Abstract
The deregulation of miR-101 has been implicated in multiple cancer types including lung cancer, but the exact role, mechanisms and how silencing of miR-101 remain elusive. Here we confirmed miR-101 downregulation in lung cancer cell lines and patient tissues. Restored miR-101 expression remarkably sensitized lung cancer cells to chemotherapy and inhibited invasion. Mechanistically, we indicated that miR-101 inversely correlated with RUNX1 expression, and identified RUNX1 as a novel target of miR-101. RUNX1 impaired the effects of miR-101 on chemotherapeutic sensitization and invasion inhibition. Moreover, RUNX1 knockdown resulted into increase of miR-101 expression and elevation of luciferase activity driven by miR-101 promoter in lung cancer cells, suggesting RUNX1 negatively transcriptionally regulated miR-101 expression via physically binding to miR-101 promoter. These findings support that miR-101 downregulation accelerates the progression of lung cancer via RUNX1 dependent manner and suggest that miR-101/RUNX1 feedback axis may have therapeutic value in treating refractory lung cancer.
Keywords: miR-101, lung cancer, RUNX1, chemotherapy, invasion
Introduction
Lung cancer is the most common type of malignant tumor and the leading cause of cancer-related mortality worldwide [1]. Non-small cell lung cancer (NSCLC) accounts for ~80% of primary lung cancer, and approximately two-thirds of NSCLC patients are diagnosed in advanced stages, which contribute to the high mortality levels. Because the majority of patients present with invasive and metastatic disease, understanding the basis of lung cancer progression is vital [2]. Chemotherapy mostly based on cDDP is useful treatment for patients with NSCLC, but NSCLC usually is insensitive to chemotherapy in a clinical setting, which causes the treatment failure. Many factors, including oncogenes and tumor suppressors have been reported to be involved in lung tumorigenesis or progression, but the underlying mechanisms remain to be fully determined [3].
MicroRNAs (miRNAs) are small non-coding RNAs that usually repress gene expression through partially complementary binding to the target mRNA. The discovery of miRNAs has revealed a new regulatory layer of gene expression that affects the development and progression of various diseases, especially including cancer [4]. Numerous studies have demonstrated that miRNAs can function as oncogenes or tumor suppressor as results of their fundamental roles in diverse cellular processes, such as cell differentiation, proliferation, apoptosis, invasion and metastasis [5]. The aberrant expression of specific miRNAs has been found to be associated with development and clinical outcomes of various cancers, but mechanisms of deregulated miRNAs and their roles in tumorigenesis are still largely unknown. In regard to lung cancer, although there are several reports imply the involvement of deregulated miRNAs in lung cancer, such as miR-148a [6] and miR-21 [7]. Studies investigating miR-101 as a tumor suppressor have recently attracted much attention. Varambally et al. reported the decrease of miR-101 expression during prostate cancer progression by targeting polycomb group protein EZH2 [8]. The regulatory axis of miR-101 targeting EZH2 was subsequently confirmed in aggressive endometrial cancer cells [9], glioblastoma [10] and lung cancer [11]. MiR-101 is able to target multiple oncogenes, including myeloid cell leukemia sequence 1 in hepatocellular carcinoma [12] and cyclooxygenase 2 in gastric cancer [13], Recently, although miR-101 also was reported to suppress lung tumorigenesis through inhibition of DNMT3a [14], the roles of miR-101 and mechanisms of miR-101 deregulation in lung cancer are far from full understanding. In the present study, we confirmed the decrease of miR-101 expression in lung cancer and indicated that miR-101 sensitized lung cancer cells to chemotherapy and inhibited invasion via directly targeting RUNX1 in lung cancer cells. Reciprocally, we identified RUNX1 physically binds to the miR-101 promoter and negatively transcriptionally regulates miR-101 expression. These findings suggest the miR-101/RUNX1 feedback regulatory loop in lung cancer as valuable biomarker and potential therapeutic target.
