;1 | 1010 | 756 | 2 | 0 | Yes | Yes | Yes | Yes | Yes | AT2G25810 | Pt_0006s25620 | |||
| HbTIP5;1 | 1181 | 759 | 2 | 0 | Yes | Yes | AT3G47440 | Pt_0001s00690 | ||||||
| HbTIP5;2 | 1119 | 744 | 2 | 0 | AT3G47440 | Pt_0001s00690 | ||||||||
| NIP | HbNIP1;1 | 1582 | 861 | 4 | 0 | Yes | Yes | Yes | RT-PCR | AT4G18910 | Pt_0011s06770 | |||
| HbNP1;2 | 1939 | 864 | 4 | 0 | Yes | Yes | Yes | Yes | Yes | Yes | RT-PCR | AT4G18910 | Pt_0011s06770 | |
| HbNIP2;1 | 2930 | 858 | 4 | 0 | Yes | Yes | Yes | Yes | Pt_0017s11960 | |||||
| HbNP3;1 | 1249 | 849 | 4 | 0 | Yes | Yes | RT-PCR | Pt_0002s09740 | ||||||
| HbNIP4;1 | 1237 | 804 | 4 | 0 | Yes | AT5G37810 | Pt_0010s12330 | |||||||
| HbNIP4;2 | 1268 | 846 | 4 | 0 | Yes | AT5G37820 | Pt_0017s03060 | |||||||
| HbNIP5;1 | 1932 | 897 | 3 | 0 | Yes | Yes | Yes | Yes | Yes | AT4G10380 | Pt_0001s45920 | |||
| HbNIP6;1 | 3371 | 927 | 4 | 0 | Yes | Yes | Yes | Yes | Yes | Yes | RT-PCR | AT1G80760 | Pt_0001s14850 | |
| HbNIP7;1 | 1359 | 897 | 4 | 0 | AT3G06100 | Pt_0008s20750 | ||||||||
| XIP | HbXIP1;1 | 973 | 870 | 1 | 0 | Pt_0009s13090 | ||||||||
| HbXIP1;2 | 1022 | 891 | 1 | 0 | Yes | Pt_0009s13090 | ||||||||
| HbXIP1;3 | 959 | 831 | 1 | 0 | Pt_0009s13090 | |||||||||
| HbXIP1;4 | 959 | 831 | 1 | 0 | Pt_0009s13090 | |||||||||
| HbXIP2;1 | 1163 | 918 | 2 | 0 | Yes | Yes | Yes | Yes | Yes | RT-PCR | Pt_0009s13110 | |||
| HbXIP3; 1 | 771 | 771 | 0 | 0 | Pt_0009s13070 | |||||||||
| SIP | HbSIP1;1 | 720 | 720 | 0 | 0 | Yes | Yes | Yes | Yes | AT3G04090 | Pt_0014s15250 | |||
| HbSIP1;2 | 4572 | 720 | 2 | 0 | Yes | Yes | Yes | Yes | Yes | Yes | RT-PCR | AT3G04090 | Pt_0019s04640 | |
| HbSP1;3 | 5438 | 720 | 2 | 0 | Yes | Yes | Yes | Yes | Yes | AT3G04090 | Pt_0019s04640 | |||
| HbSIP2;1 | 13833 | 723 | 2 | 0 | Yes | Yes | Yes | Yes | Yes | Yes | RT-PCR | AT3G56950 | Pt_0016s02560 | |
aBased on the 454 transcriptome data under the NCBI SRA accession number of DRX000223; b Based on the 454 transcriptome data of SRX451708 and Illumina transcriptome data of SRX206128, SRX206129, SRX206130, SRX203083, SRX203085, SRX203117, SRX203118 and SRX278515; c Based on the 454 transcriptome data of SRX451705 and Illumina transcriptome data of SRX037405, SRX206131, SRX206132 and SRX278514; d Based on the 454 transcriptome data of SRX451707 and Illumina transcriptome data of SRX278513; e Based on the 454 transcriptome data of SRX451710; f Based on the 454 transcriptome data of SRX451709. Read-mapping was carried out using Bowtie2 with default parameters, and mapped read number of more than one was counted as “Yes” representing detected genes
Fig. 1.

Phylogenetic analysis of deduced amino acid sequences of the 51 HbAQPs with Arabidopsis and poplar homologs. Amino acid sequences were aligned using ClustalX and the phylogenetic tree was constructed using bootstrap maximum likelihood tree (1000 replicates) method and MEGA6 software. The distance scale denotes the number of amino acid substitutions per site. The name of each subfamily is indicated next to the corresponding group. Species and accession numbers are listed in Table 1 and Additional file 2
Homology search showed that 11 out of 51 HbAQP genes have EST hits in GenBank (as of June 2015), i.e. HbPIP1;1, HbPIP1;2, HbPIP1;4, HbPIP1;5, HbPIP2;1, HbPIP2;2, HbPIP2;3, HbPIP2;7, HbPIP2;8, HbTIP1;1 and HbTIP1;5. Eight full-length HbAQP cDNAs, including HbPIP1;1 (GenBank accession number GQ903902), HbPIP1;4 (denoted as HbPIP1 under the accession number of GQ479823), HbPIP2;1 (FJ851079), HbPIP2;2 (KP990544), HbPIP2;5 (denoted as HbPIP2;3 under the accession number of KF921089), HbPIP2;7 (denoted as HbPIP2 under the accession number of GQ479824), HbTIP1;1 (FJ851080) and HbTIP1;2 (denoted as HbTIP1 under the accession number of KP990545) have been reported [21–23, 26–29]. Read alignments against RNA sequencing data of rubber tree shoot apex, leaf, laticifer, bark, root and somatic embryogenesis [30–35] indicated that the expression of 44 HbAQP genes was observed in at least one of the examined tissues (Table 1). Whereas, seven genes coding for two TIPs (HbTIP1;4 and HbTIP5;2), one NIP (HbNIP7;1), four XIPs (HbXIP1;1, HbXIP1;3, HbXIP1;4 and HbXIP3;1) might be expressed exclusively in response to a specific stimulus or in a very specific part of the plant and thus are excluded in the available datasets. In Arabidopsis, the orthologs of HbTIP1;4 (AtTIP1;3) and HbNIP7;1 (AtNIP7;1) were also shown to be pollen or anther-specific, respectively [37, 38]. Besides supported by ESTs and/or RNA sequencing reads, the exon-intron structures of laticifer-expressed HbAQP genes (see below) were also confirmed with cloned cDNAs (Table 1).
Analysis of exon-intron structure
The exon-intron structures of the 51 HbAQP genes were analyzed based on the gene models. Although the ORF (open reading frame) length of each gene is similar (684–927 bp), the gene size (from start to stop codons) is distinct (720–13833 bp, Table 1; Fig. 2). The introns of HbAQP genes harbor an average length of 404 bp, with the minimum of 71 bp in HbNIP2;1 and the maximum of 13000 bp in HbSIP2;1. Genes in different subfamilies harbor distinct exon-intron structures. All members of the HbPIP subfamily feature three introns (92–736 bp, 78–1650 bp and 80–186 bp, respectively). Except for HbTIP1;1, HbTIP1;2, HbTIP1;3 and HbTIP1;4 that contain only one intron, other HbTIP genes contain two introns instead. HbNIP genes usually have four introns except for HbNIP5;1 containing three introns. Most HbSIP genes contain two introns except for HbSIP1;1 without any intron. Subgroups of HbXIP genes vary in the number of introns: one intron for subgroup one, two or zero for subgroups two and three, respectively (Fig. 2).
Fig. 2.

Exon-intron structures of the 51 HbAQP genes. Shown is a graphic representation of the gene models of all 51 HbAQPs identified in this study using GSDS. UTRs are shown as gray boxes, exons are shown as white boxes and introns are shown as black lines
Structural features of HbAQPs
Sequence analysis showed that the 51 deduced HbAQPs consist of 227–305 amino acids, with a theoretical molecular weight of 23.78–32.28 kDa and a pI value of 4.59–9.74. Homology analysis revealed a high sequence diversity existing within and between the five subfamilies. The sequence similarities of 66.4–99.3 % were found within HbPIPs, 49.8–98.4 % within HbTIPs, 45.3–93.1 % within HbXIPs, 44.6–90.6 % within HbNIPs, and 40.6–95.0 % within HbSIPs. HbPIPs share the highest sequence similarity of 35.2–49.0 % with HbTIPs, 34.0–41.8 % with HbXIPs, 30.2–36.8 % with HbNIPs, and the lowest of 22.9–32.5 % with HbSIPs. HbTIPs show 28.7–46.4 %, 28.1–39.9 % and 24.2–41.1 % sequence similarities with HbNIPs, HbXIPs and HbSIPs, respectively. HbNIPs share sequence similarities of 26.0–35.0 % and 23.0–32.2 % with HbXIPs and HbSIPs, whereas HbSIPs share the lowest similarity of 20.8–33.7 % with HbXIPs (Additional file 4).
