Abstract
Background: Clinical significance of Notch1 activation in gastric cancer has been elucidated in our previous study, but the role of its ligands remains obscure. This study aims to evaluate the prognostic value of Jagged1 expression in patients with gastric cancer. Methods: We examined Jagged1 expression in tumor and nontumor tissues in retrospectively enrolled 302 patients with gastric cancer undergoing gastrectomy at Zhongshan Hospital of Fudan University in 2008 by immunohistochemical staining. The Kaplan-Meier method and Cox regression models were used to evaluate the prognostic value of Jagged1 expression and its association with clinicopathological features. We created a predictive nomogram by integrating Jagged1 expression with the TNM staging system for overall survival of gastric cancer patients. Results: Jagged1 expression in gastric cancer was decreased compared with that in nontumor tissues. Low expression of Jagged1 in tumor and nontumor both predicted a dismal outcome. The Jagged1 risk derived from Jagged1 expression in tumor/nontumor tissue gave a further discrimination for the prognosis of gastric cancer patients. By Cox multivariate analysis, the Jagged1 risk was defined as an independent prognostic factor. The generated nomogram performed well in predicting the 3- and 5-year overall survival of gastric cancer patients. Conclusion: Jagged1 is a potential prognostic biomarker for overall survival, which could be integrated with TNM stage to give a better risk stratification for gastric cancer patients.
Keywords: Gastric cancer, Jagged1, overall survival, prognosis, biomarker, nomogram
Introduction
Although the incidence and mortality rates have been dramatically decreased since 1930s, gastric cancer remains the fourth most common malignant neoplasm worldwide and accounts for the second cancer-related death in both sex. The incidence of gastric cancer in China and some East Asia countries is much higher than that in America or Europe countries [1]. Furthermore, due to atypical symptoms at the early stage, over 80% of patients with gastric cancer were diagnosed at an advanced stage, which usually indicates a poor prognosis [2]. Thus, elucidation of key molecules in carcinogenesis of gastric cancer and targeting the underlying mechanisms may provide novel therapeutic targets for gastric cancer.
Notch signaling is an evolutionally conserved cell interaction mechanism, which could play crucial roles in diverse cellular processes including cell-fate determination, regulation of apoptosis and self-renewal of stem cells [3]. Four Notch molecules (Notch1 to Notch4) were identified as receptors for five ligands (Jagged1, Jagged2, DLL1, DLL3 and DLL4) in mammals. Aberrant expression of Notch1 has been identified correlated with the prognosis of patients in some malignant tumors. Being the main ligand for Notch1, aberrant expression of Jagged1 was found to be related to a poor prognosis in human renal clear cell carcinoma, breast cancer and papillary bladder transitional cell carcinoma [4-6]. Activated Notch1, accomplished by up-regulated Jagged1 was also found critical in development of colorectal cancer [7]. However, the expression of Jagged1 and its clinical significances in gastric cancer remain largely unknown and need to be further studied.
Since Dufraine et al first discovered that activated Notch1 mutations in human T cell could lead to acute lymphoblastic leukemia, increasing studies have unraveled the role of Notch signaling in human malignant tumors [8-10]. Given the critical role of Notch signal in carcinogenesis, it seems to be a potential therapeutic target for the treatment of malignant tumor. Previous studies have identified gamma-secretase inhibitors, which could prevent the generation of the intracellular domain of Notch molecules by targeting Notch receptors to suppress Notch activity [11]. However, targeting the binding of the ligands to Notch receptors could also have therapeutic value for blocking the activation of Notch signaling in malignant tumor.
Previous studies have proved that the activation of Notch1 signaling in gastric cancer was related to rapid disease progression, and high expression of intracellular domain of Notch1 (NICD), which represented activated Notch1 in gastric tumor tissue, predicted a poor clinical outcome in patients with gastric cancer [12,13]. Although DLL1 was reported to be involved in the activation of Notch1 [14], whether Jagged1 is involved in Notch1 activation in gastric cancer remains unknown.
In this study, we sought to investigate the expression of Jagged1 in gastric cancer and determine whether Jagged1 is responsible for Notch1 activation. Furthermore, we also explored its relation with the clinicopathological characteristics and clinical outcomes. In addition, a nomogram integrated Jagged1 expression with TNM stage was developed to predict the 3- and 5-year overall survival in patients with gastric cancer after surgery.
