Abstract
X-ray repair cross-complementing group 1 (XRCC1) plays an important role in the base excision repair. Many studies have reported the association of XRCC1 Arg399Gln, Arg194Trp and Arg280His polymorphisms with lung cancer risk, but the results remained controversial. In this meta-analysis, we performed a meta-analysis of ten published case-control studies in Caucasian populations to investigate the associations between lung cancer risk and XRCC1 Arg399Gln (2187 cases and 3453 controls from ten studies), Arg194Trp (857 cases and 2108 controls from six studies) and Arg280His (894 cases and 1133 controls from five studies). The results in total population showed that XRCC1 codon 399 polymorphism (OR=0.93, 95% CI=0.82-1.04) and codon 194 (OR=0.94, 95% CI=0.73-1.21) was significantly associated with lung cancer risk. However, no association was found between lung cancer risk and codon 280 (OR=1.17, 95% CI=0.89-1.54). In conclusion, this meta-analysis has demonstrated that codon 399 and codon 194 might have contributed to individual susceptibility to lung cancer in Caucasian populations. To further evaluate effect of XRCC1 polymorphisms, large studies with thousands of subjects are required to get conclusive results.
Keywords: X-ray repair cross-complementing group 1, lung cancer, polymorphism, meta-analysis
Introduction
Lung cancer is a major cause of cancer-related death in the worldwide and the overall survival rate has still an extremely poor [1]. According to cancer statistics 2012, lung cancer is expected to account for 26% of all female cancer deaths and 29% of all male cancer deaths [2]. Although cigarette smoking remains the predominant cause of lung cancer, it cannot fully explain epidemiologic characteristics of lung cancer in nonsmokers [3]. Currently, genetic susceptibility to environmental or occupational diseases is believed to play an important role in determining individual differences in the development of lung cancer [4,5]. Moreover, genetic variations in DNA repair genes have been reported to be associated with the genomic instability and increasing risk of genomic damages [6].
X-ray repair cross-complementing group 1 (XRCC1), one of the >20 genes that participate in base excision repair (BER) pathway, encodes a protein that function in the repair of single-strand breaks [7]. XRCC1 plays a central role in the BER pathway by interacting with other DNA repair proteins [8,9], giving the possibility that XRCC1 has some relationship with the response to therapy and the overall survival of lung cancer.
Three single nucleotide polymorphisms (SNPs) in XRCC1, Arg194Trp (exon 7), Arg280His (exon 10) and Arg399Gln (exon 11), are common and the most-studied SNPs in the XRCC1 gene. Some studies have reported the relationship between polymorphisms in XRCC1 gene and the risk of lung cancer patients [10-12], however the results were inconsistent. For Arg399Gln, many studies have shown that Arg399Gln is obvious associated with increased risk of lung cancer [13,14]; while some non-significant or negative association are also reported by other studies [15,16]. Furthermore, research has suggested that the effect of the XRCC1 SNPs on lung cancer may be dependent on ethnicity [17]. So we performed a systemic review and meta-analysis to assess the association of XRCC1 polymorphisms on the risk of lung cancer.
Materials and methods
Identification and eligibility of relevant studies
We conducted a comprehensive literature search using the database of PubMed, Springer and Elsevier for relevant articles published in English between December 2003 and January 2012. We retrieved the relevant articles using the following terms: “XRCC1”, “X-ray repair cross complementing protein 1”, “lung cancer” and “polymorphism”. Only full-text articles and the most recent studies were included in this meta-analysis.
Criteria for inclusion
The inclusion criteria were as follows: 1) the paper should be case-control or cohort association studies of lung cancer in Caucasian people with XRCC1 polymorphisms; 2) the paper should be included at least one of the three polymorphisms, Arg399Gln, Arg194Trp, and Arg280His; 3) the results were expressed as odds ratio (OR) and corresponding 95 percent confidence interval (95% CI); and 4) genotype distribution of control for a certain polymorphism must be in Hardy-Weinberg equilibrium (HWE).