Materials and methods
Clinical specimens and cell culture
17 paired fresh lung cancer tissues and adjacent morphologically normal lung tissues, and 57 formalin-fixed paraffin-embedded tissues were collected at Renmin Hospital of Wuhan University (Hubei, China). Fresh tissue samples were immediately snap-frozen in liquid nitrogen used for mRNA and miRNA extraction. The study was approved by each of the patients and by the ethics committee of Renmin Hospital of Wuhan University. Human lung cancer cell lines (A549, H460, H1299, H1975), and a normal diploid human cell line of lung fibroblasts MRC5 were obtained from and maintained as recommended by the American Type Culture Collection (ATCC, Manassas, VA, USA). The human lung cancer cell lines 95C and 95D were obtained from the Cell Bank of the Chinese Academy of Science (Shanghai, China). The A549, H460, H1299, H1975, 95C and 95D cells were maintained in RPMI-160 medium supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA). The MRC5 cells were maintained in DMEM medium (Gibco) supplemented with 10% fetal bovine serum. The cells were incubated in an atmosphere of 5% CO2 at 37°C. cDDP was from Sigma-Aldrich (Steinheim, Germany).
Real-time PCR (qRT-CPR) for mRNAs and miRNAs
The miRNAs and mRNAs were extracted simultaneously and purified with miRNA isolation system (Exiqon, Vedbaek, Denmark). For miRNA qRT-PCR, cDNA was generated with the miScript II RT Kit (QIAGEN, Hilden, Germany) and the quantitative real-time PCR (qRT-PCR) was done by using the miScript SYBR Green PCR Kit (QIAGEN) following the manufacturer’s instructions. The miRNA sequence-specific qRT-PCR primers for miR-101 and endogenous control RNU6 were purchased from QIAGEN, and the qRT-PCR analysis was carried out using 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). For mRNA qRT-PCR, cDNAs from the mRNAs were synthesized with the first-strand synthesis system (Thermo Scientific, Glen Brunie, MA, USA). Real-time PCR was carried out according to standard protocols using an ABI 7500 with SYBR Green detection (Applied Biosystems). GAPDH was used as an internal control and the qRT-PCR was repeated three times. The primers for GAPDH were: forward primer 5’-ATTCCATGGCACCGTCAAGGCTGA-3’, reverse primer 5’-TTCTCCATGGTGGTGAAGACGCCA-3’; primers for RUNX1 were: forward primer 5’-CTGCCCATCGCTTTCAAGGT-3’, reverse primer 5’-GCCGAGTAGTTTTCATCATTGCC-3’. The gene expression threshold cycle (CT) values of miRNAs or mRNAs were calculated by normalizing with internal control RNU6 or GAPDH and relative quantization values were calculated.
Western blot
Total proteins were extracted from corresponding cells using the RIPA buffer (Thermo Scientific, Rockford, IL, USA) in the presence of Protease Inhibitor Cocktail (Thermo Scientific). The protein concentration of the lysates was measured using a BCA Protein Assay Kit (Thermo Scientific). Equivalent amounts of protein were resolved and mixed with 5× Lane Marker Reducing Sample Buffer (Thermo Scientific), electrophoresed in a 10% SDS-acrylamide gel and transferred onto Immobilon-P Transfer Membrane (Merck Millipore, Schwalbach, Germany). The membranes were blocked with 5% non-fat milk in Tris-buffered saline and then incubated with primary antibodies followed by secondary antibody. The signal was detected using an ECL detection system (Merck Millipore). The RUNX1 antibody and β-Actin antibody were from Cell Signaling Technology (Danvers, MA, USA). HRP-conjugated secondary antibody was from Thermo.