Topological analysis showed that all HbAQPs were predicted to harbor six transmembrane helical domains (Table 2), which is consistent with the results from multiple alignments with structure proven AQPs (see Additional file 5). The subcellular localization of each HbAQP was also predicted (Table 2). HbPIPs with an average pI value of 8.47 are localized to plasma membranes. HbTIPs with an average pI value of 5.81 are mainly localized to vacuoles (known as lutoids in laticifers with a natural pH of about 6), though several members were predicted to be localized to endoplasmic reticulum (ER), chloroplast and cytosol. HbNIPs with an average pI value of 7.60 are mostly localized to plasma membranes, but HbNIP2;1 and HbNIP3;1 were predicted to be localized to the membrane of vacuole and chloroplast, respectively. Two members (HbSIP1;2 and HbSIP1;3) of the SIP subfamily (with an average pI value of 9.06) were predicted to be localized to plasma membranes, whereas HbSIP1;1 and HbSIP2;1 are localized to the membrane of vacuole and chloroplast, respectively. Although the XIP subfamily harbors only six members (with an average pI value of 7.95), the predicted localizations are diverse, including the vacuole, chloroplast, plasma membrane and cytosol. To learn more about the putative function of HbAQPs, the conserved residues typical of dual NPA motifs, the ar/R filter, five Froger’s positions and nine SDPs were also identified (Tables 2 and 3).
Table 2.
Structural and subcellular localization analysis of the HbAQPs
| Name | Len | Mw (KDa) | pI | TMa | TMb | TMc | Locd | Ar/R selectivity filter | NPA motifs | Froger's positions | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H2 | H5 | LE1 | LE2 | LB | LE | PI | P2 | P3 | P4 | P5 | ||||||||
| HbPIP1;1 | 287 | 30.80 | 8.59 | 6 | 6 | 6 | Plas | F | H | T | R | NPA | NPA | E | S | A | F | W |
| HbPIP1;2 | 287 | 30.80 | 8.59 | 6 | 6 | 6 | Plas | F | H | T | R | NPA | NPA | E | S | A | F | W |
| HbPIP1;3 | 287 | 30.74 | 8.24 | 6 | 6 | 6 | Plas | F | H | T | R | NPA | NPA | E | S | A | F | W |
| HbPIP1;4 | 287 | 30.78 | 8.62 | 6 | 6 | 6 | Plas | F | H | T | R | NPA | NPA | E | S | A | F | W |
| HbPIP1;5 | 269 | 28.92 | 8.60 | 5 | 6 | 5 | Plas | F | H | T | R | NPA | NPA | Q | S | A | F | W |
| HbPIP2;1 | 288 | 30.71 | 7.61 | 6 | 6 | 6 | Plas | F | H | T | R | NPA | NPA | Q | S | A | F | W |
| HbPIP2;2 | 288 | 30.71 | 8.20 | 6 | 6 | 6 | Plas | F | H | T | R | NPA | NPA | Q | S | A | F | W |
| HbPIP2;3 | 286 | 30.58 | 8.50 | 6 | 6 | 6 | Plas | F | H | T | R | NPA | NPA | Q | S | A | F | W |
| HbPIP2;4 | 286 | 30.61 | 8.19 | 6 | 6 | 6 | Plas | F | H | T | R | NPA | NPA | Q | S | A | F | W |
| HbPIP2;5 | 285 | 30.34 | 9.18 | 6 | 6 | 6 | Plas | F | H | T | R | NPA | NPA | Q | S | A | F | W |
| HbPIP2;6 | 286 | 30.39 | 9.06 | 6 | 6 | 6 | Plas | F | H | T | R | NPA | NPA | Q | S | A | F | W |
| HbPIP2;7 | 278 | 29.56 | 9.11 | 6 | 6 | 6 | Plas | F | H | T | R | NPA | NPA | M | S | A | F | W |
| HbPIP2;8 | 280 | 29.76 | 8.97 | 6 | 6 | 6 | Plas | F | H | T | R | NPA | NPA | M | S | A | F | W |
| HbPIP2;9 | 280 | 29.84 | 6.51 | 6 | 6 | 6 | Plas | F | H | T | R | NPA | NPA | M | S | A | F | W |
| HbPIP2;10 | 285 | 30.45 | 9.05 | 6 | 6 | 6 | Plas | F | H | T | R | NPA | NPA | M | S | A | F | W |
| HbTIP1;1 | 252 | 25.91 | 5.91 | 7 | 6 | 7 | Vacu | H | I | A | V | NPA | NPA | T | S | A | Y | W |
| HbTIP1;2 | 252 | 26.07 | 5.70 | 7 | 6 | 7 | Vacu | H | I | A | V | NPA | NPA | T | S | A | Y | W |
| HbTIP1;3 | 253 | 26.44 | 6.27 | 6 | 6 | 6 | Cyto | H | I | A | V | NPA | NPA | T | S | A | Y | W |
| HbTIP1;4 | 227 | 23.78 | 6.18 | 3 | 6 | 4 | Vacu | H | I | A | V | NPA | NPA | T | S | A | Y | W |
| HbTIP1;5 | 252 | 25.88 | 4.96 | 7 | 6 | 6 | Vacu/Cyto | H | I | A | V | NPA | NPA | T | S | A | Y | W |
| HbTIP1;6 | 252 | 25.79 | 5.70 | 7 | 6 | 7 | Cyto | H | I | A | V | NPA | NPA | T | S | A | Y | W |
| HbTIP1;7 | 252 | 25.90 | 4.97 | 6 | 6 | 6 | Vacu/Cyto | H | I | A | V | NPA | NPA | T | S | A | Y | W |
| HbTIP1;8 | 252 | 25.72 | 4.79 | 6 | 7 | 6 | Vacu/Cyto | H | I | A | V | NPA | NPA | T | S | A | Y | W |
| HbTIP2;1 | 248 | 25.40 | 5.33 | 7 | 6 | 6 | Vacu | H | I | G | R | NPA | NPA | T | S | A | Y | W |
| HbTIP2;2 | 248 | 25.34 | 5.59 | 7 | 6 | 7 | Vacu | H | I | G | R | NPA | NPA | T | S | A | Y | W |
| HbTIP2;3 | 250 | 25.31 | 4.87 | 6 | 6 | 6 | Vacu | H | i | G | R | NPA | NPA | T | S | A | Y | W |
| HbTIP2;4 | 250 | 25.31 | 4.59 | 4 | 6 | 5 | Vacu/Plas | H | I | G | S | NPA | NPA | A | S | A | Y | W |
| HbTIP3;1 | 257 | 27.38 | 6.43 | 6 | 6 | 6 | Cyto | H | I | A | R | NPA | NPA | T | A | A | Y | W |
| HbTIP3;2 | 243 | 25.66 | 9.74 | 6 | 6 | 6 | Cyto | H | I | A | R | NPA | NPA | T | A | A | Y | W |
| HbTIP4;1 | 251 | 26.24 | 5.91 | 6 | 6 | 7 | Vacu | H | I | A | R | NPA | NPA | T | S | A | Y | W |
| HbTIP5;1 | 252 | 25.95 | 6.71 | 5 | 6 | 6 | ER | N | V | G | C | NPA | NPA | I | A | A | Y | W |
| HbTIP5;2 | 247 | 25.32 | 5.13 | 6 | 6 | 6 | Chlo | N | V | G | C | NPA | NPA | I | A | A | Y | W |
| HbNIP1;1 | 286 | 30.41 | 7.57 | 6 | 6 | 6 | Plas | W | V | A | R | NPA | NPA | F | S | A | Y | L |
| HbNIP1;2 | 287 | 30.39 | 8.94 | 6 | 6 | 6 | Plas | W | V | A | R | NPA | NPA | F | S | A | Y | I |
| HbNIP2;1 | 285 | 29.88 | 8.72 | 6 | 6 | 6 | Vacu | G | S | G | R | NPA | NPA | L | S | A | Y | I |
| HbNIP3;1 | 282 | 30.20 | 8.34 | 6 | 6 | 5 | Chlo | W | V | A | R | NPA | NPA | F | S | A | F | I |
| HbNIP4;1 | 267 | 28.80 | 5.28 | 6 | 6 | 6 | Plas | W | V | G | R | NPA | NPV | L | S | A | Y | I |
| HbNIP4;2 | 281 | 29.60 | 5.71 | 6 | 6 | 6 | Plas | W | V | A | R | NPA | NPA | F | S | A | Y | I |
| HbNIP5;1 | 298 | 30.98 | 8.65 | 6 | 6 | 5 | Plas | A | I | G | R | NPS | NPV | F | T | A | Y | L |
| HbNIP6;1 | 305 | 31.58 | 8.68 | 6 | 6 | 5 | Plas | T | I | A | R | NPS | NPV | F | T | A | Y | L |
| HbNIP7;1 | 298 | 31.85 | 6.50 | 6 | 6 | 6 | Plas | A | V | G | R | NPA | NPA | Y | S | A | Y | I |
| HbSIP1;1 | 239 | 25.35 | 9.54 | 5 | 6 | 5 | Vacu | A | V | P | N | NPT | NPA | M | A | A | Y | W |
| HbSIP1;2 | 239 | 25.97 | 7.74 | 6 | 6 | 6 | Plas | F | V | P | N | NPT | NPA | M | A | A | Y | W |
| HbSIP1;3 | 239 | 25.83 | 9.36 | 6 | 6 | 5 | Plas | V | V | P | N | NPT | NPA | M | A | A | Y | W |
| HbSIP2;1 | 240 | 26.40 | 9.60 | 4 | 6 | 5 | Chlo | S | H | G | S | I NPl | NPA | F | V | A | Y | W |
| HbXIP1;1 | 289 | 31.23 | 6.36 | 6 | 6 | 6 | Cyto | V | V | V | R | SPV | NPA | M | C | A | F | W |
| HbXIP1;2 | 296 | 32.13 | 8.06 | 6 | 7 | 6 | Chlo | V | V | V | R | SPI | NPA | M | F | A | F | W |
| HbXIP1;3 | 276 | 29.63 | 7.50 | 5 | 6 | 5 | Cyto | V | I | P | R | NPT | NPA | M | C | A | F | W |
| HbXIP1;4 | 276 | 29.50 | 8.31 | 5 | 6 | 5 | Vacu | V | I | A | R | NPT | NPA | M | C | A | F | W |
| HbXIP2;1 | 305 | 32.28 | 8.61 | 6 | 6 | 6 | Plas | V | V | R | NPV | NPA | V | C | A | F | W | |
| HbXIP3;1 | 256 | 27.16 | 8.87 | 6 | 7 | 5 | Vacu | V | V | A | R | NPL | NPA | V | C | A | F | W |
a,b,cRepresenting the numbers of transmembrane helices predicted by TOPCONS, TMPRED and TMHMM, respectively; dBest possible subcellular localization prediction by the WoLF PSORT. (Ar/R aromatic/arginine, Chlo chloroplast, Cyto cytosol, ER endoplasmic reticulum, H2 transmembrane helix 2, H5 transmembrane helix 5, LE loop E, Loc subcellular localization, NPA Asn-Pro-Ala, Plas plasma membrane, TM transmembrane helix, Vacu vacuolar membrane)
Table 3.