Materials and methods
Patients
A total of 302 patients who received gastrectomy from the same surgical team in Zhongshan Hospital of Fudan University (Shanghai, China) in 2008 were enrolled in the study. Six of these patients had distant metastasis, whose surgery was more focused on relieving symptoms while the others all received a radical (R0) resection. None of these patients received antitumor therapy before surgery. All the clinicopathological and baseline demographic characteristics were collected retrospectively, including age, gender, tumor size, tumor location, Lauren classification, tumor differentiation and tumor stage. Tumor stage and tumor differentiation were reassessed by two independent gastroenterology pathologists according to the 2010 International Union against Cancer TNM classification system. The average age of these patients is 59.5 years old with a range from 29 to 88, and 69.2% of these patients are male. Referred to Lauren classification, 63.9% are intestinal type, 22.8% are diffuse type, and the rest are mixed type. 63.9% of the patients have lymph nodes metastasis. All the patients were followed up until April 2014 with a median follow-up time of 45 months. Overall survival was defined as the time from the date of surgery to the date of death or last visit. Written informed consent from each patient was achieved. The use of human specimens was approved by the Clinical Research Ethics Committee of Zhongshan Hospital.
Tissue microarray, immunohistochemistry and evaluation
The construction of tissue microarray and the immunohistochemical protocols were as previously described [5,12]. Primary antibody was rabbit polyclonal anti-Jagged1 antibody (1.00 mg/ml, ab7771, Abcam, Cambridge, MA, USA). The density of positive staining was measured using the computerized image system composed of an Olympus CCD camera connected to a Nikon eclipse Ti-s microscope. The stained sections were scanned at high-power magnification (×200) and captured by NIS-Elements F3.2 software to identify three independent microscopic fields with the strongest staining to ensure representativeness and homogeneity. Each photo used an identical setting.
The density was counted by Image-Pro Plus version 6.0 software (Media Cybernetics Inc., Bethesda, MD, USA). Integrated optical density (IOD) of all the positive staining in each photograph was measured to give a quantitative assessment for the staining. The mean IOD of the three captured computerized microscopic fields was regarded as the density of Jagged1 expression in the represented tissue. The evaluation of immunostaining was performed by two independent gastroenterology pathologists who were blinded to the patient outcomes and clinicopathological characteristics. For immunohistochemical density, the cut-points for the definition of high/low expression subgroups were determined by X-tile software [15].
Statistical analysis
Analyses were performed with SPSS 19.0 (SPSS Inc., Chicago, IL) and R software version 3.0.2 and the “rms” package (R Foundation for Statistical Computing, Vienna, Austria). Pearson χ2 test or Fisher’s exact test was used to compare categorical variables Continuous variables were analyzed by one-way ANOVA. Kaplan-Meier analysis was used to determine the survival and Log-rank test was used to compare the patient survival between subgroups. Cox proportional hazards model was used to perform multivariate analysis. Nomogram was created by R software using “rms” package. Calibration plots were generated to examine the performance characteristics of the predictive nomogram. The Harrell’s concordance indices (c-indices) were used to measure the prognostic accuracy. All statistical analyses were two-sided and P<0.05 was regarded as statistically significant.
Results
Jagged1 expression in human gastric cancer
The positive staining of Jagged1 was seen on the membrane and/or in the cytoplasm of cells in both tumor and matched nontumor tissues (Figure 1A-D). By evaluating the integrated optical density (IOD) of each specimen, we found that IHC staining of Jagged1 varied greatly in the gastric cancer tissues and matched nontumor tissues. The measured IOD of IHC staining was 15.71±34.91 (median, 4.915; range, 0-401.58) in tumor tissue and 109.23±134.70 (median, 32.51; range, 0-627.22) in nontumor tissue respectively. The expression of Jagged1 in human gastric cancer tissue was decreased compared with that in nontumor tissue (P<0.001).
Figure 1.

Immunohistochemical analysis of Jagged1 expression in gastric cancer. A. High Jagged1 expression in nontumor tissue; B. Low Jagged1 expression in nontumor tissue; C. High Jagged1 expression in tumor tissue; D. Low Jagged1 expression in tumor tissue. Magnification 200×.