Immunohistochemistry
The sections were dried at 55°C for 2 h and then deparaffinized in xylene and rehydrated using a series of graded alcohol washes. The tissue slides were then treated with 3% hydrogen peroxide in methanol for 15 min to quench endogenous peroxidase activity and antigen retrieval then performed by incubation in 0.01 M sodium citrate buffer (pH 6.0) and heating using a microwave oven. After a 1 h preincubation in 10% goat serum, the specimens were incubated with primary antibody overnight at 4°C. The tissue slides were treated with a non-biotin horseradish peroxidase detection system according to the manufacturer’s instruction (DAKO, Glostrup, Denmark). Two different pathologists evaluated the immunohistological samples.
Transfection
miR-101 mimics and relative control were purchased from Exiqon (Vedbaek, Denmark). Cells were trypsinised, counted and seeded onto 6-well plates the day before transfection to ensure 70% cell confluence on the day of transfection. The transfection was carried out using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s procedure. The mimics and control were used at a final concentration of 100 nM. At 36 h post-transfection, follow-up experiments were performed. The siRNAs target to RUNX1 and control were purchased from Santa Cruz Biotechnology (Dallas, Texas, USA). The transfection of 50 nM siRNA or control, and 4 μg of the pcDNA3.1-control and pcDNA3.1-RUNX1 plasmids were performed as above, 48 h later, RUNX1 was determined by western blot, and the experiment was repeated four times.
Luciferase reporter assay
For miRNA luciferase reporter assay: The DNA sequences with each 50 base at up-and downstream of miR-101 binding site in the 3’UTR of RUNX1 were synthesized with restriction sites for SpeI and HindIII located at both ends of the oligonucleotides for further cloning, and subsequently cloned into pMir-Report plasmid downstream of firefly luciferase reporter gene. Cells were seeded in 96 well-plates and co-transfected with pMir-Report luciferase vector, pRL-TK Renilla luciferase vector and miR-101 mimics using Lipofectamine 2000 (Invitrogen). For promoter activity assay: To determine whether RUNX1 regulates the promoter activity of miR-101, a two kilobase (kb) region upstream of the miR-101 precursor starting site was cloned into the pGL4-reporter vector upstream of the luciferase gene. Cells were seeded in 96-well plates and co-transfected with the pGL4-reporter vector and the pRL-TK Renilla luciferase vector with or without the si-RUNX1 using Lipofectamine 2000. After transfection of 48 h, luciferase activity was determined using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) on the BioTek Synergy 2. The Renilla luciferase activity was used as internal control and the firefly luciferase activity was calculated as the mean ± SD after being normalized by Renilla luciferase activity.
MTS assay
The CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI, USA) was used to determine the sensitivity of cells to cDDP. Briefly, cells were seeded in 96-well plates at a density of 4×103 cells/well (0.2 ml/well) for 24 h before use. The culture medium was replaced with fresh medium containing cDDP at different concentrations and cells were then incubated for a further 72 h. Then, MTS (0.02 ml/well) was added. After a further 2 h incubation, the absorbance at 490 nm was recorded for each well on the BioTek Synergy 2. The absorbance represented the cell number and was used for the plotting of dose-cell number curves and IC50 values were calculated.
Cell invasion assay
Invasion of cells was assessed using the Cell Invasion Assay Kit (BD Biosciences) according to the manufacturer’s instructions. Briefly, at 36 h post-transfection, 3×104 cells in 300 μL serum-free medium were added to the upper chamber precoated with ECMatrix™ gel. Then, 0.5 ml of 10% FBS-containing medium was added to the lower chamber as a chemoattractant. Cells were incubated for 24 h at 37°C, and then non-invading cells were removed with cotton swabs. Cells that migrated to the bottom of the membrane were fixed with pre-cold methanol and stained with 2% Giemsa solution. Stained cells were visualized under a microscope. To minimize the bias, at least three randomly selected fields with 100× magnification were counted, and the average number was taken.