Summary of typical SDPs and those identified in the HbAQPsa

aThe SDPs (specificity-determining positions) in rubber tree aquaporins differing from typical SDPs determined in this study are highlighted in red
HbPIP subfamily
All HbPIPs were identified to have similar sequence length, however, HbPIP2s (278–288 residues) can be distinguished from HbPIP1s (269–287 residues) by harboring relatively shorter N-terminal and longer C-terminal sequences (Additional file 5). The five HbPIP1s have sequence similarities of 79.2–99.3 %, whereas the similarity percents of ten HbPIP2s are 77.4–98.3 %. Between HbPIP1 and HbPIP2 members, sequence similarities of 59.1–65.9 % are observed (Additional file 4). The dual NPA motifs, ar/R filter (F-H-T-R), and four out of five Froger’s positions are highly conserved in HbPIPs (Table 2). In contrast, the P1 position is more variable with the appearance of an E, Q or M residue (Table 2). In addition, two phosphorylation sites corresponding to S115 and S274 in SoPIP2;1 [13] are invariable in HbPIP2s, and the former one is even highly conserved in all HbPIPs, HbTIPs and HbXIPs except for the S → T substitution in several members (Additional file 5), implying their regulation by phosphorylation.
HbTIP subfamily
HbTIPs consist of 227–257 residues. Those belonging to HbTIP1s (227–253 residues) share 73.1–98.0 % sequence similarities, whereas HbTIP2s (248–250 residues) have sequence similarities of 83.2–98.4 % (Table 2). Members of the HbTIP1 subgroup exhibit sequence similarities of 56.1–76.6 %, 59.1–70.6 %, 52.7–67.3 % and 49.8–64.3 % with subgroups HbTIP2, HbTIP3, HbTIP4 and HbTIP5, respectively. HbTIP2s share sequence similarities of 61.1–68.9 %, 59.6–65.1 % and 63.1–68.4 % with HbTIP3s, HbTIP4 and HbTIP5s, respectively. HbTIP3s share sequence similarities of 61.1–63.8 % and 58.0–60.5 % with HbTIP4 and HbTIP5s, respectively (Additional file 4). And HbTIP4 shares 59.0 % and 60.2 % sequence similarities with HbTIP5;1 and HbTIP5;2, respectively. Dual NPA motifs and P3, P4 and P5 positions are highly conserved in HbTIPs (Table 2). Residue substitution is observed at the P1 and P2 positions: T is replaced by A in HbTIP2;4 or I in HbTIP5s at the P1 position, and S is replaced by A in HbTIP3s and HbTIP5s (Table 2). Of the ar/R filter, H at H2 and I at H5 positions are replaced by N and V in HbTIP5s, respectively; A is found to be conserved in HbTIP1s, HbTIP3s, and HbTIP4 and G in HbTIP2s and HbTIP5s at the LE1 position, respectively; and residues at the LE2 position are more variable, mainly V, R, S or C (Table 2).
HbNIP subfamily
HbNIPs consist of 267–305 residues (Table 2). With the exception of HbNIP1 and HbNIP4 subgroups that contain two members, each of the other five subgroups harbors a single member. HbNIP1;1 shares the highest sequence similarity of 90.6 % with HbNIP1;2, whereas HbNIP4;1 shares a similarity of 69.4 % with HbNIP4;2. HbNIP1s show sequence similarities of 54.5–55.7 %, 56.5–57.0 %, 59.9–63.8 %, 51.8 %, 50.0–50.6 % and 47.0 % within the subgroups of HbNIP2, HbNIP3, HbNIP4, HbNIP5, HbNIP6 and HbNIP7, respectively. HbNIP2 shows 54.8 %, 52.2–53.7 %, 49.1 %, 48.0 % and 45.4 % sequence similarities with HbNIP3, HbNIP4s, HbNIP5, HbNIP6 and HbNIP7, respectively. The HbNIP3 shows 52.1–55.0 %, 48.4 %, 47.3 % and 45.2 % sequence similarities with HbNIP4s, HbNIP5, HbNIP6 and HbNIP7, respectively. HbNIP4s show 45.1–47.2 %, 44.6–47.7 % and 44.7–46.3 % sequence similarities with HbNIP5, HbNIP6 and HbNIP7, respectively. HbNIP5 shows 75.1 % and 46.4 % sequence similarities with HbNIP6 and HbNIP7, respectively. HbNIP6 shows 46.3 % sequence similarity with HbNIP7. The HbNIPs have typical dual NPA motifs except for HbNIP4;1, HbNIP5;1 and HbNIP6;1 (Table 2). A is replaced by V in the second NPA motif of HbNIP4;1 and by S or V in the first or second NPA motif of HbNIP5;1 and HbNIP6;1. Compared with other subfamilies, HbNIPs are highly variable in the ar/R filter and Froger’s positions: W/G/A/T at the H2 position, F/L/V/I at the H5 position, A/G at the LE1 position, F/L/Y at the P1 position, S/T at the P2 position, F/Y at the P4 position and L/I at the P5 position (Table 2). In addition, one CDPK phosphorylation site corresponding to S262 in GmNOD26 [39] was also found in the C-terminus of most HbNIPs (Additional file 5).
HbSIP subfamily
There are only four members in the HbSIP subfamily. Three HbSIP1s consist of 239 residues, while HbSIP2;1 has 240 residues (Table 2). HbSIP2;1 shares sequence similarities of 40.6–43.0 % with HbSIP1s. Within the HbSIP1 subgroup, HbSIP1;2 shares the highest sequence similarity of 95.0 % with HbSIP1;3, whereas HbSIP1;1 shows the lowest similarity of 70.0 % with HbSIP1;2 (Additional file 4). The three HbSIP1s harbor the same NPT/NPA motifs, ar/R filter (A/FV-V-P-N) and Froger’s positions (M-A-A-Y-W), whereas HbSIP2;1 exhibits NPL/NPA motifs, S-H-G-S ar/R filter and F-V-A-Y-W Froger’s positions (Table 2).
HbXIP subfamily
HbXIPs vary from 256 to 305 residues in length (Table 2). HbXIP1s share sequence similarities of 45.3–50.3 % and 45.5–47.4 % with HbXIP2;1 and HbXIP3;1, respectively, whereas HbXIP2;1 shows 66.6 % sequence similarity with HbXIP3;1. Within the HbXIP1 subgroup, HbXIP1;4 shares the highest sequence similarity of 93.1 % with HbXIP1;3, whereas HbXIP1;2 shows the lowest similarity of 55.3 % with HbXIP1;3 (Additional file 4). In HbXIP1s, the second NPA motif and the LE2, P3, P4 and P5 positions are highly conserved. SPV/SPI/NPT/NPV/NPL in the first NPA motif, V/I at the H2 position, I/V at the H5 position, V/P/A at the LE1 position, M/V at the P1 position and F/C at the P2 position were observed (Table 2). Similar to most XIPs [7], two highly conserved C residues in the LGGC motif of LC and the NPARC motif of LE were also found in HbXIPs except for HbXIP1;2, in which the F residue is located at the corresponding position of LE (Additional file 5).
Transcriptional profiles of HbAQP genes in the laticifer and their response to ethephon stimulation
The rubber tree laticifer is a single-cell-type tissue specifically for natural rubber biosynthesis. To identify the AQP genes expressed in the laticifer and determine the most important members in the laticifer water balance, the latex representing the laticifer cytoplasm was collected and high-quality total RNAs (260/280 value between 1.95 and 2.00, 28S/18S value between 3.0 and 3.2 and RIN value between 8.9 and 9.1) were isolated from three biological replicates, respectively. Then, RNAs were pooled and subjected to Illumina RNA sequencing. Approximate 5.49 gigabase pairs of raw data (100 nt paired-end reads) were generated. After cleaning and quality checks, about 44.5 million high-quality clean reads with an average length of 95 nt were retained and assembled into 74,102 Unigenes longer than 200 bp, with an average length of 775 bp and an N50 of 1260 bp (i.e. 50 % of the assembled bases were incorporated into Unigenes of more 1260 bp). Expression profiling showed that 19 out of the 51 identified HbAQP genes were detected in the laticifer transcriptome, including genes coding for 10 PIPs (sort by abundance, HbPIP2;7, HbPIP1;4, HbPIP2;5, HbPIP1;3, HbPIP2;3, HbPIP2;1, HbPIP1;1, HbPIP2;4, HbPIP1;5 and HbPIP1;2, the same as follows), 3 TIPs (HbTIP1;2, HbTIP2;2 and HbTIP1;6), 4 NIPs (HbNIP1;2, HbNIP3;1, HbNIP6;1 and HbNIP1;1) and 2 SIPs (HbSIP2;1 and HbSIP1;2) (Fig. 3). Based on the RPKM value, the total expression level of PIP members was 1306, 225, 104 folds more than the NIP, TIP or SIP members, respectively, indicating a crucial role of the PIP subfamily in the laticifer water balance. Among ten laticifer-expressed PIP genes, HbPIP2;7, HbPIP1;4, HbPIP2;5, HbPIP1;3 and HbPIP2;3 were considerably more abundant, counting about 418-, 306-, 204-, 30- and 15-folds higher than the well-studied HbPIP1;1. Whereas, HbSIP2;1, the sixth laticifer-abundant AQP gene, expressed relatively more than any other non-PIP members (Fig. 3).
