Abstract
Background: There is no consensus regarding the association between polymorphisms in the myosin IXB (MYO9B) gene and celiac disease (CD) risk. In this study, we performed a meta-analysis to evaluate genetic variants in MYO9B with CD. Methods: Four MYO9B polymorphisms (rs1545620, rs1457092, rs2305767 and rs2305764) were assessed. A literature search was conducted using PubMed, Scopus, and Web of Science databases until June 2015. Odds ratio (OR) and 95% confidence interval (CI) were used to investigate the strength of the association under dominant, recessive, homozygote and allelic comparison models. Results: Seven case-control studies with a total of 1965 CD patients and 4894 controls were included in this meta-analysis. The results showed that rs1545620 was associated with CD risk in Europeans in dominant (OR=1.31, 95% CI: 1.10-1.58, Pz=0.003), recessive (OR=1.36, 95% CI: 1.08-1.72, Pz=0.009), homozygote (OR=1.55, 95% CI: 1.20-2.01, Pz=0.001), and allelic comparison models (OR=1.24, 95% CI: 1.10-1.40, Pz=0.001), whereas in a Latin American group there were significant associations of CD with rs1457092 in dominant (OR=15.30, 95% CI: 3.51-66.67, Pz<0.001), homozygote (OR=16.55, 95% CI: 3.62-75.65, Pz<0.001), and allelic comparison models (OR=1.95, 95% CI: 1.31-2.91, Pz=0.001), and rs2305767 in dominant (OR=5.35, 95% CI: 2.42-11.86, Pz<0.001) and allelic comparison models (OR=1.65, 95% CI: 1.11-2.45, Pz=0.013). There was no association between rs2305764 and CD risk in either Europeans or the Latin American group. Conclusion: rs1545620 is associated with CD risk in Europeans, whereas rs1457092 and rs2305767 are associated with CD risk in a Latin American group.
Keywords: Celiac disease, meta-analysis, myosin IXB, polymorphism
Introduction
Celiac disease (CD) is a chronic immune-mediated disorder characterized by a gluten-sensitive enteropathy resulting in damage to the mucosa in the small intestine [1]. It is the commonest immunological gastrointestinal disorder in the western world, affecting 0.6 to 1.0% of the population in the USA and most European countries [2]. CD can present at any age, but most adult cases occur in the 4th and 5th decades of life. The majority of patients present with ‘classical’ symptoms of malabsorption: weight loss, diarrhea or failure to thrive [1]. However, it has become increasingly clear that the clinical features of CD are varied and may include ‘non-classical’ presentations, including anaemia, abdominal pain, glossitis and irritable bowel syndrome type symptoms [2]. Lifelong adherence to a strict gluten-free diet is the only current effective treatment for the disease [3].
Genetic factors play a critical role in predisposition to CD. The strongest and best-characterized genetic susceptibility factor in CD is the human leukocyte antigen (HLA)-DQ locus [4,5]. HLA-DQ is responsible for presentation of toxic cereal derived peptides to intestinal immune cells. The HLA-DQ2 haplotype (DQA1*0501/DQB1*0201) is found in greater than 90% of Northern European CD patients; however, it is also found in one third of the general population, implicating that the HLA association is necessary but not sufficient to explain the hereditary nature of the disease [6]. The HLA-DQ locus is estimated to account for 35% of the heritable risk of CD [6]. Recent genome-wide association (GWA) and multilineage studies have identified a number of common non-HLA genes which also contribute to the development of CD, including cytotoxic T-lymphocyte antigen 4 (CTLA-4), interleukin-2 (IL-2), protein tyrosine phosphatase nonreceptor 22 (PTPN22), and SH2B adaptor protein 3 (SH2B3) [7-10]. Identifying non-HLA genetic factors for CD will not only lead to a greater understanding of CD pathogenesis, but also allow the generation of newer therapeutic modalities.
The Myosin IXB (MYO9B) gene is located at 19p13.1 and was recently considered as a susceptibility gene to CD [11]. MYO9B encodes a Ras homologous (Rho) family guanosine-triphosphatase (GTPase) activating protein, which is involved in epithelial cell cytoskeletal organization and influences tight junction assembly [12,13]. Human MYO9B is expressed in intestinal epithelial cells, and increased MYO9B gene expression is correlated with increased intestinal permeability in vivo [14]. MYO9B is also highly expressed in immune cells, suggesting a direct role in immune function [15]. Many single nucleotide polymorphisms (SNPs) have been identified in the MYO9B gene. Among them, four polymorphisms are most frequently analyzed: rs1545620 A>C (exon 20), rs1457092 C>A (intron 20), rs2305767 A>G (intron 14), and rs2305764 A>G (intron 28). Several association studies have evaluated the relationship of these polymorphisms with CD, but the results of these studies remain contradictory rather than convincing, possibly because single studies may have been underpowered. Given the amount of accumulated data and the still equivocal role of MYO9B in the etiology of CD, we aim to perform a meta-analysis of published case-control studies to investigate potential associations between the MYO9B polymorphisms and CD risk.