ChIP-qPCR
The ChIP assay was performed using the EZ-CHIPTM chromatin immunoprecipitation kit (Merck Millipore). Briefly: Chromatin proteins were cross-linked to DNA by addition of formaldehyde to the culture medium to a final concentration of 1%. After a 10 min incubation at room temperature, the cells were washed and scraped off in ice-cold phosphate-buffered saline (PBS) containing Protease Inhibitor Cocktail II. Cells were pelleted and then resuspended in lysis buffer containing Protease Inhibitor Cocktail II. The resulting lysate was subjected to sonication to reduce the size of DNA to approximately 200-1000 base pairs in length. The sample was centrifuged to remove cell debris and diluted ten-fold in ChIP dilution buffer containing Protease Inhibitor Cocktail II. Then 5 µg of anti-RNA Polymerase antibody (positive control, included with the kit), or anti-RUNX1 antibody (cell signal technology) were added to the chromatin solution and incubated overnight at 4°C with rotation. After antibody incubation, protein G agarose was added and the sample incubated at 4°C with rotation for an additional 2 h. The protein/DNA complexes were washed with Wash Buffers four times and eluted with ChIP Elution Buffer. Cross-links were then reversed to free DNA by the addition of 5 M NaCl and incubation at 65°C for 4 h. The DNA was purified according to the manufacturer’s instructions. 50 μL of DNA was obtained for each treatment. 2 μL of DNA from each group was used as a template for PCR. Primers for the miR-101 promoter containing putative RUNX1 binding sites were as follows, sense: 5’-GATACAATGATTACTCTTATAAGACC-3’, antisense: 5’-GGAAAGCCAACAAAAGAGAGGA-3’ (for site A); sense: 5’-CCTCCTCTCTTTTGTTGGCTTTC-3’, antisense: 5’-CCTGCTGTCCAAGGGTCATG-3’ (for site B); sense: 5’-CTGGACTAGTAGGGAATTGTAGAAG-3’, antisense: 5’-CACAGTGTCATTTATAATTCAAGTC-3’ (for site C). Primers for the human GAPDH gene: sense, 5’-TACTAGCGGTTTTACGGGCG-3’, antisense, 5’-TCGAACAGGAGGAGCAGAGAGCGA-3’. The PCR conditions were as follows: 1 cycle of 95°C for 5 min; 40 cycles of 95°C for 20 s, 60°C for 30 s, and 72°C 30 s; and 1 cycle of 72°C for 10 min. The results were calculated by normalizing to the positive control, and relative quantization values were calculated using % positive control = 2^(-ΔCt [(Ct [RUNX1]-(Ct [positive control]]) method.
Statistical analysis
All data are expressed as means ± standard deviation from three independent experiments. Statistical analyses were performed using SPSS16.0 software (SPSS, Chicago, IL). The differences between groups were analyzed using Student’s t-test with only two groups or one-way analysis of variance (ANOVA) when more than two groups were compared. Pearson’s correlation analysis was used to determine the correlation between miR-101 expression and RUNX1 mRNA level in the tissues. P values less than 0.05 were considered statistically significant.
Results
MiR-101 levels are inversely correlated with RUNX1 expression in lung cancer
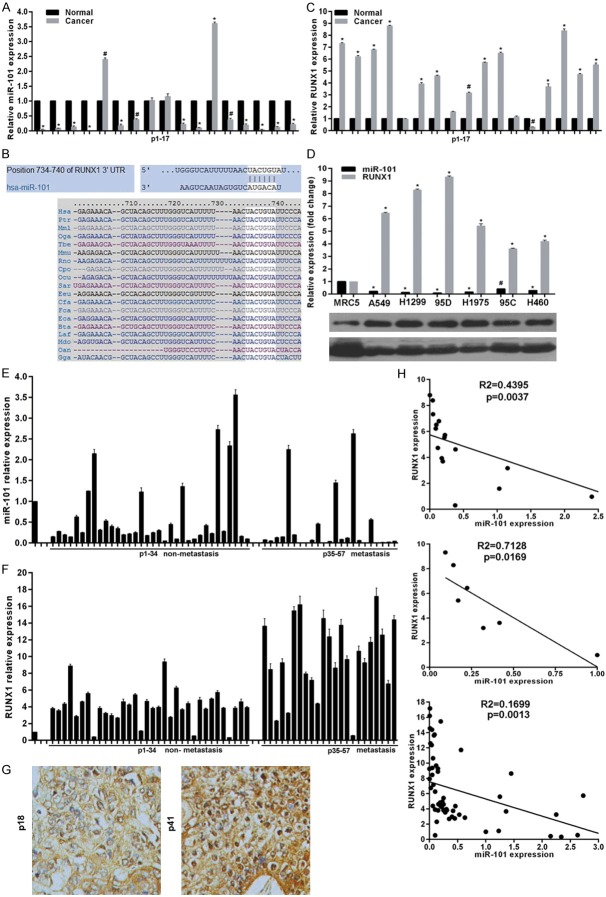