Fig. 3.

Expression profiles of 19 HbAQP genes detected in the laticifer based on Illumina sequencing
Given the important role and wide application of ethephon stimulation on rubber yield promotion, the response of the above 19 laticifer-expressed HbAQP genes subjected to ethephon treatment was analyzed using qRT-PCR over a time course (6–40 h). As described before, treating the rubber tree bark with ethephon was shown to induce a huge increase in latex yield, starting as early as 6 h after the treatment, and the yield increase was about 2.2-, 3.1-, 2.9- and 4.4-folds higher than the control at 6, 16, 24, and 40 h after the treatment, respectively; the TSC was significantly decreased at the time points of 24 and 40 h; and the latex flow duration was significantly prolonged from the time point of 16 h [22]. As shown in Fig. 4, except for HbNIP6;1, ethephon treatment had significant effects on all other tested HbAQP genes at one or more time points, implying their regulation by ethylene. At the early stage of ethephon treatment, i.e. 6 h, the transcriptional levels of 13 HbAQP genes were significantly affected, including two up-regulated (HbPIP1;2 and HbSIP2;1) and eleven down-regulated (HbPIP1;1, HbPIP1;4, HbPIP1;5, HbPIP2;1, HbPIP2;4, HbPIP2;7, HbTIP1;2, HbNIP1;1, HbNIP1;2, HbNIP3;1 and HbSIP1;2) genes. At 16 h post treatment, 16 HbAQP genes were significantly regulated, including eight up-regulated (HbPIP1;4, HbPIP1;5, HbPIP2;3, HbPIP2;5, HbTIP1;6, HbNIP1;2, HbNIP3;1 and HbSIP2;1) and eight down-regulated (HbPIP1;1, HbPIP1;3, HbPIP2;1, HbPIP2;4, HbTIP1;2, HbTIP2;2, HbNIP1;1 and HbSIP1;2) genes. At 24 h post treatment, 17 HbAQP genes were significantly regulated, including thirteen up-regulated (HbPIP1;1, HbPIP1;2, HbPIP1;3, HbPIP1;4, HbPIP1;5, HbPIP2;3, HbPIP2;5, HbTIP1;2, HbTIP1;6, HbTIP2;2, HbNIP1;1, HbNIP1;2 and HbSIP2;1) and four down-regulated (HbPIP2;1, HbPIP2;7, HbNIP3;1 and HbSIP1;2) genes. At 40 h post treatment, 14 HbAQP genes were significantly regulated, including seven up-regulated (HbPIP1;5, HbPIP2;5, HbTIP1;2, HbTIP1;6, HbNIP1;1, HbNIP1;2 and HbSIP2;1) and seven down-regulated (HbPIP1;1, HbPIP1;2, HbPIP1;3, HbPIP2;1, HbPIP2;7, HbNIP3;1 and HbSIP1;2) genes. Although the time points of 16 and 24 h post treatment harbored similar significantly regulated genes, the later had relatively more genes (especially PIP subfamily members) that were up-regulated. Although the expression patterns of the regulated genes were diverse, they could be classified into seven groups: the cluster 1 that includes HbPIP2;5 was gradually increased upon ethephon stimulation; the cluster 2 including HbPIP2;3 and HbSIP2;1 were firstly increased and then decreased, which is like a clock; the cluster 3 including HbTIP1;6 was firstly increased, subsequently decreased and increased at the last time point tested; the cluster 4 including HbPIP1;2 was firstly increased, subsequently decreased, then increased and finally decreased; the cluster 5 including HbNIP1;1 and HbSIP1;2 were firstly decreased and then increased; the cluster 6 that includes 9 genes (i.e. HbPIP1;1, HbPIP1;3, HbPIP1;4, HbPIP2;1, HbPIP2;4, HbPIP2;7, HbTIP1;2, HbTIP2;2 and HbNIP3;1) were firstly decreased, subsequently increased and finally decreased; the cluster 7 that includes HbPIP1;5 and HbNIP1;2 were firstly decreased, subsequently increased, then decreased and finally increased. At 24 h post ethephon stimulation, eight genes (i.e. HbPIP1;1, HbPIP1;2, HbPIP1;3, HbPIP2;3, HbPIP2;4, HbTIP1;2, HbTIP2;2 and HbSIP2;1) exhibited the highest expression levels, whereas the highest expression of six genes (i.e. HbPIP1;5, HbPIP2;5, HbTIP1;6, HbNIP1;1, HbNIP1;2 and HbNIP6;1) occurred at 40 h. Moreover, the transcript abundance of HbPIP2;5 and HbNIP1;1 were similar at the time points of 24 h and 40 h (Fig. 4). As describe above, HbPIP2;5, HbPIP2;3 and HbPIP1;3 were among the top 5 highly abundant AQP genes expressed in laticifers (Fig. 3). In addition, another highly abundant AQP genes (i.e. HbPIP1;4) was expressed most at 16 h post ethephon stimulation (Fig. 4).
Fig. 4.
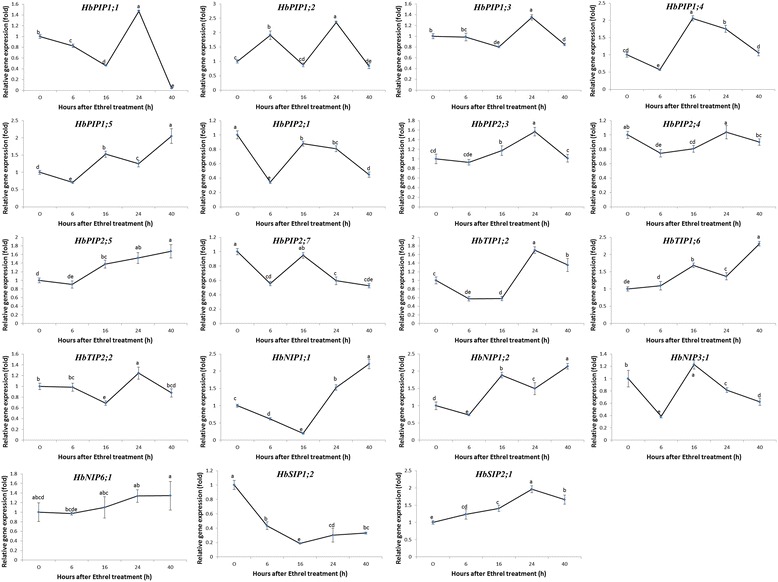
qRT-PCR analysis of 19 laticifer-expressed HbAQP genes upon ethephon stimulation. Data are mean ± SD (n = 9). Different letters mean significant difference over the time course
Discussion
High abundance and diversity of HbAQPs
A total of 51 full-length AQP genes were identified from the rubber tree genome, which is comparable to 55 members reported in poplar (a tree species also belongs to Malpighiales) [7, 36]; more than 23 in grapevine [6], 33 in rice [5], 35 in Arabidopsis [3], 36 in maize [4], 41 in potato [10] and 47 in tomato [9]; less than 66 in soybean [11] and 71 in cotton [8]. Since the AQP genes in Arabidopsis and poplar were well characterized, their deduced proteins were added in the phylogenetic analysis of HbAQPs, which assigned them to five subfamilies. With the exception of the XIP subfamily, the further classification of HbAQP subfamilies into subgroups is consistent with Arabidopsis, i.e. two PIP subgroups, five TIP subgroups, seven NIP subgroups and two SIP subgroups. Nevertheless, classing AtNIP2;1 and AtNIP3;1 into the NIP1 subgroup was proposed. As shown in Fig. 1, AtNIP2;1 and AtNIP3;1 were clustered with the NIP1 subgroup, sharing the highest similarity with AtNIP1;2 in Arabidopsis, HbNIP1;2 or HbNIP1;1 in rubber tree, PtNIP1;2 or PtNIP1;1 in poplar, respectively. Thereby, no NIP2s and NIP3s were retained in Arabidopsis as seen in rubber tree and poplar (Fig. 5). Since no XIP homologs were found in the Arabidopsis genome, the nomenclature for poplar proposed by Lopez et al. [36] was adopted to divide HbXIPs into three subgroups. Besides supported by high bootstrap values, XIP1s are characterized by the ar/R filter of V-M-V/P/A-R, XIP2s by I-I-V-R and XIP3s by V-K-A-R.
Fig. 5.
