Abstract
Spermatogenesis is an essential reproductive process that is regulated by many Y chromosome specific genes. Most of these genes are located in a specific region known as the azoospermia factor region (AZF) in the long arm of the human Y chromosome. AZF microdeletions are recognized as the most frequent structural chromosomal abnormalities and are the major cause of male infertility. Assisted reproductive techniques (ART) such as intra-cytoplasmic sperm injection (ICSI) and testicular sperm extraction (TESE) can overcome natural fertilization barriers and help a proportion of infertile couples produce children; however, these techniques increase the transmission risk of genetic defects. AZF microdeletions and their associated phenotypes in infertile males have been extensively studied, and different AZF microdeletion types have been identified by sequence-tagged site polymerase chain reaction (STS-PCR), suspension array technology (SAT) and array-comparative genomic hybridization (aCGH); however, each of these approaches has limitations that need to be overcome. Even though the transmission of AZF microdeletions has been reported worldwide, arguments correlating ART and the incidence of AZF microdeletions and explaining the occurrence of de novo deletions and expansion have not been resolved. Using the newest findings in the field, this review presents a systematic update concerning progress in understanding the functions of AZF regions and their associated genes, AZF microdeletions and their phenotypes and novel approaches for screening AZF microdeletions. Moreover, the transmission characteristics of AZF microdeletions and the future direction of research in the field will be specifically discussed.
Keywords: Male Infertility, azoospermia factor region (AZF), microdeletion, assisted reproductive techniques (ART), vertical transmission
Introduction
Infertility can be defined as the inability to conceive after one year of unprotected intercourse. This problem affects approximately 10% to 15% of married couples globally and male infertility accounts for 50% of the cases [1,2]. Genetic factors play well-recognized roles in male infertility, and genetic alterations of the Y chromosome are especially important [3,4]. The major genetic factors in male infertility are chromosomal abnormalities and Y chromosomal microdeletions (YCMs) [5]. YCMs occur in approximately 10 to 15% of azoospermic patients and 5 to 10% of severe oligospermic patients, and are commonly found at the azoospermia factor (AZF) locus in the q11.23 band [2,4,6]. Detailed molecular analyses further subdivide the AZF locus into three (possibly four) subregions: AZFa, AZFb and AZFc, along with a fourth possible AZFd region [7,8]. Microdeletions in these regions result in different degrees of spermatogenetic failure; however, AZF gene function and the exact genotype-phenotype relationship of microdeletions and infertility in the AZF locus have not been fully explored. Sequence-tagged site polymerase chain reaction (STS-PCR) is recognized as the gold-standard method for laboratory AZF microdeletion diagnosis [9,10]; however, multi-analyte suspension array (MASA) technology also provides a rapid, sensitive and high-throughput method for detecting Y chromosome microdeletions [11,12].
The rapid development of assisted reproductive technologies (ART) such as in vitro fertilization, intracytoplasmic sperm injection (ICSI) and testicular sperm extraction (TESE) make reproduction possible for millions of infertile couples; however, these techniques elevate the risk of transmitting genetic defects, including vertical transmission of AZF microdeletions and the related infertility problems to their sons [13,14]. Thus, routine screening for AZF microdeletions before undergoing ART treatment is a critical diagnostic test.
In this review, we will systematically update the progress that has been made in AZF region identification, describe novel approaches for AZF microdeletion screening and summarize the current understanding of associated gene functions, AZF microdeletion types and AZF microdeletion phenotypes. Furthermore, we will specifically discuss the transmission characteristics of AZF microdeletions and outline the future goals of research in this field.
AZFs in Y chromosome
The Y chromosome plays a unique role in the human genome due to its size, organization and function. The human Y chromosome consists of short (Yp) and long (Yq) arms [15,16] (Figure 1A). Cytogenetic analysis has identified several different Y chromosome regions: the pseudo autosomal regions (including PAR1 and PAR2), the euchromatic and the heterochromatic regions. The two autosomal regions are located on the telomeric ends of the chromosome and cover approximately 5% of the chromosome [3,16] (Figure 1A). Yp and Yq11 consist of euchromatin regions, while the distal part of the long arm (Yq12) is made of heterochromatin [3,16,17]. The male-specific region of the Y chromosome (MSY), previously called the non-recombining region of Y, harbors both euchromic and heterochromatic sequences and accounts for 95% of the Y chromosome length [3,16] (Figure 1A). The euchromatic MSY sequences contain 8 Mb of Yp and 14.5 Mb of Yq. These sequences are subdivided into three discrete classes: X-transposed (3.4 Mb), X-degenerate (8.6 Mb) and ampliconic (10.2 Mb) [3,4].
Figure 1.
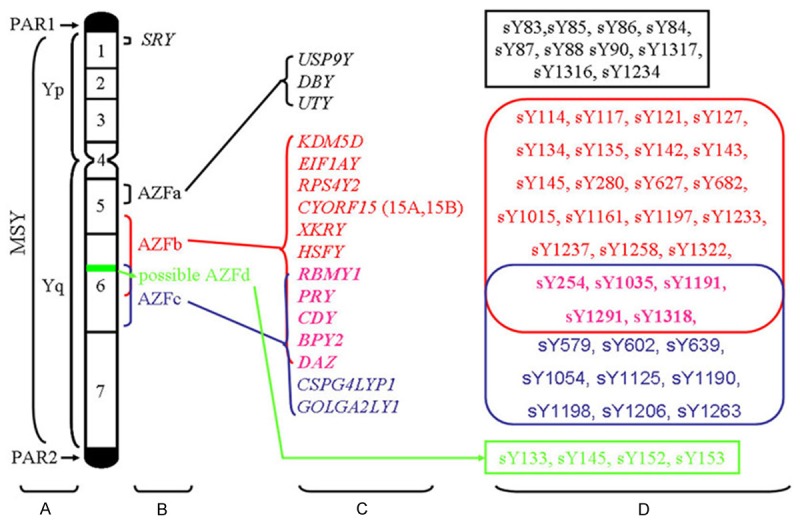
Diagram of the human Y chromosome showing the AZF loci, associated genes and STS markers. A. Schematic representation of the structure of the human Y chromosome showing the pseudo autosomal regions (PAR1, PAR2), Yp (short arm), Yq (long arm) and MSY (male specific region). B. Schematic locations of the four AZF loci. C. Corresponding candidate genes in the AZFa, AZFb and AZFc regions. The black, red, blue and pink colors represent the genes in the AZFa, AZFb, AZFc and overlapping AZFb/AZFc regions, respectively. D. Conventional STS markers associated with the AZFa, AZFb, AZFc and AZFd regions. The black, red, blue, pink and green colors represent the STS markers for the genes in the AZFa, AZFb, AZFc, overlapping AZFb/AZFc and AZFd regions, respectively.
Characterized by eight massive palindromes, ampliconic sequences constitute a major portion of the MSY euchromatic sequences. Six of the eight palindromes contain testis-specific protein coding genes [3,4,16]. Moreover, eight of nine multicopy protein coding gene families in the MSY have members contained within the palindromes and six of those families are located exclusively within the palindromes [18]. The Y chromosome harbors many of the genes that control spermatogenesis, and most of the known chromosomal aberrations related to azoospermia or oligozoospermia have been identified in the long arm of this chromosome [3,16]. Genetic analysis of patients with idiopathic azoospermia or severe idiopathic oligozoospermia identified three AZF subregions: AZFa (approximately 800 kb); AZFb (approximately 3.2 Mb); and AZFc (approximately 3.5 Mb) [19,20] (Figure 1B). Most of the Y chromosome specific genes are located in these AZF regions.