Association between clinicopathological characteristics and Jagged1 expression
The relationship between clinicopathological characteristics and Jagged1 expression is shown in Table 1. Jagged1 expression in tumor and nontumor tissue were both associated with tumor invasion depth (P=0.003 and P=0.004, respectively), lymph nodes metastasis (P=0.016 and P=0.004, respectively) and TNM stage (P=0.008 and P=0.011, respectively). Moreover, Jagged1 expression in nontumor tissue was also correlated with Lauren’s classification (P=0.048). To further discriminate patients with different prognosis, we combined Jagged1 expression features in tumor/nontumor tissue to generate four subgroups: Jagged1 tumor/nontumor both high subgroup, either high subgroups and both low subgroup. Significant association was found between the immunohistochemical feature and tumor invasion depth (P=0.002), lymph nodes metastasis (P=0.002), TNM stage (P=0.009). No association between Jagged1 expression and other clinicopathological characteristics was found.
Table 1.
Association between the clinicopathological characteristics and Jagged1 expression (n=302)
| Characteristics | Jagged1 expression in tumor tissue | Jagged1 expression in nontumor tissue | Jagged1 expression in tumor/nontumor tissue | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| High | Low | P value* | High | Low | P value | High/High | High/Low | Low/High | Low/Low | P value | |
| Age (years) | 0.385 | 0.086 | 0.218 | ||||||||
| Mean ± SD † | 58.3±11.3 | 59.8±11.6 | 57.3±11.1 | 60.1±11.6 | 54.6±11.2 | 60.2±11.0 | 58.5±10.9 | 60.1±11.7 | |||
| Gender | 0.377 | 0.736 | 0.751 | ||||||||
| Male | 36 | 173 | 44 | 165 | 12 | 24 | 32 | 141 | |||
| Female | 20 | 73 | 18 | 75 | 7 | 13 | 11 | 62 | |||
| Tumor size (cm) | 0.169 | 0.407 | 0.120 | ||||||||
| Mean ± SD | 3.41±2.46 | 3.83±1.98 | 3.56±2.16 | 3.80±2.05 | 3.91±3.28 | 3.14±1.91 | 3.40±1.45 | 3.92±2.06 | |||
| Lauren’s classification | 0.568 | 0.048 | 0.162 | ||||||||
| Intestinal | 39 | 154 | 32 | 161 | 10 | 29 | 22 | 132 | |||
| Diffuse | 10 | 59 | 21 | 48 | 6 | 4 | 15 | 44 | |||
| Mixed | 7 | 33 | 9 | 31 | 3 | 4 | 6 | 27 | |||
| Histology‡ | 0.103 | 0.982 | 0.124 | ||||||||
| Differentiated | 18 | 58 | 16 | 60 | 4 | 14 | 12 | 46 | |||
| Undifferentiated | 38 | 188 | 46 | 180 | 15 | 23 | 31 | 157 | |||
| Tumor depth | 0.003 | 0.004 | 0.002 | ||||||||
| T1+2 | 27 | 69 | 29 | 67 | 10 | 17 | 19 | 50 | |||
| T3+4 | 29 | 177 | 33 | 173 | 9 | 20 | 24 | 153 | |||
| Lymph nodes metastasis | 0.016 | 0.004 | 0.002 | ||||||||
| Absent | 28 | 81 | 32 | 77 | 12 | 16 | 20 | 61 | |||
| Present | 28 | 165 | 30 | 163 | 7 | 21 | 23 | 142 | |||
| Distant metastasis | 0.597 | 0.352 | 0.673 | ||||||||
| Absent | 56 | 240 | 62 | 234 | 19 | 37 | 43 | 197 | |||
| Present | 0 | 6 | 0 | 6 | 0 | 0 | 0 | 6 | |||
| pTNM stage | 0.008 | 0.011 | 0.009 | ||||||||
| I | 22 | 47 | 22 | 47 | 9 | 13 | 13 | 34 | |||
| II | 14 | 59 | 18 | 55 | 4 | 10 | 14 | 45 | |||
| III | 20 | 134 | 22 | 132 | 6 | 14 | 16 | 118 | |||
| IV | 0 | 6 | 0 | 6 | 0 | 0 | 0 | 6 | |||
χ2 test, Fisher’s exact test or one-way ANOVA was performed. P<0.05 was regarded as statistically significant.
SD: standard deviation.