Materials and methods
Literature search
The literature searches were conducted using PubMed, Scopus, and Web of Science until June 2015 for eligible studies. The search strategy included using the keywords “celiac disease, CD, case-control studies, polymorphism, risk, genetics, myosin IXB, and MYO9B”. All relevant publications identified through the search were scanned on the basis of title and abstract by one of us and were rejected in the initial screening if the study clearly did not meet the inclusion criteria. The remaining studies were then evaluated in their entirety. Only papers published in the English language were considered for inclusion. The reference lists of identified studies were also searched by hand for relevant papers.
Inclusion and exclusion criteria
Qualified studies had to meet the following criteria: (1) case-control study, regardless of sample size; (2) sufficient data for calculating an odds ratio (OR) with 95% confidence interval (CI); (3) studies published as full-length articles or letters in English. The major exclusion criteria were as follows: (1) no control subjects; (2) evaluation of other variants; (3) family-based study; (4) insufficient data for genotype distribution; (5) duplicate data.
Data extraction
For each study, the following information was extracted using standard forms: first author, publication year, country, ethnicity, sample size, age of subjects, genotyping method, and genotype distribution. Two reviewers extracted data from the published studies. Disagreements were resolved by discussion and consensus.
Statistical analysis
All statistical analyses were performed using Stata version 11.0. Raw data without adjustment were used for calculation of the study-specific estimates of ORs and 95% CIs. The presence of heterogeneity between studies was explored with the Cochran’s Q statistic; P<0.10 indicated significant heterogeneity. The pooled estimation of the OR of each study was calculated by the Mantel-Haenszel fixed effects model or the Dersimonian-Laird random effects model [16]. The random effects model uses weights that incorporate both the within-study and between-study variance. The overall pooled ORs were calculated assuming dominant, recessive, homozygote and allelic comparison models. The significance of the pooled OR was determined by the Z test and P<0.05 was considered as statistically significant. Subgroup analysis was performed according to ethnicity. Considering that the number of included studies was small, sensitivity analysis was not conducted. Publication bias was evaluated through the Begg’s test, with P<0.05 being considered statistically significant. Funnel plot displaying the log odds ratios of individual studies on the vertical axis and the standard errors of the log odds ratios on the horizontal axis was also constructed to assess publication bias.
Results
Study characteristics
We retrieved 97 published studies using our search criteria (Figure 1). Seven studies published between 2006 and 2012 met our inclusion criteria with a total of 1965 CD patients and 4894 controls (Figure 1) [17-23]. In terms of ethnicity, six studies were performed in Europeans [17-22], whereas one study was undertaken in a Latin American mixed group [23]. In terms of age, two studies were performed in children [19,23], three studies were undertaken in both children and adults [17,21,22], and two studies did not report any information on age [18,20]. Table 1 summarizes the characteristics of the seven studies included in our meta-analysis.
Figure 1.
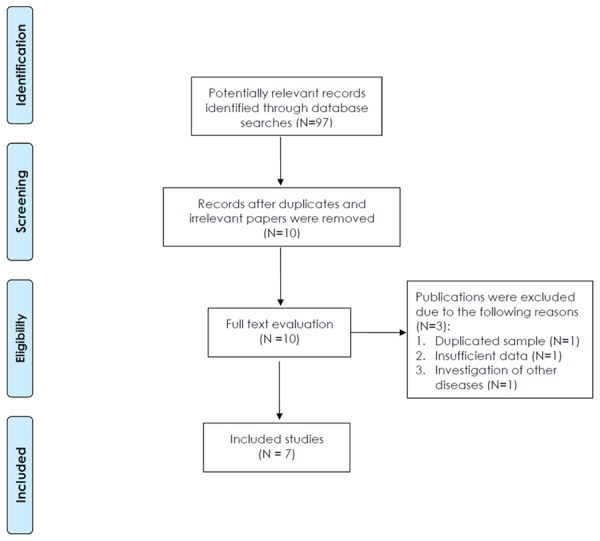
Prisma flow diagram.
Table 1.
Characteristics of eligible studies in meta-analysis
| First author | Year | Country | Ethnicity | Number | Age of subjects | Genotyping method | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Patients | Controls | ||||||
| Loeff | 2012 | Chile | Latin Americans | 104 | 104 | Children | Taqman assays |
| Wolters | 2007 | Netherlands | Europeans | 421 | 1624 | Children and adults | Taqman assays |
| Latiano | 2007 | Italy | Europeans | 337 | 452 | Children and adults | Taqman assays |
| Sánchez | 2007 | Spain | Europeans | 90 | 345 | NR | Taqman assays |
| Cirillo | 2007 | Italy | Europeans | 223 | 600 | Children | PCR-RFLP |
| Núñez | 2006 | Spain | Europeans | 415 | 433 | NR | Taqman assays |
| Hunt | 2006 | UK | Europeans | 375 | 1336 | Children and adults | Taqman and Sequenom methods |
NR, not reported; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; UK, united kingdom.