AZFa and candidate genes
The AZFa subregion is located in the proximal region of deletion interval 5 of Yq 11.21 (subinterval 5C of the human Y chromosome) [21,22]. The region spans about 800 kb and encodes single copy genes with homologues in the X chromosome [4]. The genes have been found to be necessary for normal spermatogenesis. Candidate genes in the AZFa locus include USP9Y (Ubiquitin specific peptidase 9, Y-linked), DBY (Dead box on Y) and UTY (Ubiquitously transcribed tetratricopeptide repeat gene, Y-linked) [3,4] (Figure 1C).
USP9Y consists of 46 exons and spans 159 kb of genomic DNA [23]. The USP9Y gene encodes a ubiquitin-specific protease and belongs to the peptidase C19 family [23]. Molecular analysis revealed USP9Y deletions or mutations in infertile patients [24]; however, a recent study reported that normal spermatogenesis was observed in an individual with a complete AZFa region USP9Y deletion [25]. Thus, USP9Y is not required for normal spermatogenesis, but may exert its effects on fertility in combination with other potential candidate genes in the AZFa region. DBY consists of 17 exons spanning 16 kb [26] and it is specifically expressed in testis tissue [27]. This gene encodes an ATP dependent RNA helicase DEAD box protein in humans and plays a significant role in the pre-meiotic spermatogonia phase of spermatogenesis [27]. Molecular analysis revealed a high prevalence of deletions or mutations in the DBY gene in infertile patients when compared to the USP9Y and UTY genes [25,26]. All available data support the hypothesis that DBY is an important spermatogenesis gene. UTY encodes a protein that is rich in tetratricopeptide repeats [28]. This gene is mapped to the 5C interval corresponding to the AZFa region [28]. Deletion analysis revealed the presence of a UTY deletion in one infertile male patient, but this deletion was also accompanied by a DBY deletion [26]. No infertility associated UTY specific deletion has been found yet.
AZFb and candidate genes
The AZFb locus is located in the middle region of Yq11 (approximately between the 5 M to 6B subintervals) [22]. This region spans 6.2 to 7.7 Mb of MSY sequences, but overlaps with the AZFc region by 1.5 Mb [29]. The AZFb region contains several single copy genes as well as multicopy gene families. Single copy protein coding genes found within the AZFb region include KDM5D (lysine (K)-specific demethylase 5D), EIF1AY (eukaryotic translation initiation factor 1A, Y-linked), RPS4Y2 (Ribosomal protein S4 Y isoform 2) and CYORF15 (chromosome Y open reading frame 15A and 15B) [3,4] (Figure 1C).
KDM5D encodes a histone H3 lysine 4 (H3K4) demethylase that forms a protein complex with the MSH5 (MutS protein homolog 5) DNA repair factor during spermatogenesis, suggesting KDM5D involvement in male germ cell chromatin remodeling [30,31]. KDM5D is located in the germ cell nucleus during prophase I, and KDM5D disruption may contribute to the AZFb deletion phenotype [3]. EIF1AY is a ubiquitously expressed Y-linked member of the EIF-1A family and is involved in translation initiation [32], but EIF1AY’s function in male fertility has not been well established. RPS4Y2 expression has been demonstrated to be is testis-specific, so it is reasonable to hypothesize that RPS4Y2 may potentially play a role in posttranscriptional regulation of the spermatogenic program [33]. The CYORF15A and CYORF15B sequences belong to the taxilin family and are involved in transcriptional regulation in osteoblasts [3,34]. Despite their presence in the AZFb region, the role of the CYORF15 genes in normal spermatogenesis is largely unknown.
Due to the presence of ampliconic sequences, the AZFb region also contains a set of seven multicopy gene families: XKRY (XK, Kell blood group complex subunit-related, Y-linked), HSFY (heat shock transcription factor, Y-linked), RBMY1A1 (RNA binding motif protein, Y-linked, family 1, member A1), PRY (PTPN13-like, Y-linked), CDY (Chromodomain Y, Y-linked), BPY2 (Basic protein Y2, Y-linked) and DAZ (deleted in azoospermia) [3,19] (Figure 1C). Twenty members from these gene families are located in AZFb, but several genes also map to the AZFc region (Figure 1C). Only the genes that are exclusively located in the AZFb region are discussed in this section.
The XKRY gene is expressed specifically in the testis and maps to the yellow-coded amplicon family [3,35], but no role for XKRY in spermatogenesis has been validated. HSFY encodes a member of the heat shock factor family of transcriptional activators and displays testis-predominant expression [36]. HSFY’s functions in male fertility have been extensively studied. First, the HSFY gene is found in Sertoli cells and in spermatogenic cells up to the spermatid stage, with predominant expression in round spermatids [37,38]. Second, HSFY protein levels are low in spermatogenic cell samples from patients with maturation arrest [39]. Finally, Vinci et al. detected a partial AZFb deletion that was only found to affect the functional copies of HSFY in an azoospermic man [40]. The PRY gene encodes two full-length functional units in the AZFb region (PRY1 and PRY2) and two shorter versions in the AZFc region (PRY3 and PRY4) [41]. PRY1 and PRY2 exhibit testis-specific expression, but PRY expression in germ cells is irregular and is only detected in a few individual sperm and spermatids [42]. Interestingly, both gene and protein expression of PRY are higher in the defective germ cell fraction of the ejaculate and in sperm obtained from men with abnormal semen parameters. This expression pattern suggests that PRY could be a useful biomarker for defective spermatogenesis [42]. Furthermore, it has been suggested that PRY plays a role in male germ cell apoptosis because approximately 40% of PRY-positive cells show DNA fragmentation [42]. RBMY1A1 is part of the RBM gene family and is solely expressed in male germ cells [43]. This gene has been linked to mRNA storage and transport from the nucleus during spermatogenesis [3]. Furthermore, RBMY1A1 disruption plays a significant role in the AZFb deletion phenotype [44].
AZFc and candidate genes
The AZFc region has been extensively studied due to its important role in male infertility. The AZFc locus is located in the distal part of Yq (deletion subintervals 6C through 6E) [22]. The AZFc region spans 4.5 Mb and resides among three large palindromic sequences that are derived from six distinct amplicon families [3,22]. The AZFc locus encodes 21 candidate genes and 11 families of transcription units that are specifically expressed in the testis [3,4]. Seven of these families are located in the AZFc deletion interval, including GOLGA2LY1 (Golgi autoantigen, golgin subfamily a2-like, Y-linked 1) and CSPG4LYP1 (chondroitin sulfate proteoglycan 4-like, Y-linked pseudogene 1) [4] (Figure 1C). Important candidate genes in this deletion interval include DAZ, BPY2 and CDY1 (CDY1a and CDY1b, Y chromosome 1) [4,45,46] (Figure 1C). Interestingly, restriction mapping has also identified three PRY and TTY2 genes proximal to the AZFc region [47]; however, specific deletions of these genes have not yet been identified and their status as possible AZFc candidate genes remains unconfirmed.