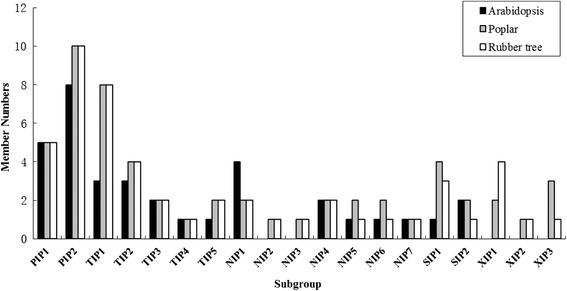
Distribution of the 51 HbAQP genes and their Arabidopsis and poplar homologs in subgroups
Gene pairs were identified not only in rubber tree, but also in poplar and Arabidopsis (Fig. 1). For example, five AtPIP1s were clustered together apart from PIP1s of rubber tree and poplar; HbPIP1;1 and HbPIP1;2 were clustered with PtPIP1;1 and PtPIP1;2. These results suggest the occurrence of more than one gene duplication events. Previous studies indicated that poplar underwent one whole-genome triplication event (designated ‘γ’) and one doubling event, whereas Arabidopsis underwent the same γ event and two independent doubling events, though the Arabidopsis genome encodes relatively less AQP genes due to massive gene loss and chromosomal rearrangement after genome duplications [40–42]. The γ duplication occurred at approximate 117 million years ago, shortly before the origin of core eudicots [43]. As a core eudicot plant, the rubber tree appears to share the γ duplication. However, another one as the data suggested is likely to be a doubling event independent from both Arabidopsis and poplar, probably occurred after the divergence of Euphorbiaceae and Salicaceae. A genome-wide comparative analysis may provide more information.
Functional inference of HbAQPs
Although plant AQPs firstly raised considerable interest for their high water permeability, when heterologously expressed in Xenopus oocytes or yeast cells, increasing evidence has shown that some of them are also participated in the transport of other small molecules such as glycerol, urea, boric acid, silicic acid, NH3, CO2 and H2O2 [44]. Based on atomic resolution structures and molecular dynamics stimulations of GlpF, AqpZ, AQP1 and other MIPs, several structural features determining their transport selectivity were identified, e.g. the two opposite NPA motifs, the ar/R filter and the amino acid residues at Froger’s positions for discriminating between AQPs and GLPs [14, 45]. As shown in Table 2, most HbAQPs exhibit an AqpZ-like Froger’s positions to favor the permeability of water. In contrast, HbSIP2;1 and NIP subfamily members possess mixed key residues of GlpF for P1 and P5, and AqpZ for P2–P4. The glycerol permeability of GmNOD26 and Arabidopsis NIPs was reported [46, 47], however, the potential glycerol transport ability of HbSIP2;1-like SIPs have not be confirmed by experimental means yet.
In addition to high permeability to water, plant PIPs were reported to transport urea, boric acid, CO2 and H2O2 [48]. As shown in Table 2, all HbPIPs represent the F-H-T-R ar/R filter as observed in AqpZ which harbors an extremely narrow and hydrophilic pore (diameter 2.8 Å) [45], suggesting their high water permeability. However, when expressed in Xenopus, extremely low water permeability of HbPIP1 members such as HbPIP1;1 and HbPIP1;4 was observed [22, 26] as seen in many other plant species [49]. Based on the SDP analysis proposed by Hove and Bhave [15], all HbPIPs represent urea-type SDPs (H-P-F/L-F/L-L-P-G-G/S-N); HbPIP1s represent boric acid-type SDPs (T-I-H-P-E-L-L-T-P); HbPIP1;3 represents CO2-type SDPs (I-I-C-A-I-D-W-D-W); HbPIPs except for HbPIP2;9 represent H2O2-type SDPs (A-G-V-F/V-I-H/Q-Y-V/A-P) (Table 3), supporting their similar functionality.
Although highly variable in the ar/R filter, plant TIPs were shown to transport water as efficiently as PIPs [21]. Additionally, they also allow urea, NH3 and H2O2 through [50]. As shown in Table 3, all HbTIPs except for HbTIP2;4 and HbTIP4;1 represent urea-type SDPs (H-P-F/L-F/L-L-A/P-G-S-N), whereas HbTIP1;5, HbTIP1;6, HbTIP4;1, HbTIP5;1 and HbTIP5;2 represent H2O2-type SDPs (S-A-L-A/L/V-I-H/Q-Y-V-P), indicating similar functionality. Compared with typical NH3-SDPs (F/T-K/L/N/V-F/T-V/L/T-A-D/S-A/H/L-E/P/S-A/R/T), HbTIP2;2 seems to represent novel SDPs with the substitution of S for A/R/T at SDP9.
Besides glycerol and water, plant NIPs have been found to transport urea, boric acid, silicic acid, NH3 and H2O2 [50–52]. As shown in Table 3 and Additional file 6, HbNIP5;1 is promised to be a urea and boric acid transporter with nine SDPs of H-P-I-A-L-P-G-S-N or T-I-H-P-E-L-L-A-P. HbNIP2;1 represent typical urea SDPs (H-P-T-A-M-P-G-S-N), and SDPs of V-V-H-P-E-I-I-A-P with the substitution of V for I at SDP2 in comparison to typical boric acid SDPs (T/V-I-H-P-E-I/L-I/L-A/T-A/G/P). Compared with typical urea and boric acid SDPs, HbNIP6;1 seems to represent novel SDPs-types with the substitution of Q for A/P at SDP6 or Q for A/P/G at SDP9. Although characterized as an NIP III member, the silicic acid transport ability of HbNIP2;1 needs to be experimentally validated since it seems to represent novel SDPs (S-F-V-H-G-N-R-T-Q in contrast to typical C/S-F/Y-A/E/L-H/R/Y-G-K/N/T-R-E/S/T-A/K/P/T) similar to that of GmNIP2;1 and GmNIP2;2 (S-Y-E-R-G-N-R-T-P) [53]. Although GmNOD26 was reported to transport NH3 [54], whether its close rubber tree homologs (i.e. HbNIP1;1, HbNIP1;2, HbNIP3;1, HbNIP4;1 and HbNIP4;2) represent novel SDPs-types still needs to be tested. HbNIP3;1, HbNIP4;2 and HbNIP5;1 represent H2O2-type SDPs (A/S-A-L-L/V-I/V-L-Y-V-P) slightly different from AtNIP1;2 (S-A-L-L-V-L-Y-V-P) [50].
As a recently identified AQP subfamily, plant XIPs were shown to transport water, glycerol, urea, boric acid and H2O2 [36, 55]. According to phylogenetic relationships, XIPs are split into two independent clusters termed XIP-A and XIP-B, where XIP-A includes only XIP1 subgroup and XIP-B contains at least four subgroups, i.e. XIP2, XIP3, XIP4, and XIP5 [36]. Consistent with poplar XIPs (two XIP1, one XIP2 and three XIP3), six HbXIPs can be assigned to subgroups XIP1 (4), XIP2 (1) and XIP3 (1) (Fig. 4). When expressed in Xenopus oocytes, PtXIP2;1 and PtXIP3;3 transported water while other PtXIPs did not. Although the mechanism why PtXIP1s, PtXIP3;1 and PtXIP3;2 do not transport water is still unclear, the close homologs of PtXIP1s in Nicotiana tabacum and potato were also reported to have undetectable water permeability. In contrast, Solanaceae XIPs showed high permeability to glycerol [55]. Therefore, although exhibiting an AqpZ-like Froger’s positions, all HbXIPs maybe transport glycerol. Meanwhile, HbXIP2;1 and HbXIP3;3 are probably capable of transporting water. As shown in Table 3, HbXIP1;1, HbXIP1;2, HbXIP2;1 and HbXIP3;1 are promised to be urea transporters with nine SDPs of H-P-F/L-A-L-G-G-G-N; HbXIP1;3, HbXIP1;4 and HbXIP2;1 may represent novel boric acid SDPs-types with the substitution of Q or T for A/G/K/P at SDP9; HbXIP1;3 and HbXIP1;4 harbor H2O2-type SDPs (A-G-L-V-L-H-Y-V-P) with a slight difference from some Solanaceae XIPs (S-A-V-A-V-L-Y-V-P) [55].
A crucial role of HbPIPs in the water balance of laticifers
As a unique site for rubber biosynthesis, the laticifers are present in a wide variety of rubber tree tissues, including shoots, roots, stems, leaves, flowers, fruits, cotyledons, inner seed coats, etc., and can be divided into primary and secondary laticifers according to their origin [16]. Compared with the procambium-derivation of primary laticifers, the secondary laticifers, mainly located in the soft inner bark of the rubber tree trunk, are periodically differentiated from the vascular cambium and serve as a sole source for the commercial latex [56]. During the differentiation and maturation process, laticifer mother cells articulate with each other and further anastomose together into a successive vertical network (called rings or mantles) arranged as concentric sheaths in the secondary phloem [56]. Unlike other cells such as neighboring parenchyma cells, the mature laticiferous cells are totally devoid of plasmodesmata [25], and thus its water exchanges with surrounding cells are mainly governed by AQPs. Upon bark tapping, the laticifer cytoplasm is expelled in the form of latex due to the high turgor pressure inside [57]. Generally, latex flow can continue for several hours until coagulation processes lead to the plugging of severed laticifers [58]. During the latex flow, a progressive decrease in DRC was observed [21–23], indicating rapid water influx and latex dilution inside laticifers caused by the activity of HbAQPs. Given that of HbPIPs and HbTIPs account for more than 62.7 % of the total HbAQPs and their AqpZ-like Froger’s positions favoring the high water permeability, we initially prospect that these two subfamilies may play important roles in the laticifer water balance: the plasma membrane-targeted HbPIPs facilitate the water transport from the extracellular space to the laticifer cytoplasm, whereas the lutoid-targeted HbTIPs play an essential role in maintaining the cell osmotic balance as observed in most plant cells [59]. However, in contrast to the mature plant cells characterized by a large central vacuole which occupies 80 % or more of the intracellular space, the lutoids in laticifers are polydispersed microvacuoles occupying only 12 % of the total latex [60], arguing the central role of HbTIPs in the laticifer water balance, though their potential role in the lutoid stability and latex vessel plugging should be noted. To address this issue, the transcriptome of such a single-cell-type tissue was deeply sequenced. Results showed that PIP members were the main AQP genes expressed in the laticifer (similar results were also observed when the recently available laticifer transcriptome of clone RRIM928 was analyzed, see Additional file 7), suggesting their crucial role, especially the highly abundant HbPIP2;7, HbPIP1;4 and HbPIP2;5, in the laticifer water balance. When expressed in Xenopus oocytes, our previous study showed that HbPIP2;5 could transport water as efficiently as HbPIP2;1 [21, 23]; in contrast, HbPIP1;4 and HbPIP2;7 were shown to be less efficient [22]. In addition, as a PIP1 member, the poor efficiency of HbPIP1;1 was also observed [26]. Therefore, the exact role of HbPIP2;7, HbPIP1;4 and HbPIP2;5 in the water balance of rubber tree laticifers needs further investigations.