Differentiated: tub1, tub2, pap; Undifferentiated: por1, por2, sig, muc.
Prognostic value of Jagged1 expression in human gastric cancer
To further explore the prognostic value of Jagged1 expression in gastric cancer, we applied Kaplan-Meier analysis to determine the overall survival according to Jagged1 expression. Log rank test was used to find the difference among the subgroups. Cut-points for high and low expression of Jagged1 were determined by X-tile software by minimum p value method. Low expression of Jagged1 in tumor tissue was associated with poor overall survival (Figure 2A, P<0.001) and low expression of Jagged1 in nontumor tissue was also found to be associated with poor overall survival (Figure 2B, P<0.001).
Figure 2.

Kaplan-Meier analysis for overall survival of patients with gastric cancer according to Jagged1 expression. Kaplan-Meier analysis for overall survival according to (A) expression of Jagged1 in tumor tissue, (B) expression of Jagged1 in nontumor tissue, (C) combined expression of Jagged1 in tumor/nontumor tissue. Patients were classified into three subgroups according to Jagged1 expression feature (D). High risk, tumor low and nontumor low; Medium risk, tumor low, nontumor high and tumor high, nontumor low; Low risk, tumor high, nontumor high. Kaplan-Meier analysis was applied and the overall survival was statistically significant among the three subgroups. P value, calculated by Log rank test, <0.05 was regarded as statistically significant.
Kaplan-Meier analysis was also applied to compare overall survival according to Jagged1 expression in different tumor invasion depth, lymph nodes metastasis status and TNM stage in tumor and nontumor tissues, respectively. Jagged1 expression was associated with overall survival in most subgroups, except that in nontumor tissue in N0 group (P=0.154) and in TNM I+II group (P=0.054) (Figures S1, S2 and S3).
By Kaplan-Meier analysis, we found that significant differences did exist among the four subgroups (P<0.001) (Figure 2C). In the Jagged1 tumor/nontumor low/high expression and high/low expression subgroups, the overall survival was quite similar (hazard ratio =1.173, 95% CI, 0.471-2.921; P=0.731). Thus, we combined the two subgroups as the “Medium Jagged1 risk” subgroup, while Jagged1 tumor/nontumor low/low expression subgroup and high/high expression subgroup were defined as the “High Jagged1 risk” subgroup and the “Low Jagged1 risk” subgroup, respectively. The overall survival among the three risk classifications was significantly different (P<0.001) (Figure 2D). Further analyses found the Jagged1 risk classification was associated with overall survival in different TNM stages (Figure 3A, 3B) and histological types (Figure 3C, 3D). The constructed Jagged1 risk classification had a better capability to differentiate the clinical prognosis with a higher Harrell’s concordance index (c-index) of 0.638 (95% CI, 0.604-0.672) compared with that of Jagged1 expression in tumor (0.581; 95% CI, 0.552-0.610) or that of Jagged1 expression in nontumor (0.590; 95% CI, 0.561-0.618).
Figure 3.
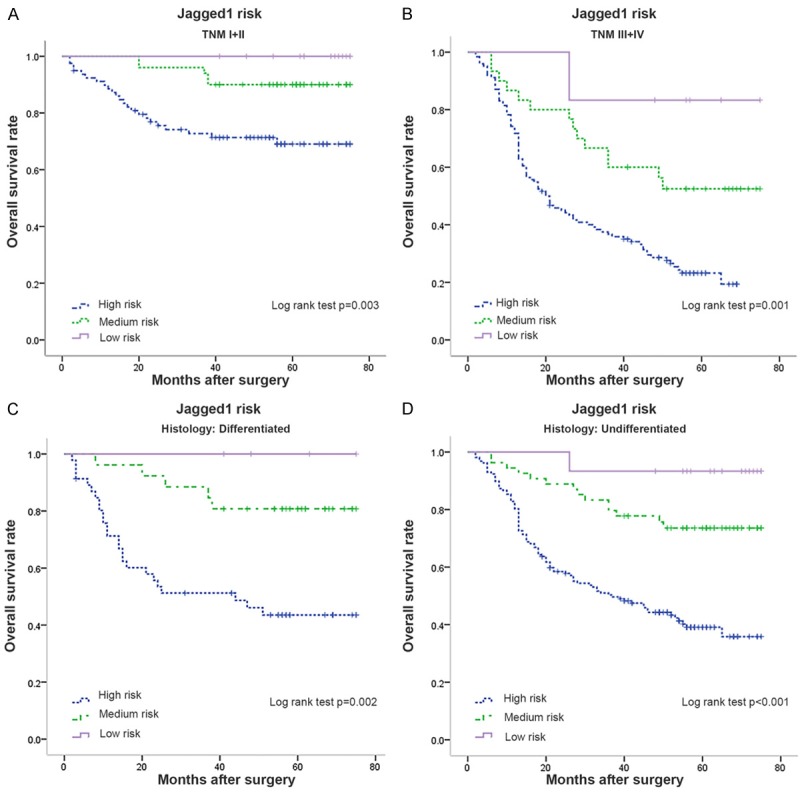
Kaplan-Meier analysis for overall survival of patients with gastric cancer according to Jagged1 risk classification. Kaplan-Meier analysis for overall survival according to Jagged1 risk classification (A) in patients with TNM I+II stage tumor, and (B) in patients with TNM III+IV stage tumor. Kaplan-Meier analysis for overall survival according to Jagged1 risk classification (C) in patients with differentiated histological type, and (D) in patients with undifferentiated histological type.