DAZ was the first identified and most important candidate gene family in AZFc region. The DAZ family consists of four RNA binding protein-encoding genes (DAZ1-4) [48]. DAZ protein is localized to the innermost layer of the male germ cell epithelium and to spermatozoa tails [48]. A high incidence of DAZ gene deletion has been observed in infertile men [49], indicating that DAZ may be critically important for spermatogenesis; however, other genes in the AZFc loci also play a vital role in normal spermatogenesis [50,51]. BPY2 encodes a highly charged testis-specific protein that has been tentatively linked to cytoskeletal regulation in spermatogenesis [45,52]. BPY2 protein exhibits nuclear localization in male germ cells at all developmental stages [53], but the exact role of BPY2 in spermatogenesis is still unclear. CDY1 genes are specifically expressed in the testis and are involved in histone replacement during spermatogenesis [54]. Two CDY1 genes are located in the AZF region, one within the DAZ cluster and other at the distal end [55]. Removal of DAZ also removes one copy of CDY1, strongly suggesting that the gene plays a role in spermatogenesis.
Possible AZFd involvement
The AZFd locus was first identified using multiplex PCR reactions and proposed to exist between AZFb and AZFc [8] (Figure 1B). However, the AZFd region remains controversial. Simoni et al. assert that the MSY sequences and microdeletion mechanism clearly indicate that AZFd locus cannot exist between the AZFb and AZFc regions [56], while Muslumanoglu et al. reported microdeletions in the putative AZFd loci by deletion analysis through multiplex PCR reactions using four STS (sY133, sY145, sY152 and sY153) [57]. Moreover, recent studies reporting AZFd deletions in infertile men add further credence to the existence and involvement of the AZFd locus in male fertility [9,58,59].
AZF region microdeletions
YCMs occur in the q11.23 band and are commonly subdivided according to microdeletion location into AZFa, AZFb, AZFc or AZFd microdeletions [3,7]. Complete AZFa deletion removes an estimated 792 kb DNA in the proximal Yq11 region, including two candidate genes [60]. Complete AZFb deletion removes 6.23 Mb DNA spanning 32 genes and all the testis-specific gene families in the AZFb region [29]. AZFc deletions are estimated to span 3.5 Mb DNA encoding 21 genes and seven gene families [45]. Furthermore, three partial AZFc deletions have been identified, including the b1/b3, b2/b3 and gr/gr deletions. These partial AZFc deletions have been associated with a high risk of dysfunctional spermatogenesis [3,4,61]. AZF microdeletions also include combined deletions, including AZFab, AZFac, AZFad, AZFabc, AZFbc, AZFbd and AZFbcd [3,4]. Each of these microdeletions may result in different degrees of spermatogenetic failure [22,62].
Mechanisms of microdeletions
YCMs occur due to errors in homologous recombination [63]. AZFa deletions result from homologous intrachromosomal recombination between two human endogenous retroviral sequences HERV15yq1 and HERV15yq2 that are located in the proximal Yq11 region [63]. Complete AZFb deletions are caused by homologous recombination between the Palindrome P5 and the proximal arm of palindrome P1 in the Yq arm [29]. AZFc deletions have been demonstrated to result from homologous recombination between the sub-amplicons b2 and b4 in palindromes P3 and P1 [45]. The proximity of AZFc to the Yq12 heterochromatic region may also trigger a high percentage of unequal intra-chromosomal recombination, increasing the chance of AZFc deletion [4,45]. Non-homologous recombination between P5/distal-P1 or between P4/distal-P1 may also lead to AZFbc deletions [29]. The mechanisms underlying other types of microdeletions require further investigation.
Prevalence of AZF microdeletions
AZF microdeletions are observed in 10 to 15% of infertile men with azoospermia or severe oligozoospermia, and are very rare in fertile men or men with a sperm density greater than 5 million/ml [7]. In a large cohort of 1,738 infertile men in Northeastern China, the YCM incidence was 8.57% [2]. The most common microdeletion observed in the 1,738 patient study was in AZFc, followed by AZFbc, AZFb, AZFabc, AZFa and AZFac [2]. Similarly, AZF microdeletions were detected in 5.06% of patients (185 cases) in another survey of 3,654 patients, including 147 patients with azoospermia and 38 patients with severe oligozoospermia [64]. The most frequent microdeletions in the 3,654 patient study were observed in the AZFc region, followed by the AZFbcd, AZFbc, AZFb, AZFa and AZFac regions [64]. AZFc deletion accounted for at least two thirds of all Yq deletions in all previous studies. Interestingly, a recent study reported that the prevalence of AZF microdeletions was much higher in men from couples with recurrent pregnancy loss than in men from fertile couples. The result of this study suggests that YCM in the AZF region may be a possible etiologic factor for recurrent pregnancy loss [65].
AZF microdeletion phenotypes
The partial removal of the AZFa region is associated with hypo-spermatogenesis, whereas the complete deletion of the AZFa region inhibits the production and maturation of germ cells in the seminiferous tubules. Testicular biopsies of AZFa deletion patients reveal a Sertoli cell-only (SCO) phenotype that is characterized by the total absence of germ cells in the seminiferous tubules or by the presence of germ cells in a very small number of tubules [4]. Consequently, it is virtually impossible to retrieve mature sperm with the TESE technique [66]. Brown et al. [67] reported two patients with USP9Y deletions who presented with SCO type 1 syndrome, and a third patient with a similar deletion who had hypospermatogenesis. Similar findings were also observed in other investigations [23,24]. Unlike AZFa microdeletions, patients with AZFb microdeletions had normal spermatogonia and primary spermatocytes in all tubules observed [21]. However, deletions in the AZFb region lead to pre-meiotic spermatogenic arrest or SCO syndrome and, eventually, azoospermia. It is difficult to recover mature sperm from AZFb deletion patients using TESE. Patients with AZFc microdeletions were found to have less severe spermatogenic disruption. Vogt et al. observed the presence of germ cells at different developmental stages in some tubules; however, the majority of tubules were devoid of all germ cells except Sertoli cells [22]. Four of the patients examined by Vogt et al. produced a small quantity of motile sperm, ranging from 0.1 to 2 million/ml [22]. Similar to a SCO type 2 phenotype, Vogt et al. observed germ cells at different stages in different tubules. All evidence supports the hypothesis that the primary cause of the spermatogenic failure in AZFc deletion patients is a post meiotic spermatid or sperm maturation defect. Partial deletions involving the DAZ genes in the AZFc region have been found to cause hypospermatogenesis [68]. Moreover, complete AZFb and AZFc deletions were associated with SCO syndrome and alterations in spermatocyte maturation [69]. Infertile men with deletions in the putative AZFd region may have a mildly depressed or even normal sperm count, but abnormal sperm morphology [8]. Muslumanoglu et al. [57] reported three azoospermic men with maturation-arrested testicular histology. These three patients were found to carry a single locus (sY152) microdeletion confined to the putative AZFd region. Interestingly, a recent study revealed that patients with expanded or de novo AZF microdeletions had significantly higher levels of FSH and lower levels of inhibin B when compared to fertile control men [69]. Therefore, low inhibin B and high FSH levels may be useful diagnostic markers that indicate potential causative mutations in the AZF region in infertile men.