To profile the AQP genes in response to ethylene stimulation in laticifers, the latex at different time points after ethephon treatment was collected from rubber tree clone PR107. Similar to PB217, PR107 clone is characterized as a relatively late mature variety which has a high TSC, short latex flow duration and low latex metabolism, however, ethephon stimulation could significantly prolong its latex flow duration and enhance latex yield [22, 23]. Our qRT-PCR analysis showed that the expression levels of most laticifer-expressed genes significantly changed at least one tested time point after ethephon application (Fig. 4), indicating their involvements in the ethephon enhanced water influx into laticifers. Among these time points, the latex collected at 24 and 40 h (especially 24 h) after ethephon treatment was shown to harbor the most abundant transcripts, which include four of the five highly abundant HbPIP1;3, HbPIP2;3, HbPIP1;4 and HbPIP2;5, corresponding to the significantly decreased TSC, the longest latex flow duration and the highest latex yield as reported by Wang et al. who utilized the same materials [22]. Besides, similar effects of ethephon on latex yield and latex TSC of the PB217 clone were also observed by Tungngoen et al., although they used mature virgin trees as materials [21].
Conclusions
To our knowledge, this is the first genome-wide study of the rubber tree AQP gene family and using systematic nomenclature assigned 51 HbAQPs into five subfamilies based on the sequence similarity and phylogenetic relationship with their Arabidopsis and poplar counterparts. Furthermore, their structural and functional properties were investigated based on the analysis of the ar/R filter, Froger’s positions and SPDs, which suggested the potentially key role of HbPIPs and HbTIPs in the laticifer water balance. Most importantly, the laticifer transcriptome was deeply sequenced to identify the most important AQPs in such a single-cell-type tissue, and qRT-PCR analysis was also performed to investigate the expression profiles of laticifer-expressed HbAQP genes upon ethephon stimulation. Our results revealed that HbPIPs were the mainly AQP genes expressed in the laticifer. Among 19 HbAQP genes detected in the laticifer, most of them were significantly regulated by ethylene. Consistent with the significantly decreased TSC and increased latex yield, most laticifer-expressed PIP genes were considerably induced at the time point of 24 h after ethephon application, supporting their crucial roles in the water balance of laticifers in the case of ethephon stimulation. This study provides an important genetic resource of HbAQP genes, which will be useful to improve the water use efficiency and latex yield of Hevea.
Methods
Identification of rubber tree aquaporin genes
The deduced amino acids of HbAQPs available in the NCBI GenBank were used as queries to search the available RRIM600 genome and our in-house RY7-33-97 genome for rubber tree homologs. Sequences with an E-value of less than 1e−5 in the tBlastn search [61] were selected for further analyses. The gene structures were firstly predicted using GeneMark.hmm [62], and the gene models were further validated with ESTs and raw RNA sequencing reads available at GenBank. The exon-intron structures of AQP genes detected in the laticifer transcriptome were also confirmed by aligning the cloned cDNAs to the corresponding gene sequences. Gene structures were displayed using GSDS [63]. Homology search for nucleotides or Sanger ESTs was performed using Blastn, and sequences with an identity of more than 98 % were taken into account. RNA sequencing reads were mapped using Bowtie 2 [64] with default parameters, and mapped read number of more than one was counted as expressed. Unless specific statements, the tools used in this study were performed with default parameters.
Sequence alignments and phylogenetic analysis
Multiple sequence alignment using deduced proteins was performed with ClustalX [65], and the unrooted phylogenetic tree was constructed by the maximum likelihood method using MEGA6 [66]. The reliability of branches in the resulting tree was supported with 1,000 bootstrap resamplings. Classification of AQPs into subfamilies and subgroups was done as described before [3, 36].
Structural features of rubber tree aquaporins
Biochemical features of HbAQPs were determined using ProtParam (http://web.expasy.org/protparam/). The subcellular localization was predicted using WoLF PSORT [67]. The transmembrane regions were detected using TOPCONS [68], TMPRED [69] and TMHMM [70]. Functional prediction was carried out based on dual NPA motifs, ar/R filters (H2, H5, LE1, LE2), Froger’s positions (P1–P5) and specificity-determining positions (SDP1–SDP9) from alignments with the structure resolved Spinacia oleracea PIP2;1 and functionally characterized AQPs as collected by Hove and Bhave [15].
Plant materials and field experiments
PR107, the male parent of rubber tree clone RY7-33-97, was planted at the experimental farm of Chinese Academy of Tropical Agricultural Sciences (Danzhou, China) in 2002. Six batches of three trees with similar growth performance and latex yield were selected for this study. The trees had been tapped for 3 years on the s/2 d 3 system (tapping every 3 days with half spiral) without ethephon stimulation. For ethephon stimulation, five batches of trees were treated with 1 g of 2.5 % (w/w) ethephon in carboxyl methyl cellulose (CMC, 1 %) for 6, 16, 24, and 40 h before the sampling. The sixth batch was treated with 1 % CMC as a control.
Latex collection and total RNA extraction
The latex was collected through tapping the bark at around 6:00 am, and samples representing three biological replicates were subjected for total RNA isolation as described by Tang et al. [71]. Briefly, the latex within the first 45 min was dropped into liquid nitrogen after discarding the first 5 drops. The frozen latex was suspended with extraction buffer (0.3 M LiCl, 0.01 M disodium salt EDTA, 10 % (W/V) SDS, 0.1 M Tris–HCl), and equal volume of water-saturated phenol/chloroform/isoamyl alcohol (PCI) (25:24:1) was added and vigorously shaken. Then, the mixture was centrifuged at 12,000 × g for 10 min at 4 °C, and the aqueous phase was collected and subjected to one more PCI and one chloroform/isoamyl alcohol (24:1) extraction. The supernatant was precipitated with 8 M LiCl solution for twice. The pellet was dissolved with H2O, and 3 M NaAc (pH 5.2) and absolute alcohol were added to precipitate the RNA. After washed with 75 % ethanol, the RNA was dissolved with H2O. The concentration and integrity of total RNA was confirmed using a 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA).
For the expression analysis, the first-strand cDNA was synthesized from 2 μg of total RNA to a final 20 μL reaction mixture using PrimeScript® RT reagent kit with gDNA Eraser (Takara, Dalian, China) according to the manufacture’s instruction, and then stored at −20 °C.
For Illumina sequencing, magnetic beads with biotin-Oligo (dT) were used to isolate poly(A) mRNA according to the manufacturer’s protocol of Illumina TruSeqTM RNA sample preparation kit (Qiagen GmbH, Hilden, Germany).
Expression analysis based on Illumina sequencing
RNA sequencing was performed as described previously [72] using Illumina HiSeq™ 2000 (Illumina Inc., San Diego, CA, USA) at Beijing Genomics Institute (Shenzhen, China). The raw data were filtered by the Illumina pipeline to remove adaptor sequences, adaptor-only reads, reads with “N” rate larger than 10 % (“N” representing ambiguous bases) and low quality reads containing more than 50 % bases with Q-value ≤ 5. Assembly of clean reads was carried out using SOAP de novo [73] (Luo et al. 2012). The trimmed reads were mapped to Unigenes using Bowtie 2 [64], and the RPKM (reads per kilo bases per million reads) method [74] was used for the expression annotation.
qRT-PCR analysis
HbYLS8, the most stably expressed genes in response to ethephon stimulation [75], was selected as the reference gene in this study. The gene-specific primers are listed in Additional file 8, and the PCR reaction was performed using the SYBR-green Mix (Takara, Dalian, China) and the Real-time Thermal Cycler (Type 5100, Thermal Fisher Scientific Oy, Finland). All qRT-PCR assays were performed in triplicate for each biological sample. The amplification efficiency of each primer pair was estimated via melting curve analysis, and PCR products were confirmed by Sanger sequencing. The relative abundance of each transcript was estimated with the 2∆∆Ct method after normalization against HbYLS8 using PikoReal2.0 software unless otherwise specified. Statistical analyses were executed using the Data Processing System software v11.0. The differences among means were tested following Duncan’s one-way multiple-range post hoc ANOVA (P < 0.05).
Acknowledgments
This work was supported by the Natural Science Foundation of Hainan Province (312026), the National Natural Science Foundation of China (31371556), the Fundamental Research Funds for Rubber Research Institute, CATAS (1630022015004 and 1630022015013) and the Earmarked Fund for Modern Agro-industry Technology Research System (CARS-34-GW5). We thank Drs. Mrinal Bhave and Runyararo Memory Hove from Swinburne University of Technology for their help on function prediction of H. brasiliensis aquaporins. The authors also appreciate those contributors who make the rubber tree transcriptome data accessible in public databases.