In the univariate Cox regression analysis of overall survival, Jagged1 risk was defined as a prognostic factor (P<0.001). The multivariate analysis demonstrated that the Jagged1 risk (P<0.001), tumor invasion depth (P=0.003), lymph nodes metastasis (P<0.001) and distant metastasis (P=0.010) were independent prognostic factors for overall survival in gastric cancer (Table 2).
Table 2.
Multivariate analysis for survival in gastric cancer patients (n=302)
| Variables | Hazard Ratio | 95% CI † | P value* |
|---|---|---|---|
| Tumor depth | 0.003 | ||
| T3+4/T1+2 | 2.214 | 1.320-3.714 | |
| Lymph nodes metastasis | <0.001 | ||
| Present/Absent | 2.634 | 1.639-4.234 | |
| Distant metastasis | 0.010 | ||
| Present/Absent | 3.005 | 1.300-6.948 | |
| Lauren’s classification | 0.368 | ||
| Diffuse/Intestinal | 1.111 | 0.742-1.664 | 0.608 |
| Mixed/ Intestinal | 1.427 | 0.869-2.345 | 0.160 |
| Jagged1 risk | <0.001 | ||
| Low/High | 12.162 | 1.691-87.486 | 0.013 |
| Medium/High | 4.451 | 0.564-31.542 | 0.146 |
CI: confidence interval;
Data obtained from the Cox proportional hazards model. P value <0.05 was regard as statistically significant.
Nomogram for predicting overall survival in gastric cancer patients
To provide a more sensitive prognostic model for outcomes of patients with gastric cancer, a quantitative nomogram was built (Figure 4A). The factors incorporated in the nomogram were independent factors for overall survival selected after multivariate analysis. In the nomogram, a higher total point predicts a worse prognosis. The total point was raised by the addition of tumor depth (rang from 0 to 34), lymph nodes metastasis (rang from 0 to 38), distant metastasis (rang from 0 to 43) and Jagged1 risk (0 for “Low”, 60 for “Medium” and 100 for “High”) for each patients correspondingly. For internal validation, calibration curves for nomogram predicted 3- and 5-year survival rate were built and performed well with the ideal model (Figure 4B, 4C). The Harrell’s c-index for the constructed nomogram was higher (0.718; 95% CI, 0.679-0.757) than that of TNM stage (0.685; 95% CI, 0.645-0.725), indicating the nomogram performed better in predicting overall survival in the gastric cancer patients.
Figure 4.