Genetic screening methods for AZF microdeletion diagnosis
Karyotyping was commonly used to identify macrodeletions in the long arm of the Y chromosome, but the technique failed to detect smaller interstitial deletions. Southern blotting was used to identify microdeletions related to azoospermia or oligozoospermia [4]. Because karyotyping and Southern blotting are both labor-intensive, time consuming, costly and complex techniques, many researchers have successfully screened for microdeletions using PCR, a relatively simple, sensitive, fast and reproducible technique that allows multiplexing. The STS-PCR technique has been in use for over two decades and has been developed into the gold-standard method for laboratory diagnosis of Y chromosomal microdeletions [3,4]. PCR-based deletion analysis is usually designed according to the following major factors: (1) Selection of the STS-PCR markers. The best and most informative markers should be highly specific, non-polymorphic, single copy markers or markers limited to a small specific region of the Y chromosome [4]. (2) Selection of DNA samples. YCMs primarily occur during the pre-fertilization stages, but they may also occur as a post-fertilization event. If microdeletions occur during pre-fertilization stages, it is highly likely that they will be transmitted to the whole body of the newborn boy. If the deletion occurs during the post-fertilization stages, the deletion will cause mosaicism characterized by normal Y chromosomes in leukocytes but deleted Y chromosomes in sperm or testicular DNA [4,70]. Although lymphocyte DNA is the cheapest and most readily available sample for basic YCM screening, sperm or testicular DNA is the most reliable sample to screen for all kinds of YCMs [4,70] PCR quality control. PCR amplification failures lead to the false screening results. Thus, high-quality DNA and appropriate internal and external positive and negative controls are necessary to minimize false negatives. (4) STS marker reliability. STS markers are short, known DNA sequences that are mapped to different locations in the genome. They offer high speed, convenience, reliability and low cost for genetic screening analysis. To date, about 1287 Y-specific STS biomarkers have been generated, including 992 single-copy and 285 multi-copy STS markers [71]. A selection of conventional STS biomarkers targeting different AZF microdeletions are listed in Figure 1D. Moreover, multiplex STS-based PCR microdeletion analysis using multiple STS markers in each AZF sub-region can detect over 95% of deletions [56].
Interestingly, Bor et al. combined fluorescent multiplex PCR with capillary electrophoresis and successfully screened for Y chromosome microdeletions in 49 azoospermic and 149 severe oligozoospermic men [72]. They concluded that the application of capillary electrophoresis to the detection of PCR products provides a useful, semi-automated and high throughput method for rapidly screening YCMs [72]; however, this approach has rarely been used to screen for YCMs in infertile patients.
Because of the rapid advancements in molecular diagnostic techniques, it has become necessary to generate a new hybridization method for the rapid and efficient detection and analysis of large quantities of genetic data. Since chemical assays utilize a solid phase that often produces high background signals and can adversely affect reaction kinetics by immobilizing a reactant [73], suspension array technology (SAT) was developed by the Luminex Corporation (Austin, TX, USA) to eliminate the need for a solid surface during clinical testing. With this technique, oligonucleotide probes are allowed to hybridize with microsphere beads bearing a unique fluorescent label. Each microsphere set is internally labeled with two spectral fluorochromes of different intensities. The unique spectral emission is recognized by a red laser, while the biotinylated amplicon bound to the surface of the microsphere is read by a green laser that quantifies the fluorescence of the reporter molecule (streptavidin R-phycoerythrin) [4,11,12]. This technology can simultaneously analyze 100 analytes by combining 100 different sets of microspheres in a single reaction. SAT has been increasingly used to detect YCMs in infertile patients [11,12]. The technique is effective, sensitive, rapid, reproducible and easily performed. However, there are a few disadvantages associated with the SAT technique, including low array size, hybridization problems and difficulties optimizing a single specific annealing temperature for the entire experiment.
Array-comparative genomic hybridization (aCGH) was developed to analyze submicroscopic Y deletions [74,75]. Yuen et al. successfully used Agilent’s Human Genome 8 × 15 K array aCGH format as a template to create a custom Y chromosome aCGH to detect microdeletion prevalence in male infertility. They concluded that their aCGH approach was a reliable, high-resolution alternative to multiplex PCR screening of pathogenic YCMs in male infertility [75]; however, additional studies are needed to comprehensively evaluate this state-of-the-art technique.
Transmission characteristics of YCMs
Men with Y deletions are generally infertile and, therefore, deletions cannot be transmitted to their offspring. In recent decades, the ICSI and TESE techniques have successfully helped men with azoospermia or severe oligozoospermia reproduce. However, these techniques greatly increase the risk of YCM transmission from infertile fathers to their male offspring. Although many studies have reported the transmission of AZF microdeletions from father to son, the data from these studies have rarely been summarized.
Prevalence of AZF microdeletion transmission
AZF microdeletions are inherited through the paternal germ line or occur as de novo events. It has been reported by many studies that more than 80% of AZF microdeletions are of de novo origin [14,76]. Most deletions occur during the pre-fertilization stage, while some deletions are post-fertilization events [77]. If a sperm with YCMs fertilizes an egg, it will transmit the YCMs to the male child. On the other hand, if the deletion occurs as a post fertilization event, it may cause mosaicism characterized by normal Y chromosomes in leukocytes and Y chromosomes with the post fertilization deletion in sperm or testicular DNA [4,77]. Since infertility is the major phenotype of men with AZF microdeletions, the natural transmission of AZF microdeletions has rarely been reported [16,76]. Dai et al. reported that YCMs in 7 of 10 infertile men were naturally transmitted from father to son [76]. Samli et al. also observed the natural transmission of an AZFb microdeletion from a father to each of his three sons [14]. In contrast, vertical transmission of AZF microdeletions from father to son through ICSI has been widely reported [78,79]. In 1996, Kent-First et al. described the first study of the sons produced by a population of 32 couples with infertile fathers who received reproductive assistance via ICSI [80]. They found that the incidence of microdeletions in the ICSI population was about 9.4%, which was close to the incidence of AZF microdeletions in infertile men.
ART have been associated with elevated incidences of sexual chromosomal aberrations, de novo chromosomal abnormalities and sperm aneuploidy [81]. Thus, one of the major concerns regarding ART is whether ART cause AZF microdeletions. A new study by Liu et al. compared the YCM occurrence in 19 candidate genes from 199 fathers and their 228 sons (Chinese, Han ethnicity) that were conceived by IVF (85 sons), ICSI (73 sons) or natural conception (70 sons). They observed that the YCM incidences of the fathers for IVF, ICSI and naturally conceived sons was 10.7%, 3.2% and 8.2%, respectively [69]. They identified one de novo YCM among the 70 naturally conceived offspring but none among the 158 ART conceived offspring. There were no statistically significant differences in incidence among the three groups or of de novo YCMs between the naturally conceived and ART conceived sons [69]. Therefore, they finally concluded that ART does not significantly increase the risk of YCM in male offspring [69]. However, this conclusion remains controversial; the correlation between the incidence of AZF microdeletions and ART needs to be verified in a large, ethnically and geographically diverse cohort of infertile men and their offspring.
Expansion of AZF microdeletions in the offspring
A significant amount of research in recent years has focused on investigating the genetic changes in offspring conceived by ICSI. Some studies have reported that ICSI can only vertically transmit YCMs without the expansion or de novo occurrence of YCMs [78,82]. Rolf et al. reported a case of what was probably an identical, partial deletion of the distal part of the AZFb region over three generations [83]. A study by Minor et al. also identified an identical and partial AZFc gr/gr deletion that was vertically transmitted over three generations via fathers receiving reproductive assistance through ICSI [84].