The exclusion criteria were: 1) reviews or conference papers; 2) without control group; 3) studies with duplicate data; and 4) genotype information couldn’t be extracted.
Quality assessment and data extraction
Two investigators independently extracted data and reached a consensus on all of the items. Any disagreement was subsequently resolved by discussion with another expert. The following information was extracted from each article: first author, year of publication, the exact data of total and exposed number in case and control groups, and genotyping information. Furthermore, we examined whether matching had been used and if the genotyping assay had been validated
Statistical analysis
The risks (ORs) of lung cancer associated with the XRCC1 polymorphisms were calculated directly from the data given in the eligible studies. We estimated the risks of the combined variant genotypes (i.e. Arg/Trp and Trp/Trp for Arg194Trp, and Arg/His and His/His for Arg280His, Arg/Gln and Gln/Gln for Arg399Gln) versus their wild genotypes (Arg/Arg). Furthermore, in the analysis of pooled data, we combined using a fixed-effects model (the inverse variance-weighted method) and a random effects model (DerSimonian and Laird method) [18,19]. The fixed-effects model is used when the effects are assumed to be homogenous, while the random effects model is used when they are heterogenous. Tests for heterogeneity between studies were performed with the Chi-square based Q test. The funnel plot and Egger’s test were used to diagnose publication bias [20].
To assess whether our results were substantially influenced by the presence of any individual study, we conducted a sensitivity analysis by systematically removing each study and recalculating the significance of the result.
All analyses were conducted in Review Manager (version 5.2, The Cochrane Collaboration). All the tests were two-sided and the significant level was 0.05.
Results
Literature search and meta-analysis databases
Relevant publications were retrieved and preliminarily screened [21-30]. Table 1 list the essential information such as first author, the publication year and the numbers of lung cancer cases and controls for three XRCC1 polymorphisms, Arg399Gln, Arg194Trp, and Arg280His, respectively. There were ten case-control studies concerning the XRCC1 polymorphisms, including 3938 cases and 6694 controls.
Table 1.
Characteristics of the studies included in the meta-analysis
| First author-published year | Lung cancer cases | Controls | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| 194 | 280 | 399 | 194 | 280 | 399 | |
| Vogel-2004 | 27/256 | 139/256 | 28/269 | 161/269 | ||
| Zienolddiny-2006 | 27/336 | 33/329 | 202/331 | 37/405 | 27/377 | 240/391 |
| Matullo-2006 | 18/116 | 65/116 | 143/1094 | 610/1094 | ||
| Ryk-2006 | 100/177 | 94/153 | ||||
| De Ruyck-2007 | 9/110 | 4/109 | 71/109 | 17/110 | 14/110 | 63/109 |
| López-Cima-2007 | 294/516 | 316/533 | ||||
| Improta-2008 | 16/94 | 52/94 | 17/121 | 68/121 | ||
| Cote-2009 | 331/387 | 360/406 | ||||
| Chang-2009 | 24/113 | 26/112 | 59/113 | 76/299 | 56/298 | 143/298 |
| Janik-2011 | 14/88 | 22/88 | 24/88 | 10/79 | 12/79 | 28/79 |
Codon 194: Arg/Trp and Trp/Trp genotypes compared with the total genotype. Codon 280: Arg/His and His/His genotypes compared with the total genotype. Codon 399: Arg/Gln and Gln/Gln genotypes compared with the total genotype.
Test of heterogeneity
All the ten case-control studies concern the Arg399Gln polymorphism, including 2187 cases and 3453 controls. Figure 1 shows the association between the Arg399Gln polymorphism and lung cancer risk. We analyzed the heterogeneity for all ten case-control studies and the test value of Chi-square was 6.35 with 9 degree of freedom (d.f.) and P=0.70 in a fixed model.
Figure 1.