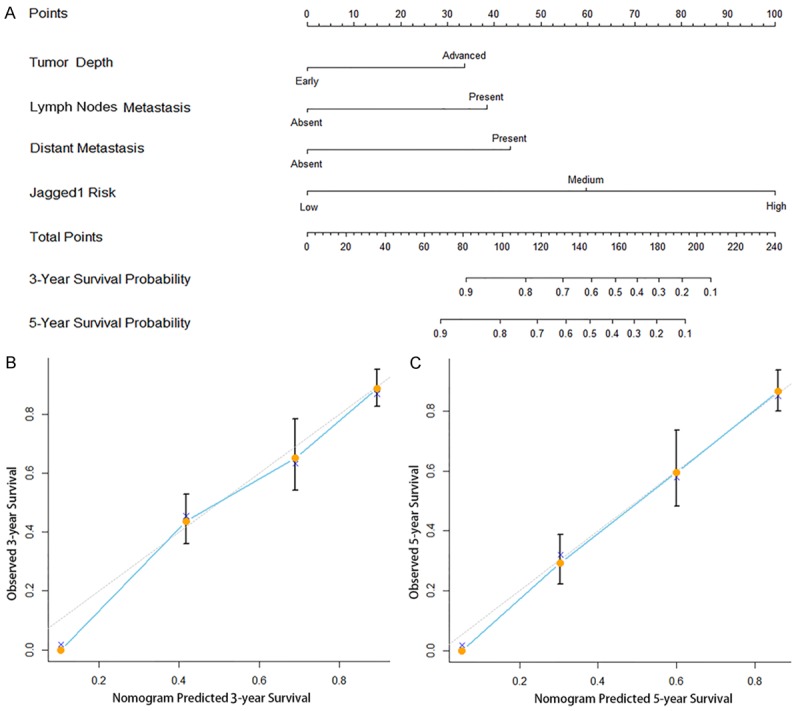
Nomogram for predicting 3- and 5-year overall survival in patients with gastric cancer. A. Nomogram for predicting clinical outcomes integrated with Jagged1 risk (High/Medium/Low), tumor depth (Early =T1+2/Advanced =T3+4), lymph nodes metastasis (Absent/Present) and distant metastasis (Absent/Present), B. Calibration plot for nomogram predicted and observed 3-year survival rate, C. Calibration plot for nomogram predicted and observed 5-year survival rate. Line of dashes: ideal model; vertical bars, 95% confident interval. In the nomogram, higher total point predicts worse prognosis. The total point was raised by the addition of tumor depth (rang from 0 to 34), lymph nodes metastasis (rang from 0 to 38), distant metastasis (rang from 0 to 43) and Jagged1 risk (0 for “Low”, 60 for “Medium” and 100 for “High”) for each patients correspondingly. The nomogram predicted 3- or 5-year survival with more accuracy than TNM stage alone shown in the calibration curves.
Discussion
Accumulating attentions have been focused on Notch signaling in solid tumors [7,9,10,16]. However, few studies have unraveled the underlying mechanisms for Notch signaling activation during carcinogenesis, including gastric cancer. In the present study, we found that Jagged1 expression in gastric cancer tissues was decreased compared with nontumor tissues and low expression of Jagged1 in both tumor and nontumor tissues correlated with poor prognosis. These findings indicated that Jagged1 might not be the main ligand for activating Notch1 signaling in gastric cancer [12,13,17].
Previous studies about Notch signaling in gastric cancer have partly unraveled its significance in the carcinogenesis of gastric cancer. Notch2-induced COX-2 expression could enhance gastric cancer progression, and Notch4 could promote gastric cancer growth through activation of Wnt1/β-catenin signaling [18,19]. Therefore, decreased expression of Jagged1 in gastric cancer might not be responsible for activation of Notch2 and Notch4. The loss of Notch3 expression was reported to be an independent prognosticator for poor prognosis in gastric cancer patients, and Notch3 expression was correlated with Jagged1 expression at mRNA level [20]. Therefore, it is conceivable that Jagged1 may be responsible for Notch3 activation. Aberrant activation of the Notch signaling can be caused by overexpression of ligands or loss of negative regulators [21]. Thus, Low expression of Jagged1 in gastric cancer may lead to the failure of Notch3 activation, causing the development and progression of gastric cancer. Piazzi et al demonstrated that the increase in DLL1 expression was associated with activation of Notch1 signaling, with an increase in cleaved Notch1 intracellular domain (NICD) and Hes1, and down-regulation in Hath1. Concordantly, Notch1 signaling was activated with the overexpression of DLL1 [14]. Whether other ligands were involved in the activation of Notch1, Notch2 and Notch4 in gastric cancer needs further investigation.