To confirm the miR-101 expression pattern in lung cancer, we initially evaluated the expression levels of miR-101 in fresh-frozen matched non-small cell lung cancer (NSCLC) and adjacent non-tumor lung tissues (n = 17) and found that miR-101 was primarily expressed in normal tissues with a 76.47% reduction in tumors (Figure 1A). To explore novel mechanisms responsible for the role of miR-101 deregulation, potential targets of miR-101 were predicted using the public database-TargetScan (http://www.targetscan.org) and RUNX1 with a critically conserved binding site in the 3’-UTR region of mRNA was selected for further expression and function confirmation (Figure 1B). Expectedly, the detection of RUNX1 expression in above fresh-frozen tissues showed RUNX1 mRNA levels in lung cancer tissues were much higher than in paired non-tumor tissues (Figure 1C). We then assessed miR-101 and RUNX1 expression in one lung fibroblast line MRC5 and six NSCLC. Consistent with the results from above mentioned fresh-frozen tissues, miR-101 levels were highly decreased, whereas RUNX1 mRNA and protein was markedly elevated in lung cancer cells, when compared with MRC5 cells (Figure 1D). We further examined miR-101 and RUNX1 expression in formalin-fixed paraffin-embedded tissues from 57 NSCLC patients among which 23 patients with metastasis. Results revealed that miR-101 expression was dramatically decreased in NSCLC tissues, especially from patients with metastasis, compared with MRC5 cells (Figure 1E). In contrast, RUNX1 mRNA levels were elevated in NSCLC tissues (Figure 1F). These findings demonstrate that miR-101 is down-regulated, whereas RUNX1 is overexpressed in lung cancer cells. To address whether miR-101 and RUNX1 have potential in-parallel relationship, we analyzed miR-101 and RUNX1 expression using Pearson correlation coefficient and found that miR-101 downregulation was accompanied by RUNX1 upregulation, whereas higher RUNX1 levels were seen in patients carrying lower miR-101 levels in fresh-frozen patient tissues, cell lines and formalin-fixed paraffin-embedded tissues (Figure 1H), indicating a negative correlation between miR-101 and RUNX1 expression, and a role of miR-101/RUNX1 axis involved in lung cancer.
Figure 1.
miR-101 negatively correlated with RUNX1 expression in lung cancer. A. Relative miR-101 expression in 17 fresh-frozen lung cancer and adjacent non-cancer tissues was determined by real-time RT-PCR. B. Schematic of the putative binding site of miR-101 in 3’-UTR of RUNX1, which is broadly conserved among vertebrates. C. RUNX1 expression in mRNA levels was determined in 17 fresh-frozen lung cancer and adjacent non-cancer tissues using real-time RT-PCR. D. Relative miR-101 and RUNX1 expression in lung cancer cell lines were determined by real-time RT-PCR and western blot. E and F. Relative miR-101 and RUNX1 expression in 57 formalin-fixed paraffin-embedded tissues were examined by real-time RT-PCR. G. Representative images of RUNX1 protein levels detected by immunohistochemical staining in formalin-fixed paraffin-embedded tissues. H. Pearson’s correlation analyses between relative miR-101 expression and RUNX1 mRNA levels in cell lines or tissues. vs related normal control, *P<0.01, #P<0.05.