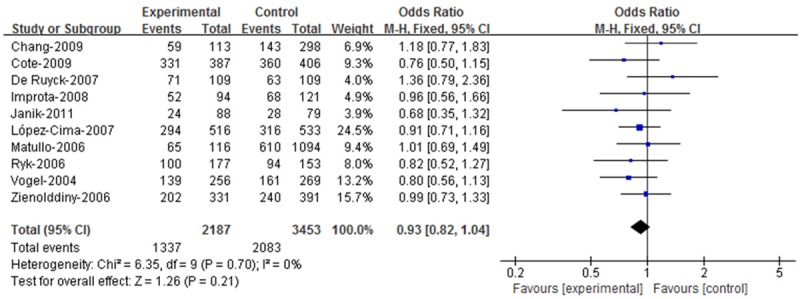
Meta-analysis with a fixed effect model for the ORs of lung cancer associated with XRCC1 Codon 399 for the Arg/Gln and Gln/Gln genotypes compared with the Arg/Arg genotype.
A total of six case-control studies concern the Arg194Trp polymorphism, including 857 cases and 2108 controls. Figure 2 showed the association between the Arg194Trp polymorphism and lung cancer risk. The Chi-square value for the heterogeneity of all six case-control studies was 4.82 with 5 d.f. and P=0.44 in a fixed model.
Figure 2.
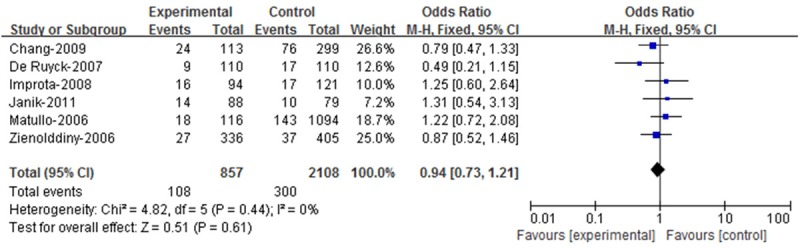
Meta-analysis with a fixed effect model for the ORs of lung cancer associated with XRCC1 codon 194 for the Arg/Trp and Trp/Trp genotypes compared with the Arg/Arg genotype.
There are five case-control studies concerning the Arg280His polymorphism, including 894 cases and 1133 controls. Figure 3 showed the association between genetic polymorphism of Arg280His and lung cancer risk. However, the Chi-square value for the heterogeneity of the five case-control studies was 8.97 with 4 d.f. and P=0.006 in a random-effect model.
Figure 3.
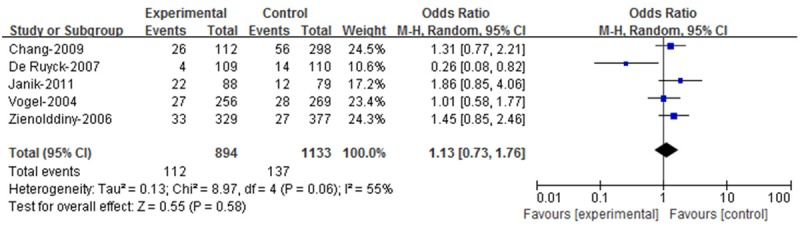
Meta-analysis with a fixed effect model for the ORs of lung cancer associated with XRCC1 codon 280 for the Arg/His and His/His genotypes compared with the Arg/Arg genotype.
Meta-analysis
The risks of lung cancer associated with XRCC1 genetic polymorphisms were estimated for each study. For the XRCC1 Arg399Gln polymorphism, the eligible studies included 1337 cases and 2083 controls which had the combined variant genotypes (Arg/Gln and Gln/Gln), and 850 cases and 1370 controls which had wild-type homozygote (Arg/Arg) of the XRCC1 Arg399Gln gene. The overall OR for the combined genotypes Arg/Gln and Gln/Gln versus Arg/Arg genotype was 0.93 (95% CI=0.82-1.04) in a fixed model (Z=1.26, P=0.21).