However, a growing number of studies have reported that YCM can be transmitted vertically from father to son via ICSI and that ICSI can contribute to YCM expansions as well as de novo YCM [79,80,85]. Dai et al. examined the expansion of AZF deletions in 10 father-son pairs, and found expansion microdeletions (S1/F1, S2/F2, S6/F6, S7/F7, S8/F8, S9/F9 and S10/F10) in seven father-son pairs and de novo microdeletions (S3/F3, S4/F4 and S5/F5) in the three remaining father-son pairs [76]. Samli et al. reported an unusual family in which an azoospermic patient (proband) and three brothers inherited a Yq microdeletion from their father through spontaneous pregnancy [14]. The brothers and their father all carried a Yq microdeletion in the AZFb subregion involving the RBM1 loci. Additionally, an uncle carried a different deletion in the AZFc region (sY1539). The proband and one of his brothers shared an identical deletion with their father, plus additional de novo deletions in the AZFa and AZFb subregions [14].
Although Liu et al. claimed that ART is not associated with the expansion or occurrence of de novo microdeletions [69], a significant number of studies have demonstrated that AZF microdeletions are capable of transmitting themselves and expanding over the generations through natural pregnancy or ART [79,80,85].
Perspectives
Human spermatogenesis is a complex process involving a series of coordinated events regulated and governed by key genes located in the AZF regions of the Y chromosome. Evidence indicates that DBY in AZFa; KDM5D and RBMY1A1 in AZFb; and DAZ and CDY in AZFb/c are key determinants in spermatogenesis [3,4]. Mutations in the AZFc region are the most common genetic cause of male infertility, and the DAZ gene is deleted in most cases involving AZFc deletions.
Despite the tremendous breakthroughs that have been achieved in recent decades, the functions of the AZF genes are still not well understood. The lack of easily accessible animal models and in vitro spermatogenic cell lines are the major obstacles investigating AZF gene functions in normal spermatogenesis. Additionally, the biological properties of the AZF regions appear to be quite complicated. This complexity is illustrated by the tremendous variability associated with the AZFb and AZFc sequences and the intricate regulation of their corresponding genetic determinants. Therefore, future research should focus on fully sequencing AZF diversity across the Y chromosome and pursuing more in-depth functional characterization of the AZF genes.
Genomic sequencing of AZF has clearly subdivided AZF into the AZFa, AZFb and AZFc regions, and may have identified a fourth proposed AZFd region. Homologous recombination is commonly accepted as the major cause of most AZF microdeletions; however, deletions via non-homologous recombination have also been reported. Patterns of AZF microdeletions have been verified by many specific STS markers. STS-PCR has become the gold-standard method for clinically screening AZF microdeletions, but this method is not appropriate for large sample sizes or identifying novel AZF microdeletions. New state-of-the-art techniques like SAT and aCGH have partially rescued the limitations of STS-PCR techniques; however, the efficacy of these approaches needs to be verified in future studies. Because the identification of novel AZF molecular microdeletions and their associated phenotypes is of great importance to the clinical screening of infertile patients, improvement of the current methods or generation of new approaches for laboratory and clinical screening of AZF microdeletions in large numbers of samples is greatly needed.
Although the prevalence of AZF microdeletions has been extensively studied, the results from various geographically and ethnically distinct populations are inconsistent and even controversial. Based on the available evidence, there are some clearly established points. First, AZF microdeletions are common in men with azoospermia and severe oligozoospermia, as well as in the offspring of these men conceived by ART treatment. Second, although most AZF microdeletions are of de novo origin, AZF microdeletions can be vertically transmitted to offspring, and vertically transmitted AZF microdeletions can be transmitted identically or with expansions through both natural pregnancy and ART treatment. Finally, because this information has diagnostic, prognostic, preventive and ethical value that benefits infertile couples, it is critical to perform screening for specific AZF deletions and other chromosomal abnormalities in infertile males. nuscript.
Acknowledgements
The work was supported by Jilin Province Science and Technology Department Project of Natural Science Foundation (20150101136JC). We thank Medjaden Bioscience Limited for assisting in the preparation of this manuscript.
Disclosure of conflict of interest
None.
References
- 1.Bushnik T, Cook JL, Yuzpe AA, Tough S, Collins J. Estimating the prevalence of infertility in Canada. Hum Reprod. 2012;27:738–746. doi: 10.1093/humrep/der465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang YS, Dai RL, Wang RX, Zhang HG, Chen S, Liu RZ. Analysis of Y chromosome microdeletion in 1738 infertile men from northeastern China. Urology. 2013;82:584–588. doi: 10.1016/j.urology.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 3.Navarro-Costa P, Plancha CE, Goncalves J. Genetic dissection of the AZF regions of the human Y chromosome: thriller or filler for male (in) fertility? J Biomed Biotechnol. 2010;2010:936569. doi: 10.1155/2010/936569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suganthi R, Vijesh VV, Vandana N, Fathima Ali Benazir J. Y choromosomal microdeletion screening in the workup of male infertility and its current status in India. Int J Fertil Steril. 2014;7:253–266. [PMC free article] [PubMed] [Google Scholar]
- 5.Poongothai J, Gopenath TS, Manonayaki S. Genetics of human male infertility. Singapore Med J. 2009;50:336–347. [PubMed] [Google Scholar]
- 6.Zhang F, Li L, Wang L, Yang L, Liang Z, Li J, Jin F, Tian Y. Clinical characteristics and treatment of azoospermia and severe oligospermia patients with Y-chromosome microdeletions. Mol Reprod Dev. 2013;80:908–915. doi: 10.1002/mrd.22226. [DOI] [PubMed] [Google Scholar]
- 7.Vineeth VS, Malini SS. A Journey on Y Chromosomal Genes and Male Infertility. Int J Hum Genet. 2011;11:203–215. [Google Scholar]