miR-101 represses RUNX1 by direct targeting its 3’-UTR region
Given the negative roles of miRNAs in regulating their targets and the inverse correlation between miR-101 and RUNX1 expression in lung cancer, combined with bioinformatics analysis we posited that miR-101 dysregulation may be, at least partially, responsible for abnormal RUNX1 expression. To this end, we transfected H1299 and 95D cells with miR-101 mimics or scramble control. Results indicated transfection successfully upregulated miR-101 levels (Figure 2A) with a significant decrease of RUNX1 mRNA and protein (Figure 1B). To determine whether miR-101 suppresses RUNX1 through a process triggered by the interaction between miR-101 and RUNX1 3’-UTR region, we constructed a reporter plasmid containing firefly luciferase fused with RUNX1 3’-UTR sequence of miR-101 binding site and cotransfected the reporter plasmid with miR-101 mimics or scramble control in 95D or H1299 cells for 48 h. As shown in Figure 2C, overexpressed miR-101 caused a remarkable decrease of luciferase activity driven by the RUNX1 3’-UTR in 95D or H1299 cells. Additionally, the luciferase reporter assay performed in HEK293T cells showed miR-101 reduced the luciferase activity of the vector driven by the RUNX1 3’-UTR (Figure 2D). These data suggest that RUNX1 is a direct target of miR-101 in human lung cancer cells.
Figure 2.
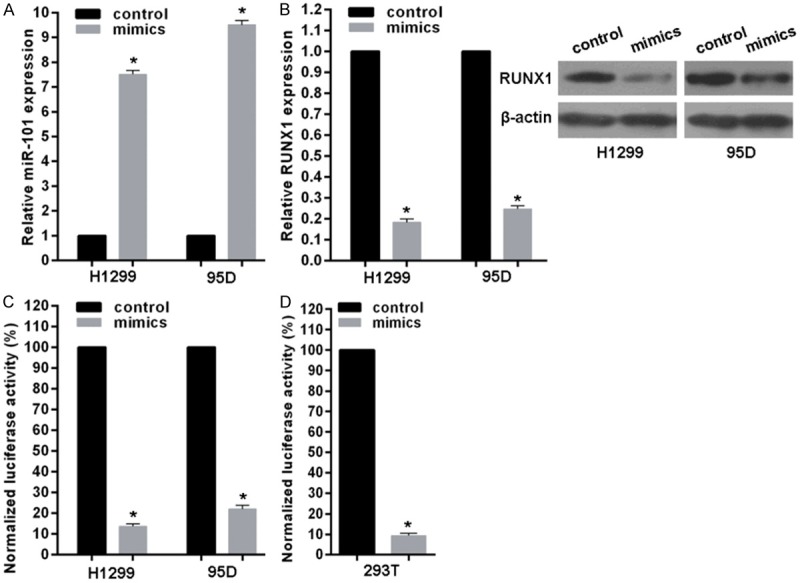
miR-101 targets RUNX1 in lung cancer cells. A. Real-time RT-PCR showed the levels of miR-101 expression in H1299 and 95D cells transfected with miR-101 mimics or scramble control. B. Real-time RT-PCR and western blotting measured RUNX1 expression in H1299 or 95D cells transfected with miR-101 mimics or scramble control. C. Co-transfection of H1299 or 95D cells with miR-101 mimics or scramble control plus firefly luciferase fused with RUNX1 3’-UTR region and pRL-TK Renilla luciferase vector. Luciferase activity was measured at 48 h after transfection, and the relative ratio of the activity in the miR-101 mimics group to that in the scramble group normalized to pRL-TK Renilla luciferase activity is presented as the mean ± S.D. from three independent experiments. D. Luciferase reporter assay in 293T cells was performed as above. vs related normal control, *P<0.01.