Quantitative data synthesis
Two studies evaluated the rs1545620 polymorphism [21,22]. All subjects were Europeans. Pooling data from these showed a significant association between CD risk and rs1545620 in dominant (OR=1.31, 95% CI: 1.10-1.58, Pz=0.003) (Table 2), recessive (OR=1.36, 95% CI: 1.08-1.72, Pz=0.009) (Table 2), homozygote (OR=1.55, 95% CI: 1.20-2.01, Pz=0.001) (Table 2), and allelic comparison models (OR=1.24, 95% CI: 1.10-1.40, Pz=0.001) (Table 2; Figure 2).
Table 2.
Meta-analysis of polymorphisms in the MYO9B gene and CD risk
| Polymorphism | No. of studies | Dominant | Recessive | Homozygote | Allelic comparison | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||
| OR (95% CI) | Phet | Pz | OR (95% CI) | Phet | Pz | OR (95% CI) | Phet | Pz | OR (95% CI) | Phet | Pz | ||
| rs1545620 | |||||||||||||
| Total | 2 | 1.31 (1.10-1.58) | 0.534 | 0.003 | 1.36 (1.08-1.72) | 0.559 | 0.009 | 1.55 (1.20-2.01) | 0.494 | 0.001 | 1.24 (1.10-1.40) | 0.415 | 0.001 |
| Europeans | 2 | 1.31 (1.10-1.58) | 0.534 | 0.003 | 1.36 (1.08-1.72) | 0.559 | 0.009 | 1.55 (1.20-2.01) | 0.494 | 0.001 | 1.24 (1.10-1.40) | 0.415 | 0.001 |
| rs1457092 | |||||||||||||
| Total | 4 | 1.35 (0.86-2.13) | <0.001 | 0.196 | 1.29 (0.86-1.95) | 0.016 | 0.222 | 1.81 (0.87-3.78) | <0.001 | 0.112 | 1.25 (0.94-1.65) | 0.001 | 0.126 |
| Europeans | 3 | 1.12 (0.84-1.48) | 0.057 | 0.441 | 1.22 (0.73-2.05) | 0.009 | 0.440 | 1.27 (0.71-2.27) | 0.006 | 0.422 | 1.11 (0.86-1.45) | 0.008 | 0.423 |
| Latin Americans | 1 | 15.3 (3.51-66.67) | NA | <0.001 | 1.62 (0.90-2.90) | NA | 0.106 | 16.55 (3.62-75.65) | NA | <0.001 | 1.95 (1.31-2.91) | NA | 0.001 |
| rs2305767 | |||||||||||||
| Total | 4 | 1.10 (0.65-1.87) | <0.001 | 0.717 | 0.82 (0.61-1.11) | 0.183 | 0.195 | 0.77 (0.48-1.24) | 0.028 | 0.284 | 0.96 (0.72-1.27) | 0.001 | 0.765 |
| Europeans | 3 | 0.80 (0.60-1.08) | 0.053 | 0.142 | 0.82 (0.58-1.16) | 0.092 | 0.254 | 0.72 (0.44-1.17) | 0.022 | 0.181 | 0.85 (0.67-1.07) | 0.021 | 0.161 |
| Latin Americans | 1 | 5.35 (2.42-11.86) | NA | <0.001 | 0.66 (0.11-4.04) | NA | 0.653 | 2.59 (0.38-17.92) | NA | 0.334 | 1.65 (1.11-2.45) | NA | 0.013 |
| rs2305764 | |||||||||||||
| Total | 7 | 1.09 (0.91-1.30) | 0.048 | 0.352 | 1.12 (0.84-1.49) | 0.002 | 0.447 | 1.16 (0.83-1.62) | 0.001 | 0.375 | 1.07 (0.92-1.25) | 0.002 | 0.383 |
| Europeans | 6 | 1.08 (0.89-1.31) | 0.026 | 0.413 | 1.15 (0.84-1.57) | 0.001 | 0.389 | 1.18 (0.82-1.69) | 0.001 | 0.375 | 1.08 (0.91-1.27) | 0.001 | 0.380 |
| Latin Americans | 1 | 1.15 (0.55-2.40) | NA | 0.708 | 0.88 (0.44-1.78) | NA | 0.720 | 1.01 (0.40-2.56) | NA | 0.979 | 1.00 (0.68-1.47) | NA | 1.000 |
CD, celiac disease; CI, confidence interval; NA, not available; OR, odds ratio; Phet, P value for heterogeneity, Pz, P value for the overall effect.
Figure 2.
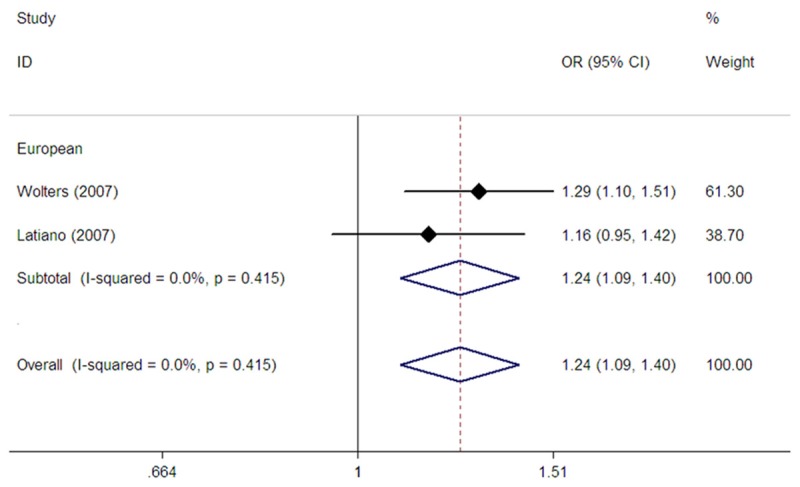
Forest plot of fixed effects meta-analysis investigating the rs1545620 polymorphism and CD risk in allele contrast.