For the XRCC1 Arg194Trp polymorphism, 108 cases and 300 controls had the combined variant genotypes (Arg/Trp and Trp/Trp) and 749 cases and 1808 controls were wild-type homozygote (Arg/Arg) for the XRCC1 Arg194Trp gene in the eligible studies. The overall OR for the combined Arg/Trp and Trp/Trp genotypes versus Arg/Arg genotype was 0.94 (95% CI=0.73-1.21) in a fixed model (Z=0.51, P=0.61).
For the XRCC1 Arg280His polymorphism, the eligible studies had 112 cases and 137 controls combing the variant genotypes (Arg/His and His/His) and 782 cases and 996 controls including the wild-type homozygote (Arg/Arg) for the XRCC1 Arg280His gene. The overall OR for the combined variant genotypes versus the wild-type genotype was 1.13 (95% CI=0.73-1.76) as estimated in a random-effect mode (Z=0.55, P=0.58).
Sensitivity analyses and publication bias
A single study included in the meta-analysis was deleted each time to reflect the influence of the individual data set to the pooled ORs, and the corresponding pooled ORs were not materially changed.
The funnel plot was used to graphically assess the publication bias. For the three XRCC1 polymorphisms, Arg399Gln, Arg194Trp and Arg280His, the shapes of the funnel plots appeared to be approximately symmetrical, suggesting that the publication bias can be neglected, and the magnitude of the main ORs was in dispersion around 1 (Figures 4, 5 and 6). This meta-analysis indicated that publication biases might not have a significant effect on the results of three XRCC1 genes.
Figure 4.
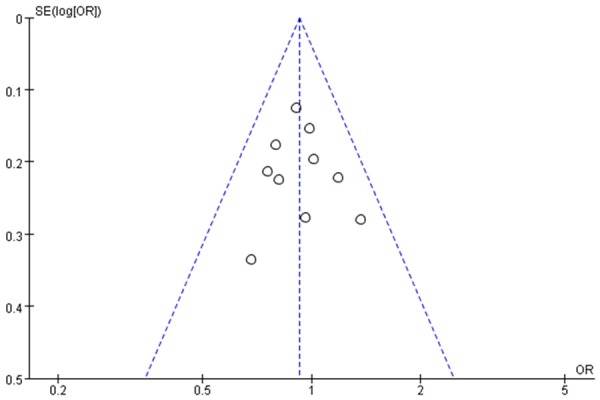
Funnel plot of the meta-analysis of lung cancer risk and the XRCC1 Arg399Gln polymorphism (Arg/Gln + Gln/Gln versus Arg/Arg).
Figure 5.
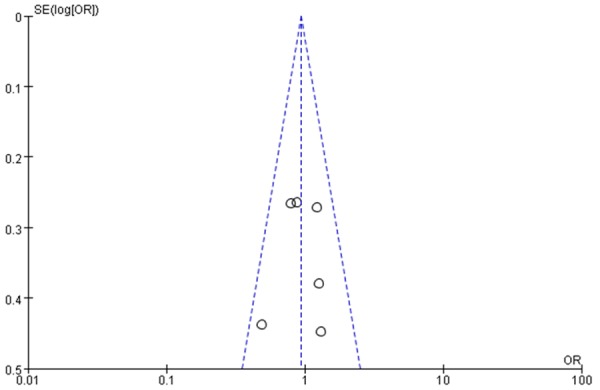
Funnel plot of the meta-analysis of lung cancer risk and the XRCC1 Arg194Trp polymorphism (Arg/Trp + Trp/Trp versus Arg/Arg).
Figure 6.
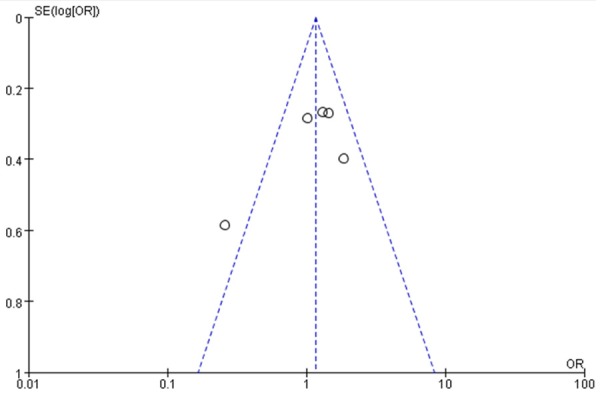
Funnel plot of the meta-analysis of lung cancer risk and the XRCC1 Arg280His polymorphism (Arg/His + His/His versus Arg/Arg).