Abbreviations
- AQP
Aquaporin
- Ar/R
Aromatic/arginine
- DRC
Dry rubber content
- EST
Expressed sequence tag
- GLP
Aquaglyceroporin
- MIP
Major intrinsic protein
- NIP
NOD26-like intrinsic protein
- NPA
Asparagine-proline-alanine
- ORF
Open reading frame
- PIP
Plasma membrane intrinsic protein
- P1-P5
Residues at P1 to P5 positions
- RPKM
Reads per kilo bases per million reads
- SDP
Specificity-determining position
- SIP
Small basic intrinsic protein
- TIP
Tonoplast intrinsic protein
- TM
Transmembrane helix
- TSC
Total solid content
- XIP
X intrinsic protein
Additional files
The gene models for the 51 HbAQP genes identified in this study. (PDF 447 kb)
List of the Phytozome accession numbers of the AQP genes identified in Arabidopsis (35) and poplar (55). (XLS 62 kb)
Percent identity of the identified HbAQP gene pairs at the nucleotide and amino acid levels. (XLS 28 kb)
Percent similarity within and between five subfamilies of the HbAQPs. (XLS 37 kb)
Alignment of predicted amino acid sequences of rubber tree aquaporins with structure determined Spinach SoPIP2;1. (PDF 275 kb)
SDP analysis of the HbAQPs based on the sequence alignment with AQPs transporting non-aqua substrates. (PDF 165 kb)
Expression profiles of the 51 HbAQP genes in the laticifer of rubber tree clone RRIM928. (PDF 36 kb)
qRT-PCR primers of the 19 HbAQP genes expressed in laticifers. (XLS 11 kb)
Footnotes
Zhi Zou, Jun Gong and Feng An contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
The study was conceived and directed by ZZ. All the experiments and analysis were directed by ZZ and carried out by ZZ, JG, FA, GX, JW and YM. ZZ and LY wrote the paper. All the authors read and approved the final manuscript.
Contributor Information
Zhi Zou, Email: zouzhi2008@126.com.
Jun Gong, Email: 378201617@qq.com.
Feng An, Email: an-f@163.com.
Guishui Xie, Email: xie23300459@163.com.
Jikun Wang, Email: kunjiwang@163.com.
Yeyong Mo, Email: 23306788@163.com.
Lifu Yang, Email: ylfri@126.com.
References
- 1.Maurel C, Verdoucq L, Luu DT, Santoni V. Plant aquaporins: membrane channels with multiple integrated functions. Annu Rev Plant Biol. 2008;59:595–624. doi: 10.1146/annurev.arplant.59.032607.092734. [DOI] [PubMed] [Google Scholar]
- 2.Abascal F, Irisarri I, Zardoya R. Diversity and evolution of membrane intrinsic proteins. Biochim Biophys Acta. 2014;1840(5):1468–81. doi: 10.1016/j.bbagen.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjövall S, Fraysse L, et al. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 2001;126(4):1358–69. doi: 10.1104/pp.126.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaumont F, Barrieu F, Wojcik E, Chrispeels MJ, Jung R. Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 2001;125(3):1206–15. doi: 10.1104/pp.125.3.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakurai J, Ishikawa F, Yamaguchi T, Uemura M, Maeshima M. Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 2005;46(9):1568–77. doi: 10.1093/pcp/pci172. [DOI] [PubMed] [Google Scholar]
- 6.Shelden M, Howitt S, Kaiser B, Tyerman S. Identification and functional characterisation of aquaporins in the grapevine, Vitis vinifera. Funct Plant Biol. 2009;36(12):1065–78. doi: 10.1071/FP09117. [DOI] [PubMed] [Google Scholar]
- 7.Gupta AB, Sankararamakrishnan R. Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant Biol. 2009;9:134. doi: 10.1186/1471-2229-9-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park W, Scheffler BE, Bauer PJ, Campbell BT. Identification of the family of aquaporin genes and their expression in upland cotton (Gossypium hirsutum L.) BMC Plant Biol. 2010;10:142. doi: 10.1186/1471-2229-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reuscher S, Akiyama M, Mori C, Aoki K, Shibata D, Shiratake K. Genome-wide identification and expression analysis of aquaporins in tomato. PLoS One. 2013;8(11):e79052. doi: 10.1371/journal.pone.0079052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkatesh J, Yu JW, Park SW. Genome-wide analysis and expression profiling of the Solanum tuberosum aquaporins. Plant Physiol Biochem. 2013;73:392–404. doi: 10.1016/j.plaphy.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 11.Zhang DY, Ali Z, Wang CB, Xu L, Yi JX, Xu ZL, Liu XQ, et al. Genome-wide sequence characterization and expression analysis of major intrinsic proteins in soybean (Glycine max L.) PLoS One. 2013;8(2):e56312. doi: 10.1371/journal.pone.0056312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderberg HI, Kjellbom P, Johanson U. Annotation of Selaginella moellendorffii major intrinsic proteins and the evolution of the protein family in terrestrial plants. Front Plant Sci. 2012;3:33. doi: 10.3389/fpls.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Törnroth-Horsefield S, Wang Y, Hedfalk K, Johanson U, Karlsson M, Tajkhorshid E, et al. Structural mechanism of plant aquaporin gating. Nature. 2006;439(7077):688–94. doi: 10.1038/nature04316. [DOI] [PubMed] [Google Scholar]
- 14.Froger A, Tallur B, Thomas D, Delamarche C. Prediction of functional residues in water channels and related proteins. Protein Sci. 1998;7(6):1458–68. doi: 10.1002/pro.5560070623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hove RM, Bhave M. Plant aquaporins with non-aqua functions: deciphering the signature sequences. Plant Mol Biol. 2011;75(4–5):413–30. doi: 10.1007/s11103-011-9737-5. [DOI] [PubMed] [Google Scholar]
- 16.de Faÿ E, Jacob JL. Anatomical organisation of the laticiferous system in the bark. In: d’Auzac J, Jacob JL, Chrestin L, editors. Physiology of rubber tree latex. Boca Raton: CRC Press; 1989. pp. 3–14. [Google Scholar]
- 17.d’Auzac J, Jacob JL. The composition of latex from Hevea brasiliensis as a laticiferous cytoplasm. In: d’Auzac J, Jacob JL, Chrestin L, editors. Physiology of rubber tree latex. Boca Raton: CRC Press; 1989. pp. 59–96. [Google Scholar]
- 18.Chang C, Bleecker AB. Ethylene biology. More than a gas. Plant Physiol. 2004;136(2):2895–9. doi: 10.1104/pp.104.900122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YF, Etheridge N, Schaller GE. Ethylene signal transduction. Ann Bot. 2005;95(6):901–15. doi: 10.1093/aob/mci100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, Zhang Z. Ethylene stimulation of latex production in Hevea brasiliensis. Plant Signal Behav. 2009;4(11):1072–4. doi: 10.4161/psb.4.11.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tungngoen K, Kongsawadworakul P, Viboonjun U, Katsuhara M, Brunel N, Sakr S, et al. Involvement of HbPIP2;1 and HbTIP1;1 aquaporins in ethylene stimulation of latex yield through regulation of water exchanges between inner liber and latex cells in Hevea brasiliensis. Plant Physiol. 2009;151:843–56. doi: 10.1104/pp.109.140228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, An F, Cai XQ, Zou Z, Zhang W, Lin WF. Function characterization and expression analysis of aquaporins (HbPIP1 and HbPIP2) in Hevea brasiliensis. Scientia Silvae Sinicae. 2014;50:69–75. [Google Scholar]
- 23.An F, Zou Z, Cai XQ, Wang J, Rookes J, Lin W, et al. Regulation of HbPIP2;3, a latex-abundant water transporter, is associated to the latex dilution and latex yield in rubber trees (Hevea brasiliensis Muell. Arg.) PLoS One. 2015;10:e0125595. doi: 10.1371/journal.pone.0125595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacob JL, Prévôt JC, Roussel D, Lacrotte R, Serres E, d’Auzac J, et al. Yield limiting factors, latex physiological parameters, latex diagnosis, and clonal typology. In: d’Auzac J, Jacob JL, Chrestin L, et al., editors. Physiology of rubber tree latex. Boca Raton: CRC Press; 1989. pp. 345–82. [Google Scholar]
- 25.de Faÿ E, Sanier C, Hebant C. The distribution of plasmodesmata in the phloem of Hevea brasiliensis in relation to laticifer loading. Protoplasma. 1989;149:155–62. doi: 10.1007/BF01322987. [DOI] [Google Scholar]
- 26.Tungngoen K, Viboonjun U, Kongsawadworakul P, Katsuhara M, Julien JL, Sakr S, et al. Hormonal treatment of the bark of rubber trees (Hevea brasiliensis) increases latex yield through latex dilution in relation with the differential expression of two aquaporin genes. J Plant Physiol. 2011;168:253–62. doi: 10.1016/j.jplph.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 27.Zhuang HY, An F, He Z, Ding X, Wu HP, Xu ZJ, et al. Molecular cloning and sequence analysis of cDNAs encoding aquaporins in Hevea brasiliensis. Acta Botanica Sinica. 2010;30:861–8. [Google Scholar]
- 28.Zou Z, Wang DH, Mo YY, An F, Yang LF. Molecular cloning and sequence analysis of a tonoplast aquaporin gene TIP1 associated with tapping panel dryness occurence in Hevea brasiliensis. Agr Biotechnol. 2014;3:17–20. [Google Scholar]
- 29.Zou Z, Mo YY, Wang DH, An F, Yang LF. Molecular cloning and sequence analysis of an aquaporin gene related to tapping panel dryness in the rubber tree (Hevea brasiliensis Muell. Arg.) Agr Biotechnol. 2014;3:37–9,43. [Google Scholar]