Four studies assessed the association between the rs1457092 SNP and CD risk [17,20,22,23]. Among them, three studies were conducted in Europeans [17,20,22], whereas one study was performed in Latin Americans [23]. Pooling the data from these studies showed no association between the polymorphism and CD in all study subjects in dominant (OR=1.35, 95% CI: 0.86-2.13, Pz=0.196) (Table 2), recessive (OR=1.29, 95% CI: 0.86-1.95, Pz=0.222) (Table 2), homozygote (OR=1.81, 95% CI: 0.87-3.78, Pz=0.112) (Table 2), and allelic comparison models (OR=1.25, 95% CI: 0.94-1.65, Pz=0.126) (Table 2; Figure 3). In subgroup analysis according to ethnicity, we did not find an association between rs1457092 and CD risk in Europeans in dominant (OR=1.12, 95% CI: 0.84-1.48, Pz=0.441) (Table 2), recessive (OR=1.22, 95% CI: 0.73-2.05, Pz=0.440) (Table 2), homozygote (OR=1.27, 95% CI: 0.71-2.27, Pz=0.422) (Table 2), and allelic comparison models (OR=1.11, 95% CI: 0.86-1.45, Pz=0.423) (Table 2; Figure 3). However, a single Latin American study showed an association between rs1457092 and CD risk in dominant (OR=15.30, 95% CI: 3.51-66.67, Pz<0.001) (Table 2), homozygote (OR=16.55, 95% CI: 3.62-75.65, Pz<0.001) (Table 2), and allelic comparison models (OR=1.95, 95% CI: 1.31-2.91, Pz=0.001) (Table 2; Figure 3).
Figure 3.
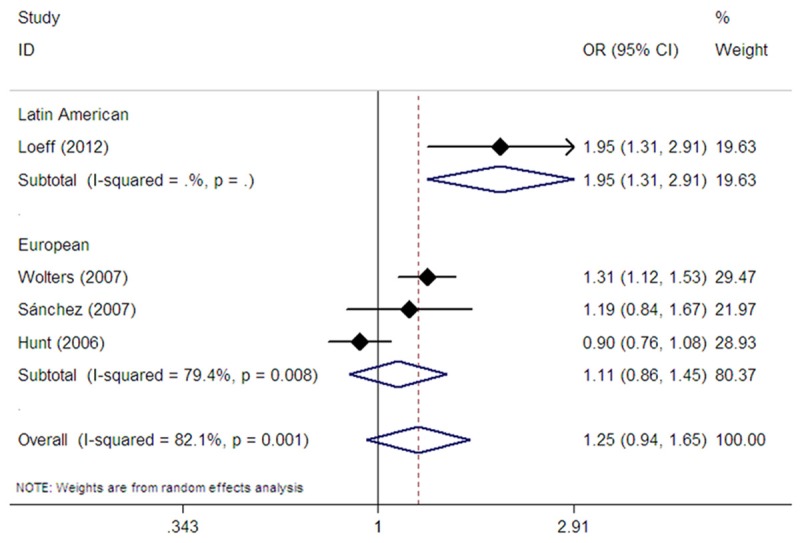
Forest plot of random effects meta-analysis investigating the rs1457092 polymorphism and CD risk in allele contrast.
The MYO9B rs2305767 polymorphism was evaluated in four studies [17,20,22,23]. Among them, three studies were undertaken in Europeans [17,20,22], whereas one study was conducted in Latin Americans [23]. The pooled analysis showed no association between rs2305767 and CD risk in all study subjects in dominant (OR=1.10, 95% CI: 0.65-1.87, Pz=0.717) (Table 2), recessive (OR=0.82, 95% CI: 0.61-1.11, Pz=0.195) (Table 2), homozygote (OR=0.77, 95% CI: 0.48-1.24, Pz=0.284) (Table 2), and allelic comparison models (OR=0.96, 95% CI: 0.72-1.27, Pz=0.765) (Table 2; Figure 4). In subgroup analysis based on ethnicity, no association between rs2305767 and CD risk was found in Europeans in dominant (OR=0.80, 95% CI: 0.60-1.08, Pz=0.142) (Table 2), recessive (OR=0.82, 95% CI: 0.58-1.16, Pz=0.254) (Table 2), homozygote (OR=0.72, 95% CI: 0.44-1.17, Pz=0.181) (Table 2), and allelic comparison models (OR=0.85, 95% CI: 0.67-1.07, Pz=0.161) (Table 2; Figure 4). However, the single Latin American study revealed an association between rs2305767 and CD risk in dominant model (OR=5.35, 95% CI: 2.42-11.86, Pz<0.001) (Table 2) and allelic comparison model (OR=1.65, 95% CI: 1.11-2.45, Pz=0.013) (Table 2; Figure 4).