Discussion
DNA repair mechanisms are important for maintaining genome integrity and preventing carcinogenesis [31]. XRCC1, an important component of the BER pathway, has multiple roles in repairing DNA base damage and single-strand DNA breaks [32,33]. A large number of molecular epidemiological studies have been conducted to evaluate the role of polymorphisms in the XRCC1 gene on lung cancer risk [34-36]; However, these original results are inconsistent and until now the lack of systematic review evaluation failed to give further insights on this issue. In this study, we carried out a meta-analysis of codon 194, codon 280 and codon 399 polymorphisms in XRCC1 gene to estimate the association of these three polymorphisms with lung cancer risk. Our results of this meta-analysis indicate that genetic variations of XRCC1 Arg399Gln and Arg194Trp may contribute to inter-individual susceptibility to lung cancer in Caucasian populations.
Three polymorphisms in XRCC1 Arg194Trp, Arg280His and Arg399Gln have been frequently examined in the studies on cancer susceptibility. The XRCC1 Arg399Gln polymorphism was the most common sequence variant among the three XRCC1 polymorphisms. Studies have shown that genetic polymorphism of XRCC1 Arg399Gln was associated with risk of lung cancer [37,38], and might be a candidate for contributing inter-individual difference in the overall survival of gemcitabine/platinum-treated advanced Non-Small-Cell Lung cancer patients [39]. However, several studies found no association, and one reported a protective effect of the variant allele [40-42]. Furthermore, two meta-analyses have demonstrated that codon 399 polymorphisms of XRCC1 gene might have contributed to individual susceptibility to lung cancer [36,43]; While two other meta-analyses have shown that there is no association between XRCC1 Arg399Gln polymorphism and lung cancer risk [36,44]. In our meta-analysis of the XRCC1 gene in Caucasian populations, the combined variant genotype (Arg/Gln and Gln/Gln) of the XRCC1 Arg399Gln polymorphism was significantly associated with lung cancer risk, as obtained from ten studies.
Previous meta-analysis of XRCC1 Arg194Trp polymorphisms on the risk of lung cancer showed that homozygous Trp/Trp variant genotype could increase lung cancer risk in total population, especially in Asians; However, the heterozygote Arg/Trp variant genotype might decrease the risk of lung cancer, especially in whites [35]. Our result showed that the combined variant genotype (Arg/Trp and Trp/Trp) of XRCC1 Arg194Trp polymorphism was also significantly associated with lung cancer risk, as obtained from six studies, whereas no statistically significant associations with lung cancer risk was observed for the XRCC1 Arg280His polymorphisms from five studies. However, the association between XRCC1 polymorphism and lung cancer risk was not consistent with previously reported meta-analyses [17,45]: one has demonstrated that the XRCC1 Arg399Gln polymorphism was associated with an increased risk of lung cancer among Asians, but not among Caucasians; the other has demonstrated that the XRCC1 Arg280His polymorphisms may be biomarkers of cancer susceptibility and may play a role in cancer development [46].
Although we have put considerable effort and resources into testing possible association between XRCC1 gene polymorphisms and lung cancer risk, there are still some limitations inherited from the published studies. Firstly, lung cancer is a complex disease, and there are complex interactions between genetic background and environmental factors especially tobacco smoking. Secondly, gene-gene interactions were also possible in the association between XRCC1 polymorphism and lung cancer risk. Finally, selection bias could have played a role because all of the studies selected in this meta-analysis were published in English. Therefore, further studies are needed to assess the possible associations.