Notch signaling begins after the binding of its ligands to the Notch receptor, resulting in the release of the active intracellular domain. Meanwhile, following the Notch ligand/receptor interaction, the Notch ligands, including Jagged1, undergo proteolytic cleavage and release an intracellular domain as well [22]. It was reported that the Jagged-1 intracellular domain (JICD) inhibited Notch1 signaling via a reduction in the protein stability of the Notch1 intracellular domain (NICD) [23]. Thus, JICD functioned as a negative regulator in Notch1 signaling via the promotion of NICD degradation. Since activating Notch1 signaling in gastric cancer was related to a poor prognosis [12,13,17], the reduced detection by immunohistochemical staining would be much more possible to be the result of loss of membranous Jagged1 expression in advanced tumors rather than the result of Jagged1 endocytosis following Jagged1/Notch interaction.
Moreover, it is becoming a consensus that the biological effect of a membrane protein is determined not only by its biochemical characteristics, but also by the microenvironment it presented. Interactions between Notch signaling and TGFβ, Wnt signaling have been reported in some tumors, such as breast cancer and colorectal cancer [7,24]. Therefore, further studies about activation of Notch signaling and interactions with tumor microenvironment are needed.
As given the critical role of Notch signaling in gastric cancer, targeting the components of the pathway could open a novel avenue for treatment of gastric cancer. The ongoing studied gamma-secretase inhibitors could block the activation of all the four Notch receptors. Since Notch3 activation could have tumor-suppressive effect, inhibitors targeting the activation of Notch1, Notch2 and Notch4 signaling may be more specific and reliable.
In this study, we have proved the prognostic value of Jagged1 expression in gastric cancer. The patients can be stratified into three-risk classification according to Jagged1 expression in tumor/nontumor tissue, which proved to be an independent prognostic factor. In the stratification analysis according to different tumor invasion depth, lymph nodes metastasis and TNM stage, the overall survival in each subgroup differs significantly, indicating that Jagged1 expression in the tissue could have additional prognostic value in each tumor stage. Further, we generated a nomogram by integrating Jagged1 risk and TNM stage to predict the 3- and 5-year overall survival for gastric cancer patients, and the nomogram performed better in discriminating patients with different clinical outcomes than using TNM stage alone.
Being a complex family in regulating initiation, progression and metastasis of malignant tumor, studies about Notch family were far beyond sufficient. Our present study focused on the decreased Jagged1 expression in gastric cancer and partially unraveled its prognostic significance. The study is retrospective in nature and the number of patients enrolled is relatively small. A larger, multi-centered, prospective data is needed to validate these results. Besides, the prognostic significance of other Notch ligands and receptors needs further studies.
In conclusion, we have identified that Jagged1 may be not responsible for activation of Notch1 in gastric cancer. The decreased Jagged1 expression in gastric cancer was identified as an independent prognostic factor and could be incorporated with TNM stage to generate a nomogram to better stratify patients with different prognosis.
Acknowledgements
This study was funded by grants from National Natural Science Foundation of China (31100629, 31270863, 31300671, 31470794, 81471621, 81472227), Program for New Century Excellent Talents in University (NCET-13-0146) and Shanghai Rising-Star Program (13QA1400300).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Mayer RJ, Venook AP, Schilsky RL. Progress against GI cancer during the American Society of Clinical Oncology’s first 50 years. J. Clin. Oncol. 2014;32:1521–1530. doi: 10.1200/JCO.2014.55.4121. [DOI] [PubMed] [Google Scholar]
- 3.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 4.Shi TP, Xu H, Wei JF, Ai X, Ma X, Wang BJ, Ju ZH, Zhang GX, Wang C, Wu ZQ, Zhang X. Association of low expression of notch-1 and jagged-1 in human papillary bladder cancer and shorter survival. J Urol. 2008;180:361–366. doi: 10.1016/j.juro.2008.02.037. [DOI] [PubMed] [Google Scholar]