Figure 4.
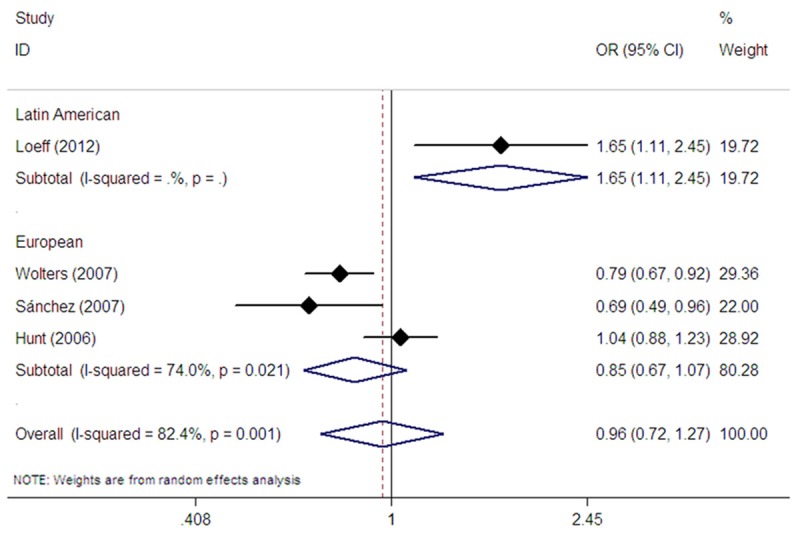
Forest plot of random effects meta-analysis investigating the rs2305767 polymorphism and CD risk in allele contrast.
Seven studies evaluated the rs2305764 polymorphism [17-23]. Among them, six studies were conducted in Europeans [17-22], whereas one study was performed in Latin Americans [23]. Pooling these seven studies did not show an association between rs2305764 and CD risk in all study subjects in dominant (OR=1.09, 95% CI: 0.91-1.30, Pz=0.352) (Table 2) , recessive (OR=1.12, 95% CI: 0.84-1.49, Pz=0.447) (Table 2), homozygote (OR=1.16, 95% CI: 0.83-1.62, Pz=0.375) (Table 2), and allelic comparison models (OR=1.07, 95% CI: 0.92-1.25, Pz=0.383) (Table 2 and Figure 5). In subgroup analysis stratified by ethnicity, pooling the data from six studies provided no evidence for an association between the polymorphism and CD risk in Europeans in dominant (OR=1.08, 95% CI: 0.89-1.31, Pz=0.413) (Table 2), recessive (OR=1.15, 95% CI: 0.84-1.57, Pz=0.389) (Table 2), homozygote (OR=1.18, 95% CI: 0.82-1.69, Pz=0.375) (Table 2), and allelic comparison models (OR=1.08, 95% CI: 0.91-1.27, Pz=0.380) (Table 2; Figure 5). The single Latin American study also revealed no association between rs2305764 and CD risk (Table 2; Figure 5).
Figure 5.
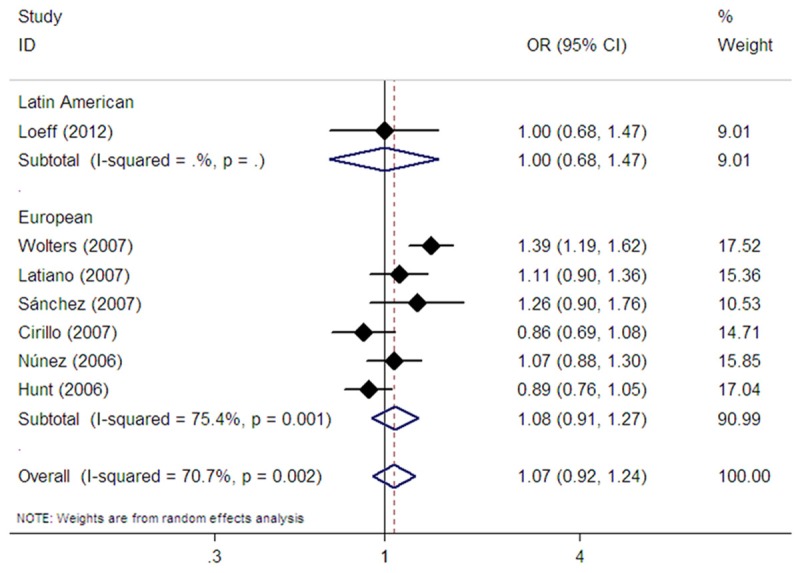
Forest plot of random effects meta-analysis investigating the rs2305764 polymorphism and CD risk in allele contrast.