In summary, our meta-analysis measured the association between genetic polymorphisms in XRCC1 and lung cancer risk, and suggested that variant genotypes of Arg399Gln and Arg194Trp, but not Arg280His, might alter inter-individual susceptibility to lung cancer in Caucasian populations. Besides, further studies are needed to assess the possible gene-gene or gene-environment interactions in the association above.
Acknowledgements
This study is supported by the Natural Science Foundation of China (81000974) and the Ph.D. Programs Foundation of Ministry of Education of China (20100141120015).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 5.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 6.Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 7.Caldecott KW, Tucker JD, Stanker LH, Thompson LH. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through proteinprotein interactions. EMBO J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu AL, Li X, Gu Y, Wright PM, Chang DY. Repair of oxidative DNA damage: mechanisms and functions. Cell Biochem Biophys. 2001;35:141–170. doi: 10.1385/CBB:35:2:141. [DOI] [PubMed] [Google Scholar]
- 10.Kiyohara C, Takayama K, Nakanishi Y. Lung cancer risk and genetic polymorphisms in DNA repair pathways: a meta-analysis. J Nucleic Acids. 2010;2010:701760. doi: 10.4061/2010/701760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo S, Li X, Gao M, Li Y, Song B, Niu W. The relationship between XRCC1 and XRCC3 gene polymorphisms and lung cancer risk in northeastern Chinese. PLoS One. 2013;8:e56213. doi: 10.1371/journal.pone.0056213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian B, Zhang H, Zhang L, Zhou X, Yu H, Chen K. Association of genetic polymorphisms in DNA repair pathway genes with non-small cell lung cancer risk. Lung Cancer. 2011;73:138–146. doi: 10.1016/j.lungcan.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Huang Y, Cao YS, Zeng J, Tong WN, Xu SL, Zhuo AS. Assessment of the association between XRCC1 Arg399Gln polymorphism and lung cancer in Chinese. Tumour Biol. 2013;34:3681–5. doi: 10.1007/s13277-013-0950-5. [DOI] [PubMed] [Google Scholar]
- 14.Zheng H, Wang Z, Shi X. XRCC1 polymorphisms and lung cancer risk in Chinese populations: a meta-analysis. Lung Cancer. 2009;65:268–273. doi: 10.1016/j.lungcan.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Hou SM, Ryk C, Kannio A, Angelini S, Falt S, Nyberg F, Husgafvel-Pursiainen K. Influence of common XPD and XRCC1 variant alleles on p53 mutations in lung tumors. Environ Mol Mutagen. 2003;41:37–42. doi: 10.1002/em.10128. [DOI] [PubMed] [Google Scholar]
- 16.Gao WM, Romkes M, Day RD, Siegfried JM, Luketich JD, Mady HH, Melhem MF, Keohavong P. Association of the DNA repair gene XPD Asp312Asn polymorphism with p53 gene mutations in tobacco-related non-small cell lung cancer. Carcinogenesis. 2003;24:1671–1676. doi: 10.1093/carcin/bgg115. [DOI] [PubMed] [Google Scholar]
- 17.Kiyohara C, Takayama K, Nakanishi Y. Association of genetic polymorphisms in the base excision repair pathway with lung cancer risk: a meta-analysis. Lung Cancer. 2006;54:267–283. doi: 10.1016/j.lungcan.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 19.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 20.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogel U, Nexo BA, Wallin H, Overvad K, Tjonneland A, Raaschou-Nielsen O. No association between base excision repair gene polymorphisms and risk of lung cancer. Biochem Genet. 2004;42:453–460. doi: 10.1023/b:bigi.0000043957.03420.7e. [DOI] [PubMed] [Google Scholar]