- 8.Kent-First M, Muallem A, Shultz J, Pryor J, Roberts K, Nolten W, Meisner L, Chandley A, Gouchy G, Jorgensen L, Havighurst T, Grosch J. Defining regions of the Y-chromosome responsible for male infertility and identification of a fourth AZF region (AZFd) by Y-chromosome microdeletion detection. Mol Reprod Dev. 1999;53:27–41. doi: 10.1002/(SICI)1098-2795(199905)53:1<27::AID-MRD4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 9.Al-Achkar W, Wafa A, Moassass F. Cytogenetic abnormalities and Y-chromosome microdeletions in infertile Syrian males. Biomed Rep. 2013;1:275–279. doi: 10.3892/br.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambulkar PS, Sigh R, Reddy M, Varma PS, Gupta DO, Shende MR, Pal AK. Genetic Risk of Azoospermia Factor (AZF) Microdeletions in Idiopathic Cases of Azoospermia and Oligozoospermia in Central Indian Population. J Clin Diagn Res. 2014;8:88–91. doi: 10.7860/JCDR/2014/7680.4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun K, Chen XF, Zhu XB, Hu HL, Zhang W, Shao FM, Li P, Miao QL, Huang YR, Li Z. A new molecular diagnostic approach to assess Y chromosome microdeletions in infertile men. J Int Med Res. 2012;40:237–248. doi: 10.1177/147323001204000124. [DOI] [PubMed] [Google Scholar]
- 12.Zhu YJ, Liu SY, Wang H, Wei P, Ding XP. The prevalence of azoospermia factor microdeletion on the Y chromosome of Chinese infertile men detected by multi-analyte suspension array technology. Asian J Androl. 2008;10:873–881. doi: 10.1111/j.1745-7262.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- 13.Kamischke A, Gromoll J, Simoni M, Behre HM, Nieschlag E. Transmission of a Y chromosomal deletion involving the deleted in azoospermia (DAZ) and chromodomain (CDY1) genes from father to son through intracytoplasmic sperm injection: case report. Hum Reprod. 1999;14:2320–2322. doi: 10.1093/humrep/14.9.2320. [DOI] [PubMed] [Google Scholar]
- 14.Samli H, Murat Samli M, Solak M. Natural transmission of AZFb Y-chromosomal microdeletion from father to his three sons. Arch Androl. 2006;52:423–426. doi: 10.1080/01485010600822655. [DOI] [PubMed] [Google Scholar]
- 15.Krausz C, McElreavey K. Y chromosome and male infertility. Front Biosci. 1999;4:E1–8. doi: 10.2741/krausz. [DOI] [PubMed] [Google Scholar]
- 16.Lahn BT, Pearson NM, Jegalian K. The human Y chromosome, in the light of evolution. Nat Rev Genet. 2001;2:207–216. doi: 10.1038/35056058. [DOI] [PubMed] [Google Scholar]
- 17.Bachtrog D, Charlesworth B. Towards a complete sequence of the human Y chromosome. Genome Biol. 2001;2:REVIEWS1016. doi: 10.1186/gb-2001-2-5-reviews1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou SF, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang SP, Waterston RH, Wilson RK, Rozen S, Page DC. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 19.Ferlin A, Moro E, Rossi A, Dallapiccola B, Foresta C. The human Y chromosome’s azoospermia factor b (AZFb) region: sequence, structure, and deletion analysis in infertile men. J Med Genet. 2003;40:18–24. doi: 10.1136/jmg.40.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- 21.Foresta C, Moro E, Ferlin A. Y chromosome microdeletions and alterations of spermatogenesis. Endocr Rev. 2001;22:226–239. doi: 10.1210/edrv.22.2.0425. [DOI] [PubMed] [Google Scholar]
- 22.Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, Kohn FM, Schill WB, Farah S, Ramos C, Hartmann M, Hartschuh W, Meschede D, Behre HM, Castel A, Nieschlag E, Weidner W, Grone HJ, Jung A, Engel W, Haidl G. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet. 1996;5:933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- 23.Sun C, Skaletsky H, Birren B, Devon K, Tang Z, Silber S, Oates R, Page DC. An azoospermic man with a de novo point mutation in the Y-chromosomal gene USP9Y. Nat Genet. 1999;23:429–432. doi: 10.1038/70539. [DOI] [PubMed] [Google Scholar]
- 24.Sargent CA, Boucher CA, Kirsch S, Brown G, Weiss B, Trundley A, Burgoyne P, Saut N, Durand C, Levy N, Terriou P, Hargreave T, Cooke H, Mitchell M, Rappold GA, Affara NA. The critical region of overlap defining the AZFa male infertility interval of proximal Yq contains three transcribed sequences. J Med Genet. 1999;36:670–677. [PMC free article] [PubMed] [Google Scholar]
- 25.Luddi A, Margollicci M, Gambera L, Serafini F, Cioni M, De Leo V, Balestri P, Piomboni P. Spermatogenesis in a man with complete deletion of USP9Y. N Engl J Med. 2009;360:881–885. doi: 10.1056/NEJMoa0806218. [DOI] [PubMed] [Google Scholar]
- 26.Foresta C, Ferlin A, Moro E. Deletion and expression analysis of AZFa genes on the human Y chromosome revealed a major role for DBY in male infertility. Hum Mol Genet. 2000;9:1161–1169. doi: 10.1093/hmg/9.8.1161. [DOI] [PubMed] [Google Scholar]
- 27.Ditton HJ, Zimmer J, Kamp C, Rajpert-De Meyts E, Vogt PH. The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum Mol Genet. 2004;13:2333–2341. doi: 10.1093/hmg/ddh240. [DOI] [PubMed] [Google Scholar]
- 28.Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, Steiner K, Tam PP, Monaco AP, Willard HF, Koopman P. The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet. 1998;7:737–742. doi: 10.1093/hmg/7.4.737. [DOI] [PubMed] [Google Scholar]
- 29.Repping S, Skaletsky H, Lange J, Silber S, Van Der Veen F, Oates RD, Page DC, Rozen S. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet. 2002;71:906–922. doi: 10.1086/342928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akimoto C, Kitagawa H, Matsumoto T, Kato S. Spermatogenesis-specific association of SMCY and MSH5. Genes Cells. 2008;13:623–633. doi: 10.1111/j.1365-2443.2008.01193.x. [DOI] [PubMed] [Google Scholar]
- 31.Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell. 2007;128:877–887. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Roll-Mecak A, Shin BS, Dever TE, Burley SK. Engaging the ribosome: universal IFs of translation. Trends Biochem Sci. 2001;26:705–709. doi: 10.1016/s0968-0004(01)02024-2. [DOI] [PubMed] [Google Scholar]
- 33.Andres O, Kellermann T, Lopez-Giraldez F, Rozas J, Domingo-Roura X, Bosch M. RPS4Y gene family evolution in primates. BMC Evol Biol. 2008;8:142. doi: 10.1186/1471-2148-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu VW, Gauthier C, St-Arnaud R. Inhibition of ATF4 transcriptional activity by FIAT/gammataxilin modulates bone mass accrual. Ann N Y Acad Sci. 2006;1068:131–142. doi: 10.1196/annals.1346.027. [DOI] [PubMed] [Google Scholar]
- 35.Ho M, Chelly J, Carter N, Danek A, Crocker P, Monaco AP. Isolation of the gene for McLeod syndrome that encodes a novel membrane transport protein. Cell. 1994;77:869–880. doi: 10.1016/0092-8674(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 36.Tessari A, Salata E, Ferlin A, Bartoloni L, Slongo ML, Foresta C. Characterization of HSFY, a novel AZFb gene on the Y chromosome with a possible role in human spermatogenesis. Mol Hum Reprod. 2004;10:253–258. doi: 10.1093/molehr/gah036. [DOI] [PubMed] [Google Scholar]