Heterogeneity and publication bias
Between-study heterogeneity was found in the pooled analyses of the rs1457092, rs2305767 and rs2305764 polymorphisms (Table 2). Publication bias were evaluated through the Begg’ test in all models. There was no significant evidence of publication bias for the rs1545620, rs1457092, rs2305767 and rs2305764 polymorphisms. Table 3 showed the results of the Begg’s test in detail. Funnel plot was generated for the rs2305764 polymorphism, and the shape seemed symmetrical (Figure 6).
Table 3.
Begg’s test for evaluating publication bias
| Polymorphism | P for dominant model | P for recessive model | P for homozygote model | P for allelic comparison model |
|---|---|---|---|---|
| rs1545620 | 1.000 | 1.000 | 1.000 | 1.000 |
| rs1457092 | 0.734 | 1.000 | 0.734 | 0.734 |
| rs2305767 | 0.308 | 0.734 | 0.734 | 0.308 |
| rs2305764 | 0.548 | 1.000 | 1.000 | 1.000 |
Figure 6.
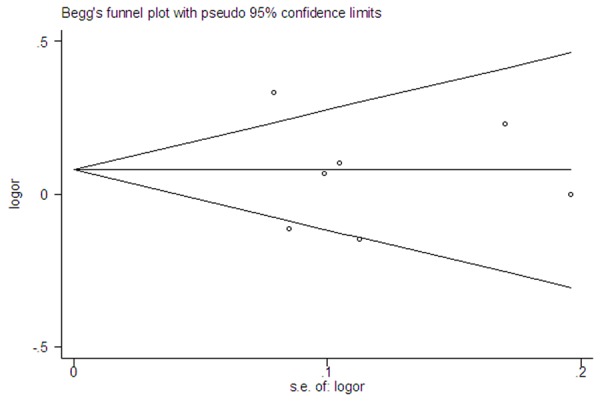
Begg’s funnel plot for the rs2305764 polymorphism and CD risk.
Discussion
The present meta-analysis of seven studies, involving a total of 1965 CD patients and 4894 controls provides the most comprehensive assessment so far of the relevance to CD of four common MYO9B polymorphisms. The main findings are as follows: (1) the rs1545620 polymorphism is associated with CD risk in Europeans; (2) there is no association between CD risk and the rs1457092, rs2305767 and rs2305764 polymorphisms in all study subjects (Europeans and a Latin American group) and Europeans; and (3) rs1457092 and rs2305767 are associated with CD risk in a Latin American group.
MYO9B encodes a single-headed molecular motor containing a Rho-family GTPase-activating protein (GAP) domain in the tail region, which is a member of the class IX myosin family [12]. It is expressed in intestinal epithelial cells and is an integral structural component of the intestinal mucosa. MYO9B acts as a negative regulator facilitating inactivation of Rho GTPases and downregulates Rho-dependent signaling pathways [15]. Rho GTPases are key regulators of actin filament remodeling and tight junction assembly, both of which lead to an increase of epithelial paracellular permeability [13,24]. The latter fact therefore leads to the hypothesis that MYO9B variants may modify the epithelial barrier function of the gut, thereby allowing gluten peptides to gain easy access to the deeper mucosal layer and be presented to the immune system. In this meta-analysis, we found that the rs1545620 polymorphism was associated with CD risk in Europeans, where as the rs1457092 and rs2305767 polymorphisms were associated with CD in a Latin American group. The rs1545620 SNP is a non-synonymous polymorphism resulting in an amino acid change (Alanine1011Serine) in the neck region of the MYO9B protein, which is involved in the binding of calmodulin [25]. Calmodulin is a protein that modulates the motor activity of MYO9B on actin filaments [26]. The conformational change of the MYO9B protein induced by the rs1545620 SNP can result in lower MYO9B activity, and may therefore lead to a defect in the intestinal barrier and contribute to the pathogenesis of CD. Moreover, MYO9B is not specifically expressed in intestinal epithelial cells. It is also abundantly expressed in immune cells including leukocytes and macrophages [15,27]. Given the pivotal role of MYO9B in immune cell shape and motility [27], it would be reasonable to hypothesize that the rs1545620 polymorphism may increase risk of CD by alternative mechanisms regulating the movement of immune cells to the site of inflammation. The rs1457092 and rs2305767 polymorphisms are intronic noncoding variations. Although part of the function of MYO9B is understood, it remains largely unknown how these two polymorphisms might influence MYO9B activity. To understand the precise mechanisms by which genetic variation of MYO9B predisposes to CD, more functional assays are needed in the future.
Ethnic differences between Europeans and Latin Americans in MYO9B association with CD were observed in this meta-analysis. It can be explained by several possible reasons. First, the two ethnic groups have distinct genetic background. Second, CD is a multifactorial disease determined by interactions between environmental and genetic factors. Individual genetic predisposition to CD may be modified by different environmental factors in different ethnic groups. Third, the discrepancy may partly be explained by clinical heterogeneity between different ethnic groups. Since the small number of included studies limited the statistical power to achieve a definitive conclusion for each ethnic group, future studies using large sample numbers are needed to confirm the associations found in this meta-analysis.