- 22.Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland L, Phillips DH, Canzian F, Haugen A. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27:560–567. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]
- 23.Matullo G, Dunning AM, Guarrera S, Baynes C, Polidoro S, Garte S, Autrup H, Malaveille C, Peluso M, Airoldi L, Veglia F, Gormally E, Hoek G, Krzyzanowski M, Overvad K, Raaschou-Nielsen O, Clavel-Chapelon F, Linseisen J, Boeing H, Trichopoulou A, Palli D, Krogh V, Tumino R, Panico S, Bueno-De-Mesquita HB, Peeters PH, Lund E, Pera G, Martinez C, Dorronsoro M, Barricarte A, Tormo MJ, Quiros JR, Day NE, Key TJ, Saracci R, Kaaks R, Riboli E, Vineis P. DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis. 2006;27:997–1007. doi: 10.1093/carcin/bgi280. [DOI] [PubMed] [Google Scholar]
- 24.Ryk C, Kumar R, Thirumaran RK, Hou SM. Polymorphisms in the DNA repair genes XRCC1, APEX1, XRCC3 and NBS1, and the risk for lung cancer in never- and ever-smokers. Lung Cancer. 2006;54:285–292. doi: 10.1016/j.lungcan.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 25.De Ruyck K, Szaumkessel M, De Rudder I, Dehoorne A, Vral A, Claes K, Velghe A, Van Meerbeeck J, Thierens H. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat Res. 2007;631:101–110. doi: 10.1016/j.mrgentox.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Cima MF, Gonzalez-Arriaga P, Garcia-Castro L, Pascual T, Marron MG, Puente XS, Tardon A. Polymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and lung cancer risk in a population of northern Spain. BMC Cancer. 2007;7:162. doi: 10.1186/1471-2407-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Improta G, Sgambato A, Bianchino G, Zupa A, Grieco V, La Torre G, Traficante A, Cittadini A. Polymorphisms of the DNA repair genes XRCC1 and XRCC3 and risk of lung and colorectal cancer: a case-control study in a Southern Italian population. Anticancer Res. 2008;28:2941–2946. [PubMed] [Google Scholar]
- 28.Cote ML, Yoo W, Wenzlaff AS, Prysak GM, Santer SK, Claeys GB, Van Dyke AL, Land SJ, Schwartz AG. Tobacco and estrogen metabolic polymorphisms and risk of non-small cell lung cancer in women. Carcinogenesis. 2009;30:626–635. doi: 10.1093/carcin/bgp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janik J, Swoboda M, Janowska B, Ciesla JM, Gackowski D, Kowalewski J, Olinski R, Tudek B, Speina E. 8-Oxoguanine incision activity is impaired in lung tissues of NSCLC patients with the polymorphism of OGG1 and XRCC1 genes. Mutat Res. 2011;709-710:21–31. doi: 10.1016/j.mrfmmm.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Chang JS, Wrensch MR, Hansen HM, Sison JD, Aldrich MC, Quesenberry CP Jr, Seldin MF, Kelsey KT, Wiencke JK. Base excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African-Americans. Carcinogenesis. 2009;30:78–87. doi: 10.1093/carcin/bgn261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furgason JM, Bahassi el M. Targeting DNA repair mechanisms in cancer. Pharmacol Ther. 2013;137:298–308. doi: 10.1016/j.pharmthera.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 32.Cappelli E, Taylor R, Cevasco M, Abbondandolo A, Caldecott K, Frosina G. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J Biol Chem. 1997;272:23970–23975. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- 33.Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang CH, Jang BG, Kim DW, Chung DH, Kim YT, Jheon S, Sung SW, Kim JH. The prognostic significance of ERCC1, BRCA1, XRCC1, and betaIII-tubulin expression in patients with nonsmall cell lung cancer treated by platinum- and taxane-based neoadjuvant chemotherapy and surgical resection. Lung Cancer. 