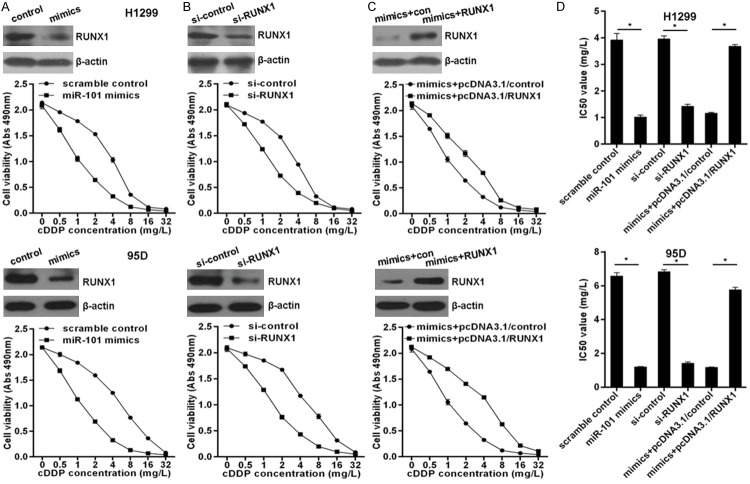
RUNX1 prevents miR-101 from sensitizing lung cancer cells to cDDP
MiR-101 is downregulated in lung cancer, so the role of miR-101 in chemotherapy should be elucidated. Here, we indicated that overexpressed miR-101 via transfection of miR-101 mimics significantly enhanced the sensitivity of H1299 and 95D cells to cDDP (Figure 3A) with marked decrease of IC50 values (Figure 3D). In agreement, RUNX1 knockdown markedly sensitized H1299 and 95D cells to cDDP (Figure 3B and 3D). To investigate whether miR-101 exerts its chemotherapy sensitizer role via suppressing RUNX1 expression, here H1299 cells and 95D were co-transfected with miR-101 mimics and RUNX-1 expressing plasmid pcDNA3.1-RUNX1 without RUNX1-3’-UTR that significantly elevated RUNX1 protein level upon miR-101 overexpressed (Figure 3C). Importantly, we found that ectopic RUNX1 expression significantly abolished miR-101 mediated chemotherapy sensitization (Figure 3C and 3D). These experimental data indicate that miR-101 sensitizes lung cancer cells to chemotherapy via directly targeting RUNX1 expression.
Figure 3.
miR-101 sensitized lung cancer cells to cDDP via targeting RUNX1. A. Overexpressed miR-101 via transfection of miR-101 mimics sensitized H1299 and 95D cells to cDDP detected by MTS assay. B. RUNX1 knockdown via transfection of RUNX1 specific siRNAs enhanced the sensitivity of H1299 and 95D cells to cDDP. C. Overexpressed RUNX1 via transfection of RUNX1 expressing plasmid pcDNA3.1-RUNX1 reversed miR-101 mediated chemotherapeutic sensitization in lung cancer cells. D. The IC50 values for related cell lines were indicated. vs related normal control, *P<0.01.
RUNX1 impairs miR-101 induced lung cancer invasion inhibition
Due to downregulation of miR-101 in lung cancer cells, here we investigated the effect of miR-101 on invasion. We found enforced miR-101 expression remarkably inhibited the invasion potential of H1299 and 95D cells determined by transwell assays (Figure 4A and 4D). Coincidentally, RUNX1 knockdown significantly decreased the invasion potential of 95D and H1299 cells (Figure 4B and 4D). Furthermore, we demonstrated that ectopic expressed RUNX1 obviously impaired miR-101 induced invasion inhibition in H1299 and 95D cells (Figure 4C and 4D), suggesting RUNX1 overexpression is responsible for the effect of miR-101 downregulation on invasion in lung cancer cells.
Figure 4.