There are several limitations to our study. First, we only included studies written in the English language, which may be a source of bias. Because significant results are more likely to be published in English. Second, because of the limitation of the published data, we were unable to conduct subgroup analysis based on age in children and adults, respectively. Third, between-study heterogeneity was found in some pooled analyses. Several potential factors, including ethnic heterogeneity, genotyping method, and age may contribute to it. Because there were insufficient data from the included studies, we did not perform meta-regression analysis to assess the sources of heterogeneity. Fourth, linkage disequilibrium among several MYO9B polymorphisms was reported in some studies [20,21]. Therefore, it may be of interest to evaluate haplotype association with the disease. However, due to discrepancy in alleles and haplotype estimation methods, we did not perform haplotype estimation in our meta-analysis.
In summary, the results of our meta-analysis demonstrate that the rs1545620 polymorphism is associated with CD risk in Europeans, whereas the rs1457092 and rs2305767 polymorphisms are associated with CD in a Latin American group.
Disclosure of conflict of interest
None.
References
- 1.Green PH, Cellier C. Celiac disease. N Engl J Med. 2007;357:1731–1743. doi: 10.1056/NEJMra071600. [DOI] [PubMed] [Google Scholar]
- 2.Gujral N, Freeman HJ, Thomson AB. Celiac disease: prevalence, diagnosis, pathogenesis and treatment. World J Gastroenterol. 2012;18:6036–6059. doi: 10.3748/wjg.v18.i42.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mulder CJ, van Wanrooij RL, Bakker SF, Wierdsma N, Bouma G. Gluten-free diet in gluten-related disorders. Dig Dis. 2013;31:57–62. doi: 10.1159/000347180. [DOI] [PubMed] [Google Scholar]
- 4.Sperandeo MP, Tosco A, Izzo V, Tucci F, Troncone R, Auricchio R, Romanos J, Trynka G, Auricchio S, Jabri B, Greco L. Potential celiac patients: a model of celiac disease pathogenesis. PLoS One. 2001;6:e21281. doi: 10.1371/journal.pone.0021281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahn R, Ding YC, Murray J, Fasano A, Green PH, Neuhausen SL, Garner C. Association analysis of the extended MHC region in celiac disease implicates multiple independent susceptibility loci. PLoS One. 2012;7:e36926. doi: 10.1371/journal.pone.0036926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trynka G, Wijmenga C, van Heel DA. A genetic perspective on coeliac disease. Trends Mol Med. 2010;16:537–550. doi: 10.1016/j.molmed.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 7.van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, Wapenaar MC, Barnardo MC, Bethel G, Holmes GK, Feighery C, Jewell D, Kelleher D, Kumar P, Travis S, Walters JR, Sanders DS, Howdle P, Swift J, Playford RJ, McLaren WM, Mearin ML, Mulder CJ, McManus R, McGinnis R, Cardon LR, Deloukas P, Wijmenga C. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, Romanos J, Dinesen LC, Ryan AW, Panesar D, Gwilliam R, Takeuchi F, McLaren WM, Holmes GK, Howdle PD, Walters JR, Sanders DS, Playford RJ, Trynka G, Mulder CJ, Mearin ML, Verbeek WH, Trimble V, Stevens FM, O’Morain C, Kennedy NP, Kelleher D, Pennington DJ, Strachan DP, McArdle WL, Mein CA, Wapenaar MC, Deloukas P, McGinnis R, McManus R, Wijmenga C, van Heel DA. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coenen MJ, Trynka G, Heskamp S, Franke B, van Diemen CC, Smolonska J, van Leeuwen M, Brouwer E, Boezen MH, Postma DS, Platteel M, Zanen P, Lammers JW, Groen HJ, Mali WP, Mulder CJ, Tack GJ, Verbeek WH, Wolters VM, Houwen RH, Mearin ML, van Heel DA, Radstake TR, van Riel PL, Wijmenga C, Barrera P, Zhernakova A. Common and different genetic background for rheumatoid arthritis and coeliac disease. Hum Mol Genet. 2009;18:4195–4203. doi: 10.1093/hmg/ddp365. [DOI] [PubMed] [Google Scholar]
- 10.Zhernakova A, Stahl EA, Trynka G, Raychaudhuri S, Festen EA, Franke L, Westra HJ, Fehrmann RS, Kurreeman FA, Thomson B, Gupta N, Romanos J, McManus R, Ryan AW, Turner G, Brouwer E, Posthumus MD, Remmers EF, Tucci F, Toes R, Grandone E, Mazzilli MC, Rybak A, Cukrowska B, Coenen MJ, Radstake TR, van Riel PL, Li Y, de Bakker PI, Gregersen PK, Worthington J, Siminovitch KA, Klareskog L, Huizinga TW, Wijmenga C, Plenge RM. Metaanalysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bähler M, Kehrer I, Gordon L, Stoffler HE, Olsen AS. Physical mapping of human myosin-IXB (MYO9B), the human orthologue of the rat myosin myr 5, to chromosome 19p13.1. Genomics. 1997;43:107–109. doi: 10.1006/geno.1997.4776. [DOI] [PubMed] [Google Scholar]