2010;68:478–483. doi: 10.1016/j.lungcan.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Jiang J, Liang X, Zhou X, Huang R, Chu Z, Zhan Q, Lin H. DNA repair gene X-ray repair cross complementing group 1 Arg194Trp polymorphism on the risk of lung cancer: a meta-analysis on 22 studies. J Thorac Oncol. 2010;5:1741–1747. doi: 10.1097/JTO.0b013e3181f0c409. [DOI] [PubMed] [Google Scholar]
- 36.Dai L, Duan F, Wang P, Song C, Wang K, Zhang J. XRCC1 gene polymorphisms and lung cancer susceptibility: a meta-analysis of 44 case-control studies. Mol Biol Rep. 2012;39:9535–9547. doi: 10.1007/s11033-012-1818-2. [DOI] [PubMed] [Google Scholar]
- 37.Butkiewicz D, Rusin M, Enewold L, Shields PG, Chorazy M, Harris CC. Genetic polymorphisms in DNA repair genes and risk of lung cancer. Carcinogenesis. 2001;22:593–597. doi: 10.1093/carcin/22.4.593. [DOI] [PubMed] [Google Scholar]
- 38.David-Beabes GL, London SJ. Genetic polymorphism of XRCC1 and lung cancer risk among African-Americans and Caucasians. Lung Cancer. 2001;34:333–339. doi: 10.1016/s0169-5002(01)00256-2. [DOI] [PubMed] [Google Scholar]
- 39.Liao WY, Shih JY, Chang GC, Cheng YK, Yang JC, Chen YM, Yu CJ. Genetic polymorphism of XRCC1 Arg399Gln is associated with survival in non-small-cell lung cancer patients treated with gemcitabine/platinum. J Thorac Oncol. 2012;7:973–981. doi: 10.1097/JTO.0b013e31824fe98c. [DOI] [PubMed] [Google Scholar]
- 40.Zhou W, Liu G, Miller DP, Thurston SW, Xu LL, Wain JC, Lynch TJ, Su L, Christiani DC. Polymorphisms in the DNA repair genes XRCC1 and ERCC2, smoking, and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2003;12:359–365. [PubMed] [Google Scholar]
- 41.Misra RR, Ratnasinghe D, Tangrea JA, Virtamo J, Andersen MR, Barrett M, Taylor PR, Albanes D. Polymorphisms in the DNA repair genes XPD, XRCC1, XRCC3, and APE/ref-1, and the risk of lung cancer among male smokers in Finland. Cancer Lett. 2003;191:171–178. doi: 10.1016/s0304-3835(02)00638-9. [DOI] [PubMed] [Google Scholar]
- 42.Park JY, Lee SY, Jeon HS, Bae NC, Chae SC, Joo S, Kim CH, Park JH, Kam S, Kim IS, Jung TH. Polymorphism of the DNA repair gene XRCC1 and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2002;11:23–27. [PubMed] [Google Scholar]
- 43.Wang Y, Yang H, Li H, Li L, Wang H, Liu C, Zheng Y. Association between X-ray repair cross complementing group 1 codon 399 and 194 polymorphisms and lung cancer risk: a meta-analysis. Cancer Lett. 2009;285:134–140. doi: 10.1016/j.canlet.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 44.Wu J, Liu J, Zhou Y, Ying J, Zou H, Guo S, Wang L, Zhao N, Hu J, Lu D, Jin L, Li Q, Wang JC. Predictive value of XRCC1 gene polymorphisms on platinum-based chemotherapy in advanced non-small cell lung cancer patients: a systematic review and meta-analysis. Clin Cancer Res. 2012;18:3972–3981. doi: 10.1158/1078-0432.CCR-11-1531. [DOI] [PubMed] [Google Scholar]
- 45.Hu Z, Ma H, Chen F, Wei Q, Shen H. XRCC1 polymorphisms and cancer risk: a meta-analysis of 38 case-control studies. Cancer Epidemiol Biomarkers Prev. 2005;14:1810–1818. doi: 10.1158/1055-9965.EPI-04-0793. [DOI] [PubMed] [Google Scholar]
- 46.Zhang K, Zhou B, Wang Y, Rao L, Zhang L. The XRCC1 Arg280His polymorphism contributes to cancer susceptibility: an update by meta-analysis of 53 individual studies. Gene. 2012;510:93–101. doi: 10.1016/j.gene.2012.08.039. [DOI] [PubMed] [Google Scholar]