- 5.Wu K, Xu L, Zhang L, Lin Z, Hou J. High Jagged1 expression predicts poor outcome in clear cell renal cell carcinoma. Jpn J Clin Oncol. 2011;41:411–416. doi: 10.1093/jjco/hyq205. [DOI] [PubMed] [Google Scholar]
- 6.Dickson BC, Mulligan AM, Zhang H, Lockwood G, O’Malley FP, Egan SE, Reedijk M. Highlevel JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol. 2007;20:685–693. doi: 10.1038/modpathol.3800785. [DOI] [PubMed] [Google Scholar]
- 7.Rodilla V, Villanueva A, Obrador-Hevia A, Robert-Moreno A, Fernandez-Majada V, Grilli A, Lopez-Bigas N, Bellora N, Alba MM, Torres F, Dunach M, Sanjuan X, Gonzalez S, Gridley T, Capella G, Bigas A, Espinosa L. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc Natl Acad Sci U S A. 2009;106:6315–6320. doi: 10.1073/pnas.0813221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng AP, Ferrando AA, Lee W, Morris JT, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 9.Dufraine J, Funahashi Y, Kitajewski J. Notch signaling regulates tumor angiogenesis by diverse mechanisms. Oncogene. 2008;27:5132–5137. doi: 10.1038/onc.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107:2223–2233. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- 11.Shih I, Wang TL. Notch signaling, gammasecretase inhibitors, and cancer therapy. Cancer Res. 2007;67:1879–1882. doi: 10.1158/0008-5472.CAN-06-3958. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Wang X, Xu J, Sun Y. Notch1 activation is a poor prognostic factor in patients with gastric cancer. Br J Cancer. 2014;110:2283–2290. doi: 10.1038/bjc.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeh TS, Wu CW, Hsu KW, Liao WJ, Yang MC, Li AF, Wang AM, Kuo ML, Chi CW. The activated Notch1 signal pathway is associated with gastric cancer progression through cyclooxygenase-2. Cancer Res. 2009;69:5039–5048. doi: 10.1158/0008-5472.CAN-08-4021. [DOI] [PubMed] [Google Scholar]
- 14.Piazzi G, Fini L, Selgrad M, Garcia M, Daoud Y, Wex T, Malfertheiner P, Gasbarrini A, Romano M, Meyer RL, Genta RM, Fox JG, Boland CR, Bazzoli F, Ricciardiello L. Epigenetic regulation of Delta-Like1 controls Notch1 activation in gastric cancer. Oncotarget. 2011;2:1291–1301. doi: 10.18632/oncotarget.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camp RL, Dolled-Filhart M, Rimm DL. Xtile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 16.Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 17.Hsu KW, Hsieh RH, Huang KH, Fen-Yau LA, Chi CW, Wang TY, Tseng MJ, Wu KJ, Yeh TS. Activation of the Notch1/STAT3/Twist signaling axis promotes gastric cancer progression. Carcinogenesis. 2012;33:1459–1467. doi: 10.1093/carcin/bgs165. [DOI] [PubMed] [Google Scholar]
- 18.Qian C, Liu F, Ye B, Zhang X, Liang Y, Yao J. Notch4 promotes gastric cancer growth through activation of Wnt1/beta-catenin signaling. Mol Cell Biochem. 2015;401:165–174. doi: 10.1007/s11010-014-2304-z. [DOI] [PubMed] [Google Scholar]
- 19.Tseng YC, Tsai YH, Tseng MJ, Hsu KW, Yang MC, Huang KH, Li AF, Chi CW, Hsieh RH, Ku HH, Yeh TS. Notch2-induced COX-2 expression enhancing gastric cancer progression. Mol Carcinog. 2012;51:939–951. doi: 10.1002/mc.20865. [DOI] [PubMed] [Google Scholar]
- 20.Kang H, An HJ, Song JY, Kim TH, Heo JH, Ahn DH, Kim G. Notch3 and Jagged2 contribute to gastric cancer development and to glandular differentiation associated with MUC2 and MUC5AC expression. Histopathology. 2012;61:576–586. doi: 10.1111/j.1365-2559.2012.04274.x. [DOI] [PubMed] [Google Scholar]
- 21.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 22.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim MY, Jung J, Mo JS, Ann EJ, Ahn JS, Yoon JH, Park HS. The intracellular domain of Jagged-1 interacts with Notch1 intracellular domain and promotes its degradation through Fbw7 E3 ligase. Exp Cell Res. 2011;317:2438–2446. doi: 10.1016/j.yexcr.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 24.Sun Y, Lowther W, Kato K, Bianco C, Kenney N, Strizzi L, Raafat D, Hirota M, Khan NI, Bargo S, Jones B, Salomon D, Callahan R. Notch4 intracellular domain binding to Smad3 and inhibition of the TGF-beta signaling. Oncogene. 2005;24:5365–5374. doi: 10.1038/sj.onc.1208528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