- 37.Shinka T, Sato Y, Chen G, Naroda T, Kinoshita K, Unemi Y, Tsuji K, Toida K, Iwamoto T, Nakahori Y. Molecular characterization of heat shock-like factor encoded on the human Y chromosome, and implications for male infertility. Biol Reprod. 2004;71:297–306. doi: 10.1095/biolreprod.103.023580. [DOI] [PubMed] [Google Scholar]
- 38.Kinoshita K, Shinka T, Sato Y, Kurahashi H, Kowa H, Chen G, Umeno M, Toida K, Kiyokage E, Nakano T, Ito S, Nakahori Y. Expression analysis of a mouse orthologue of HSFY, a candidate for the azoospermic factor on the human Y chromosome. J Med Invest. 2006;53:117–122. doi: 10.2152/jmi.53.117. [DOI] [PubMed] [Google Scholar]
- 39.Sato Y, Yoshida K, Shinka T, Nozawa S, Nakahori Y, Iwamoto T. Altered expression pattern of heat shock transcription factor, Y chromosome (HSFY) may be related to altered differentiation of spermatogenic cells in testes with deteriorated spermatogenesis. Fertil Steril. 2006;86:612–618. doi: 10.1016/j.fertnstert.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 40.Vinci G, Raicu F, Popa L, Popa O, Cocos R, McElreavey K. A deletion of a novel heat shock gene on the Y chromosome associated with azoospermia. Mol Hum Reprod. 2005;11:295–298. doi: 10.1093/molehr/gah153. [DOI] [PubMed] [Google Scholar]
- 41.Stouffs K, Lissens W, Verheyen G, Van Landuyt L, Goossens A, Tournaye H, Van Steirteghem A, Liebaers I. Expression pattern of the Y-linked PRY gene suggests a function in apoptosis but not in spermatogenesis. Mol Hum Reprod. 2004;10:15–21. doi: 10.1093/molehr/gah010. [DOI] [PubMed] [Google Scholar]
- 42.Stouffs K, Lissens W, Van Landuyt L, Tournaye H, Van Steirteghem A, Liebaers I. Characterization of the genomic organization, localization and expression of four PRY genes (PRY1, PRY2, PRY3 and PRY4) Mol Hum Reprod. 2001;7:603–610. doi: 10.1093/molehr/7.7.603. [DOI] [PubMed] [Google Scholar]
- 43.Elliott DJ, Millar MR, Oghene K, Ross A, Kiesewetter F, Pryor J, McIntyre M, Hargreave TB, Saunders PT, Vogt PH, Chandley AC, Cooke H. Expression of RBM in the nuclei of human germ cells is dependent on a critical region of the Y chromosome long arm. Proc Natl Acad Sci U S A. 1997;94:3848–3853. doi: 10.1073/pnas.94.8.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elliott DJ. RBMY genes and AZFb deletions. J Endocrinol Invest. 2000;23:652–658. doi: 10.1007/BF03343789. [DOI] [PubMed] [Google Scholar]
- 45.Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S, Page DC. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- 46.Ravel C, Chantot-Bastaraud S, El Houate B, Rouba H, Legendre M, Lorenco D, Mandelbaum J, Siffroi JP, McElreavey K. Y-chromosome AZFc structural architecture and relationship to male fertility. Fertil Steril. 2009;92:1924–1933. doi: 10.1016/j.fertnstert.2008.08.135. [DOI] [PubMed] [Google Scholar]
- 47.Huynh T, Mollard R, Trounson A. Selected genetic factors associated with male infertility. Hum Reprod Update. 2002;8:183–198. doi: 10.1093/humupd/8.2.183. [DOI] [PubMed] [Google Scholar]
- 48.Habermann B, Mi HF, Edelmann A, Bohring C, Backert IT, Kiesewetter F, Aumuller G, Vogt PH. DAZ (Deleted in AZoospermia) genes encode proteins located in human late spermatids and in sperm tails. Hum Reprod. 1998;13:363–369. doi: 10.1093/humrep/13.2.363. [DOI] [PubMed] [Google Scholar]
- 49.Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- 50.Foresta C, Ferlin A, Garolla A, Rossato M, Barbaux S, De Bortoli A. Y-chromosome deletions in idiopathic severe testiculopathies. J Clin Endocrinol Metab. 1997;82:1075–1080. doi: 10.1210/jcem.82.4.3798. [DOI] [PubMed] [Google Scholar]
- 51.Stuppia L, Gatta V, Calabrese G, Guanciali Franchi P, Morizio E, Bombieri C, Mingarelli R, Sforza V, Frajese G, Tenaglia R, Palka G. A quarter of men with idiopathic oligo-azoospermia display chromosomal abnormalities and microdeletions of different types in interval 6 of Yq11. Hum Genet. 1998;102:566–570. doi: 10.1007/s004390050741. [DOI] [PubMed] [Google Scholar]
- 52.Wong EY, Tse JY, Yao KM, Lui VC, Tam PC, Yeung WS. Identification and characterization of human VCY2-interacting protein: VCY2IP-1, a microtubule-associated protein-like protein. Biol Reprod. 2004;70:775–784. doi: 10.1095/biolreprod.103.018531. [DOI] [PubMed] [Google Scholar]
- 53.Tse JY, Wong EY, Cheung AN, O WS, Tam PC, Yeung WS. Specific expression of VCY2 in human male germ cells and its involvement in the pathogenesis of male infertility. Biol Reprod. 2003;69:746–751. doi: 10.1095/biolreprod.103.015792. [DOI] [PubMed] [Google Scholar]
- 54.Vogt PH. Azoospermia factor (AZF) in Yq11: towards a molecular understanding of its function for human male fertility and spermatogenesis. Reprod Biomed Online. 2005;10:81–93. doi: 10.1016/s1472-6483(10)60807-3. [DOI] [PubMed] [Google Scholar]
- 55.Yen PH. A long-range restriction map of deletion interval 6 of the human Y chromosome: a region frequently deleted in azoospermic males. Genomics. 1998;54:5–12. doi: 10.1006/geno.1998.5526. [DOI] [PubMed] [Google Scholar]
- 56.Simoni M, Bakker E, Krausz C. EAA/EMQN best practice guidelines for molecular diagnosis of y-chromosomal microdeletions. State of the art 2004. Int J Androl. 2004;27:240–249. doi: 10.1111/j.1365-2605.2004.00495.x. [DOI] [PubMed] [Google Scholar]
- 57.Muslumanoglu MH, Turgut M, Cilingir O, Can C, Ozyurek Y, Artan S. Role of the AZFd locus in spermatogenesis. Fertil Steril. 2005;84:519–522. doi: 10.1016/j.fertnstert.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 58.Balkan M, Tekes S, Gedik A. Cytogenetic and Y chromosome microdeletion screening studies in infertile males with Oligozoospermia and Azoospermia in Southeast Turkey. J Assist Reprod Genet. 2008;25:559–565. doi: 10.1007/s10815-008-9272-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hussein AA, Vasudevan R, Patimah I, Prashant N, Nora FA. Association of azoospermia factor region deletions in infertile male subjects among Malaysians. Andrologia. 2015;47:168–177. doi: 10.1111/and.12240. [DOI] [PubMed] [Google Scholar]
- 60.Kamp C, Hirschmann P, Voss H, Huellen K, Vogt PH. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum Mol Genet. 2000;9:2563–2572. doi: 10.1093/hmg/9.17.2563. [DOI] [PubMed] [Google Scholar]
- 61.Wan L, Cai ZM. [Partial deletions in the AZFc region of the Y chromosome are associated with male infertility] . Zhonghua Nan Ke Xue. 2009;15:165–169. [PubMed] [Google Scholar]
- 62.Cram DS, Osborne E, McLachlan RI. Y chromosome microdeletions: implications for assisted conception. Med J Aust. 2006;185:433–434. doi: 10.5694/j.1326-5377.2006.tb00641.x. [DOI] [PubMed] [Google Scholar]