- 30.Rahman AY, Usharraj AO, Misra BB, Thottathil GP, Jayasekaran K, Feng Y, et al. Draft genome sequence of the rubber tree Hevea brasiliensis. BMC Genomics. 2013;14:75. doi: 10.1186/1471-2164-14-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Triwitayakorn K, Chatkulkawin P, Kanjanawattanawong S, Sraphet S, Yoocha T, Sangsrakru D, et al. Transcriptome sequencing of Hevea brasiliensis for development of microsatellite markers and construction of a genetic linkage map. DNA Res. 2011;18:471–82. doi: 10.1093/dnares/dsr034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chow KS, Mat-Isa MN, Bahari A, Ghazali AK, Alias H, Mohd-Zainuddin Z, Hoh CC, Wan KL. Metabolic routes affecting rubber biosynthesis in Hevea brasiliensis latex. J Exp Bot. 2011;63:1863–71. doi: 10.1093/jxb/err363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chow KS, Ghazali AK, Hoh CC, Mohd-Zainuddin Z. RNA sequencing read depth requirement for optimal transcriptome coverage in Hevea brasiliensis. BMC Res Notes. 2014;7:69. doi: 10.1186/1756-0500-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duan CF, Argout X, Gébelin V, Summo M, Dufayard JF, Leclercq J, et al. Identification of the Hevea brasiliensis AP2/ERF superfamily by RNA sequencing. BMC Genomics. 2013;14:30. doi: 10.1186/1471-2164-14-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mantello CC, Cardoso-Silva CB, da Silva CC, de Souza LM, Scaloppi Junior EJ, de Souza GP, et al. De novo assembly and transcriptome analysis of the rubber tree (Hevea brasiliensis) and SNP markers development for rubber biosynthesis pathways. PLoS One. 2014;9:e102665. doi: 10.1371/journal.pone.0102665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez D, Bronner G, Brunel N, Auguin D, Bourgerie S, Brignolas F, et al. Insights into Populus XIP aquaporins: evolutionary expansion, protein functionality, and environmental regulation. J Exp Bot. 2012;63(5):2217–30. doi: 10.1093/jxb/err404. [DOI] [PubMed] [Google Scholar]
- 37.Soto G, Alleva K, Mazzella MA, Amodeo G, Muschietti JP. AtTIP1;3 and AtTIP5;1, the only highly expressed Arabidopsis pollen-specific aquaporins, transport water and urea. FEBS Lett. 2008;582(29):4077–82. doi: 10.1016/j.febslet.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Li T, Choi WG, Wallace IS, Baudry J, Roberts DM. Arabidopsis thaliana NIP7;1: an anther-specific boric acid transporter of the aquaporin superfamily regulated by an unusual tyrosine in helix 2 of the transport pore. Biochemistry. 2011;50(31):6633–41. doi: 10.1021/bi2004476. [DOI] [PubMed] [Google Scholar]
- 39.Guenther JF, Chanmanivone N, Galetovic MP, Wallace IS, Cobb JA, Roberts DM. Phosphorylation of soybean nodulin 26 on serine 262 enhances water permeability and is regulated developmentally and by osmotic signals. Plant Cell. 2003;15(4):981–91. doi: 10.1105/tpc.009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422(6930):433–8. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 41.Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16(7):1679–91. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313(5793):1596–604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 43.Jiao Y, Leebens-Mack J, Ayyampalayam S, Bowers JE, McKain MR, McNeal J, et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012;13(1):R3. doi: 10.1186/gb-2012-13-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomes D, Agasse A, Thiébaud P, Delrot S, Gerós H, Chaumont F. Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. Biochim Biophys Acta. 2009;1788(6):1213–28. doi: 10.1016/j.bbamem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 45.Savage DF, Egea PF, Robles-Colmenares Y, O’Connell JD, III, Stroud RM. Architecture and selectivity in aquaporins: 2.5 Å structure of aquaporin Z. PLoS Biol. 2003;1(3):E72. doi: 10.1371/journal.pbio.0000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dean RM, Rivers RL, Zeidel ML, Roberts DM. Purification and functional reconstitution of soybean nodulin 26. An aquaporin with water and glycerol transport properties. Biochemistry. 1999;38(1):347–53. doi: 10.1021/bi982110c. [DOI] [PubMed] [Google Scholar]
- 47.Wallace IS, Roberts DM. Distinct transport selectivity of two structural subclasses of the nodulin-like intrinsic protein family of plant aquaglyceroporin channels. Biochemistry. 2005;44(51):16826–34. doi: 10.1021/bi0511888. [DOI] [PubMed] [Google Scholar]
- 48.Bienert GP, Heinen RB, Berny MC, Chaumont F. Maize plasma membrane aquaporin ZmPIP2;5, but not ZmPIP1;2, facilitates transmembrane diffusion of hydrogen peroxide. Biochim Biophys Acta. 2014;1838(1PtB):216–22. doi: 10.1016/j.bbamem.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 49.Yaneff A, Sigaut L, Marquez M, Alleva K, Pietrasanta LI, Amodeo G. Heteromerization of PIP aquaporins affects their intrinsic permeability. Proc Natl Acad Sci U S A. 2014;111:231–6. doi: 10.1073/pnas.1316537111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dynowski M, Mayer M, Moran O, Ludewig U. Molecular determinants of ammonia and urea conductance in plant aquaporin homologs. FEBS Lett. 2008;582(16):2458–62. doi: 10.1016/j.febslet.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 51.Dynowski M, Schaaf G, Loque D, Moran O, Ludewig U. Plant plasma membrane water channels conduct the signalling molecule H2O2. Biochem J. 2008;414(1):53–61. doi: 10.1042/BJ20080287. [DOI] [PubMed] [Google Scholar]
- 52.Ma JF, Tamai K, Yamaji N, Mitani N, Konishi S, Katsuhara M, et al. A silicon transporter in rice. Nature. 2006;440(7084):688–91. doi: 10.1038/nature04590. [DOI] [PubMed] [Google Scholar]
- 53.Deshmukh RK, Vivancos J, Guérin V, Sonah H, Labbé C, Belzile F, et al. Identification and functional characterization of silicon transporters in soybean using comparative genomics of major intrinsic proteins in Arabidopsis and rice. Plant Mol Biol. 2013;83(4–5):303–15. doi: 10.1007/s11103-013-0087-3. [DOI] [PubMed] [Google Scholar]
- 54.Niemietz CM, Tyerman SD. Channel-mediated permeation of ammonia gas through the peribacteroid membrane of soybean nodules. FEBS Lett. 2000;465(2–3):110–4. doi: 10.1016/S0014-5793(99)01729-9. [DOI] [PubMed] [Google Scholar]
- 55.Bienert GP, Bienert MD, Jahn TP, Boutry M, Chaumont F. Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J. 2011;66(2):306–17. doi: 10.1111/j.1365-313X.2011.04496.x. [DOI] [PubMed] [Google Scholar]
- 56.Hao BZ, Wu JL. Laticifer differentiation in Hevea brasiliensis: induction by exogenous jasmonic acid and linolenic acid. Ann Bot. 2000;85:37–43. doi: 10.1006/anbo.1999.0995. [DOI] [Google Scholar]
- 57.Buttery BR, Boatman SG. Effect of tapping, wounding and growth regulators on turgor pressure in Hevea brasiliensis Muell. Arg J Exp Bot. 1967;18:644–59. doi: 10.1093/jxb/18.4.644. [DOI] [Google Scholar]
- 58.Yeang HY. The kinetics of latex flow from the rubber tree in relation to latex vessels plugging and turgor pressure. J Rubber Res. 2005;8:160–81. [Google Scholar]
- 59.Hunter PR, Craddock CP, Di Benedetto S, Roberts LM, Frigerio L. Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiol. 2007;145:1371–82. doi: 10.1104/pp.107.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang XC, Shi M, Wang D, Chen Y, Cai F, Zhang S, et al. Comparative proteomics of primary and secondary lutoids reveals that chitinase and glucanase play a crucial combined role in rubber particle aggregation in Hevea brasiliensis. J Proteome Res. 2013;12:5146–59. doi: 10.1021/pr400378c. [DOI] [PubMed] [Google Scholar]
- 61.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005;33:6494–506. doi: 10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu B, Jin J, Guo A, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2014;31:1296–7. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35(Web Server issue):W585–7. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bernsel A, Viklund H, Hennerdal A, Elofsson A. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 2009;37(Web Server issue):W465–8. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hofmann K, Stoffel W. TMbase-A database of membrane spanning proteins segments. Biol Chem Hoppe Seyler. 1993;374:166. [Google Scholar]
- 70.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 71.Tang CR, Qi JY, Li HP, Zhang C, Wang Y. A convenient and efficient protocol for isolating high-quality RNA from latex of Hevea brasiliensis (para rubber tree) J Biochem Biophys Methods. 2007;70:749–54. doi: 10.1016/j.jbbm.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 72.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5(7):621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 73.Li D, Deng Z, Qin B, Liu X, Men Z. De novo assembly and characterization of bark transcriptome using Illumina sequencing and development of EST-SSR markers in rubber tree (Hevea brasiliensis Muell. Arg.) BMC Genomics. 2012;13:192. doi: 10.1186/1471-2164-13-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li HP, Qin YX, Xiao XH, Tang CR. Screening of valid reference genes for real-time RT-PCR data normalization in Hevea brasiliensis and expression validation of a sucrose transporter gene HbSUT3. Plant Sci. 2011;181:132–9. doi: 10.1016/j.plantsci.2011.04.014. [DOI] [PubMed] [Google Scholar]