- 12.Post PL, Tyska MJ, O’Connell CB, Johung K, Hayward A, Mooseker MS. Myosin-IXb is a single-headed and processive motor. J Biol Chem. 2002;277:11679–11683. doi: 10.1074/jbc.M111173200. [DOI] [PubMed] [Google Scholar]
- 13.Matter K, Balda MS. Signalling to and from tight junctions. Nat Rev Mol Cell Biol. 2003;4:225–236. doi: 10.1038/nrm1055. [DOI] [PubMed] [Google Scholar]
- 14.Sapone A, de Magistris L, Pietzak M, Clemente MG, Tripathi A, Cucca F, Lampis R, Kryszak D, Cartenì M, Generoso M, Iafusco D, Prisco F, Laghi F, Riegler G, Carratu R, Counts D, Fasano A. Zonulin upregulation is associated with increased gut permeability in subjects withtype 1 diabetes and their relatives. Diabetes. 2006;55:1443–1449. doi: 10.2337/db05-1593. [DOI] [PubMed] [Google Scholar]
- 15.Wirth JA, Jensen KA, Post PL, Bement WM, Mooseker MS. Human myosin-IXb, an unconventional myosin with a chimerin-like rho/rac GTPase-activating protein domain in its tail. J Cell Sci. 1996;109:653–661. doi: 10.1242/jcs.109.3.653. [DOI] [PubMed] [Google Scholar]
- 16.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 17.Hunt KA, Monsuur AJ, McArdle WL, Kumar PJ, Travis SP, Walters JR, Jewell DP, Strachan DP, Playford RJ, Wijmenga C, van Heel DA. Lack of association of MYO9B genetic variants with coeliac disease in a Britishcohort. Gut. 2006;55:969–972. doi: 10.1136/gut.2005.086769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Núñez C, Márquez A, Varadé J, Martínez A, Polanco I, Maluenda C, Fernández-Arquero M, de la Concha EG, Urcelay E. No evidence of association of the MYO9B polymorphisms with celiac disease in the Spanish population. Tissue Antigens. 2006;68:489–492. doi: 10.1111/j.1399-0039.2006.00699.x. [DOI] [PubMed] [Google Scholar]
- 19.Cirillo G, Di Domenico MR, Corsi I, Gagliardo T, Del Giudice EM, Perrone L, Tolone C. Do MYO9B genetic variants predispose to coeliac disease? An association study in acohort of South Italian children. Dig Liver Dis. 2007;39:228–231. doi: 10.1016/j.dld.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 20.Sánchez E, Alizadeh BZ, Valdigem G, Ortego-Centeno N, Jiménez-Alonso J, de Ramón E, García A, López-Nevot MA, Wijmenga C, Martín J, Koeleman BP. MYO9B gene polymorphisms are associated with autoimmune diseases in Spanish population. Hum Immunol. 2007;68:610–615. doi: 10.1016/j.humimm.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Latiano A, Mora B, Bonamico M, Megiorni F, Mazzilli MC, Cucchiara S, Palmieri O, Valvano MR, Annese V. Analysis of candidate genes on chromosomes 5q and 19p in celiac disease. J Pediatr Gastroenterol Nutr. 2007;45:180–186. doi: 10.1097/MPG.0b013e3180616bd2. [DOI] [PubMed] [Google Scholar]
- 22.Wolters VM, Verbeek WH, Zhernakova A, Onland-Moret C, Schreurs MW, Monsuur AJ, Verduijn W, Wijmenga C, Mulder CJ. The MYO9B gene is a strong risk factor for developing refractory celiac disease. Clin Gastroenterol Hepatol. 2007;5:1399–1405. 1405.e1–2. doi: 10.1016/j.cgh.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 23.Loeff T, Araya M, Pérez-Bravo F. Frequency of MYO9B polymorphisms in celiac patients and controls. Rev Esp Enferm Dig. 2012;104:566–571. doi: 10.4321/s1130-01082012001100003. [DOI] [PubMed] [Google Scholar]
- 24.Schlegel N, Meir M, Spindler V, Germer CT, Waschke J. Differential role of Rho GTPases in intestinal epithelial barrier regulation in vitro. J Cell Physiol. 2011;226:1196–1203. doi: 10.1002/jcp.22446. [DOI] [PubMed] [Google Scholar]
- 25.Post PL, Tyska MJ, O’Connell CB, Johung K, Hayward A, Mooseker MS. Myosin-IXb is a single-headed and processive motor. J Biol Chem. 2002;277:11679–11683. doi: 10.1074/jbc.M111173200. [DOI] [PubMed] [Google Scholar]
- 26.Liao W, Elfrink K, Bähler M. Head of myosin IX binds calmodulin and moves processively toward the plus-end of actin filaments. J Biol Chem. 2010;285:24933–24942. doi: 10.1074/jbc.M110.101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanley PJ, Xu Y, Kronlage M, Grobe K, Schön P, Song J, Sorokin L, Schwab A, Bähler M. Motorized RhoGAP myosin IXb (Myo9b) controls cell shape and motility. Proc Natl Acad Sci U S A. 2010;107:12145–12150. doi: 10.1073/pnas.0911986107. [DOI] [PMC free article] [PubMed] [Google Scholar]