- 63.Sun C, Skaletsky H, Rozen S, Gromoll J, Nieschlag E, Oates R, Page DC. Deletion of azoospermia factor a (AZFa) region of human Y chromosome caused by recombination between HERV15 proviruses. Hum Mol Genet. 2000;9:2291–2296. doi: 10.1093/oxfordjournals.hmg.a018920. [DOI] [PubMed] [Google Scholar]
- 64.Totonchi M, Mohseni Meybodi A, Borjian Boroujeni P, Sedighi Gilani M, Almadani N, Gourabi H. Clinical data for 185 infertile Iranian men with Y-chromosome microdeletion. J Assist Reprod Genet. 2012;29:847–853. doi: 10.1007/s10815-012-9798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karaer A, Karaer K, Ozaksit G, Ceylaner S, Percin EF. Y chromosome azoospermia factor region microdeletions and recurrent pregnancy loss. Am J Obstet Gynecol. 2008;199:662, e1–5. doi: 10.1016/j.ajog.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 66.Hopps CV, Mielnik A, Goldstein M, Palermo GD, Rosenwaks Z, Schlegel PN. Detection of sperm in men with Y chromosome microdeletions of the AZFa, AZFb and AZFc regions. Hum Reprod. 2003;18:1660–1665. doi: 10.1093/humrep/deg348. [DOI] [PubMed] [Google Scholar]
- 67.Brown GM, Furlong RA, Sargent CA, Erickson RP, Longepied G, Mitchell M, Jones MH, Hargreave TB, Cooke HJ, Affara NA. Characterisation of the coding sequence and fine mapping of the human DFFRY gene and comparative expression analysis and mapping to the Sxrb interval of the mouse Y chromosome of the Dffry gene. Hum Mol Genet. 1998;7:97–107. doi: 10.1093/hmg/7.1.97. [DOI] [PubMed] [Google Scholar]
- 68.Ferlin A, Arredi B, Speltra E, Cazzadore C, Selice R, Garolla A, Lenzi A, Foresta C. Molecular and clinical characterization of Y chromosome microdeletions in infertile men: a 10-year experience in Italy. J Clin Endocrinol Metab. 2007;92:762–770. doi: 10.1210/jc.2006-1981. [DOI] [PubMed] [Google Scholar]
- 69.Liu XH, Yan LY, Lu CL, Li R, Zhu XH, Jin HY, Zhang Y, Zhang WX, Gao SH, Qiao J. ART do not increase the risk of Y-chromosome microdeletion in 19 candidate genes at AZF regions. Reprod Fertil Dev. 2014;26:778–786. doi: 10.1071/RD13092. [DOI] [PubMed] [Google Scholar]
- 70.Dada R, Kumar R, Shamsi MB, Kumar R, Kucheria K, Sharma RK, Gupta SK, Gupta NP. Higher frequency of Yq microdeletions in sperm DNA as compared to DNA isolated from blood. Asian J Androl. 2007;9:720–722. doi: 10.1111/j.1745-7262.2007.00274.x. [DOI] [PubMed] [Google Scholar]
- 71.Lange J, Skaletsky H, Bell GW, Page DC. MSY Breakpoint Mapper, a database of sequence-tagged sites useful in defining naturally occurring deletions in the human Y chromosome. Nucleic Acids Res. 2008;36:D809–814. doi: 10.1093/nar/gkm849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bor P, Hindkjaer J, Kolvraa S, Ingerslev HJ. A new approach for screening for Y microdeletions: capillary electrophoresis combined with fluorescent multiplex PCR. J Assist Reprod Genet. 2003;20:46–51. doi: 10.1023/A:1021215006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peterson AW, Wolf LK, Georgiadis RM. Hybridization of mismatched or partially matched DNA at surfaces. J Am Chem Soc. 2002;124:14601–14607. doi: 10.1021/ja0279996. [DOI] [PubMed] [Google Scholar]
- 74.Patrick DF, Maher E, Sharkey F, Wilkie N. Application of array-CGH for the detection of submicroscopic chromosomal Imbalances in 400 cases of children with idiopathic mental retardation and congenital malformations [Google Scholar]
- 75.Yuen RK, Merkoulovitch A, MacDonald JR, Vlasschaert M, Lo K, Grober E, Marshall CR, Jarvi KA, Kolomietz E, Scherer SW. Development of a high-resolution Y-chromosome microarray for improved male infertility diagnosis. Fertil Steril. 2014;101:1079–1085. e3. doi: 10.1016/j.fertnstert.2013.12.027. [DOI] [PubMed] [Google Scholar]
- 76.Dai RL, Sun LK, Yang X, Li LL, Zhu HB, Liu RZ. Expansion and de novo occurrence of Y chromosome microdeletions occurring via natural vertical transmission in northeastern China. J Int Med Res. 2012;40:1182–1191. doi: 10.1177/147323001204000339. [DOI] [PubMed] [Google Scholar]
- 77.Edwards RG, Bishop CE. On the origin and frequency of Y chromosome deletions responsible for severe male infertility. Mol Hum Reprod. 1997;3:549–554. doi: 10.1093/molehr/3.7.549. [DOI] [PubMed] [Google Scholar]
- 78.Cram DS, Ma K, Bhasin S, Arias J, Pandjaitan M, Chu B, Audrins MS, Saunders D, Quinn F, deKretser D, McLachlan R. Y chromosome analysis of infertile men and their sons conceived through intracytoplasmic sperm injection: vertical transmission of deletions and rarity of de novo deletions. Fertil Steril. 2000;74:909–915. doi: 10.1016/s0015-0282(00)01568-5. [DOI] [PubMed] [Google Scholar]
- 79.Page DC, Silber S, Brown LG. Men with infertility caused by AZFc deletion can produce sons by intracytoplasmic sperm injection, but are likely to transmit the deletion and infertility. Hum Reprod. 1999;14:1722–1726. doi: 10.1093/humrep/14.7.1722. [DOI] [PubMed] [Google Scholar]
- 80.Kent-First MG, Kol S, Muallem A, Ofir R, Manor D, Blazer S, First N, Itskovitz-Eldor J. The incidence and possible relevance of Y-linked microdeletions in babies born after intracytoplasmic sperm injection and their infertile fathers. Mol Hum Reprod. 1996;2:943–950. doi: 10.1093/molehr/2.12.943. [DOI] [PubMed] [Google Scholar]
- 81.Bonduelle M, Camus M, De Vos A, Staessen C, Tournaye H, Van Assche E, Verheyen G, Devroey P, Liebaers I, Van Steirteghem A. Seven years of intracytoplasmic sperm injection and follow-up of 1987 subsequent children. Hum Reprod. 1999;14(Suppl 1):243–264. doi: 10.1093/humrep/14.suppl_1.243. [DOI] [PubMed] [Google Scholar]
- 82.Buch B, Galan JJ, Lara M, Real LM, Martinez-Moya M, Ruiz A. Absence of de novo Y-chromosome microdeletions in male children conceived through intracytoplasmic sperm injection. Fertil Steril. 2004;82:1679–1680. doi: 10.1016/j.fertnstert.2004.06.039. [DOI] [PubMed] [Google Scholar]
- 83.Rolf C, Gromoll J, Simoni M, Nieschlag E. Natural transmission of a partial AZFb deletion of the Y chromosome over three generations: case report. Hum Reprod. 2002;17:2267–2271. doi: 10.1093/humrep/17.9.2267. [DOI] [PubMed] [Google Scholar]
- 84.Minor A, Wong EC, Harmer K, Ma S. Molecular and cytogenetic investigation of Y chromosome deletions over three generations facilitated by intracytoplasmic sperm injection. Prenat Diagn. 2007;27:743–747. doi: 10.1002/pd.1772. [DOI] [PubMed] [Google Scholar]
- 85.Komori S, Kato H, Kobayashi S, Koyama K, Isojima S. Transmission of Y chromosomal microdeletions from father to son through intracytoplasmic sperm injection. J Hum Genet. 2002;47:465–468. doi: 10.1007/s100380200066. [DOI] [PubMed] [Google Scholar]