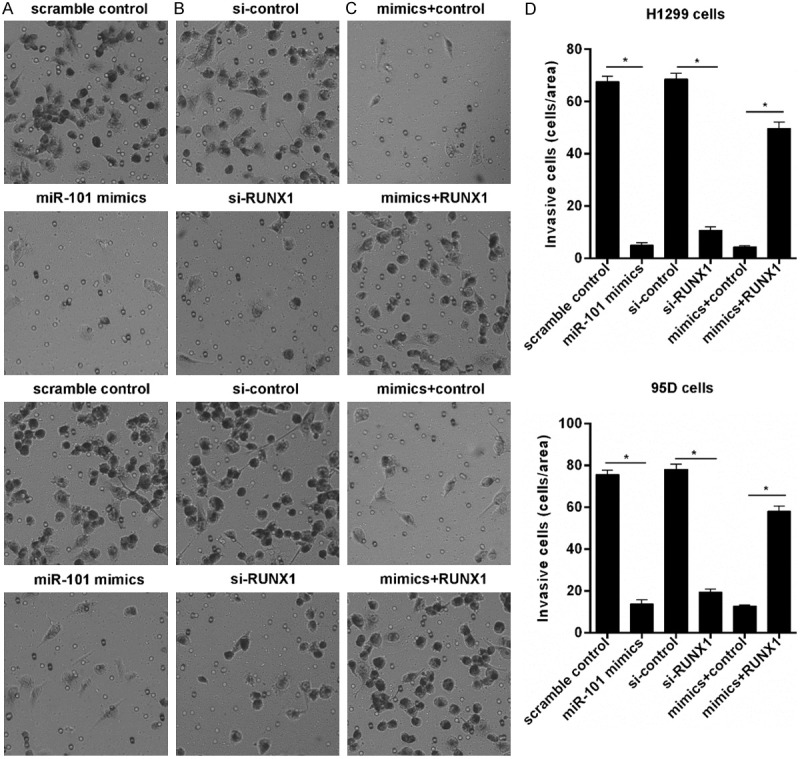
RUNX1 prevents miR-101 mediated invasion inhibition. A. Overexpressed miR-101 inhibited lung cancer cells invasion detected with transwell. B. RUNX1 knockdown inhibited lung cancer cells invasion. C. Overexpressed RUNX1 reversed miR-101 mediated lung cancer cells invasion. D. Invasive cell number was counted. *P<0.01.
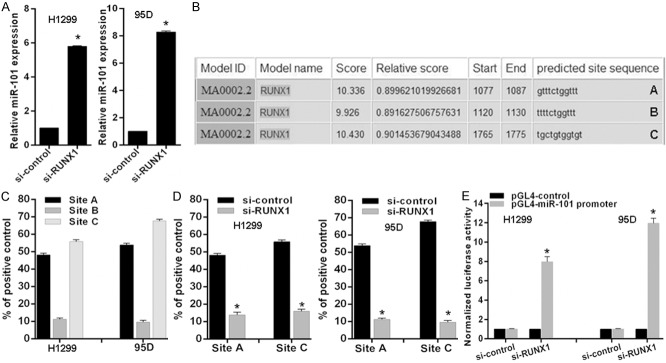
RUNX1 transcriptionally inactivates miR-101 expression
Given miR-101 directly targets RUNX1 and RUNX1 is an essential transcription factor involved in cancers, it is necessary to verify whether RUNX1 feedback transcriptionally regulates miR-101 in lung cancer cells. To address this issue, we detected miR-101 expression level in cells with RUNX1 knockdown and found miR-101 expression increased after RUNX1 knockdown in 95D and H1299 lung cancer cells (Figure 5A). Then, we analyzed the response elements of a cohort of transcription factors within a 2 kb region upstream of the miR-101 precursor start site using the online software “The JASPAR database” and found three putative RUNX1 binding sites within it (A, B and C) (Figure 5B). To confirm the direct association of RUNX1 with the miR-101 promoter, we performed ChIP-qPCR assays in 95D and H1299 cells and found that RUNX1 most significantly bound to site A and site C (Figure 5C). Expectedly, RUNX1 knockdown decreased RUNX1 binding to miR-101 promoter in 95D and H1299 cells (Figure 5D). To observe whether the 2 kb region indeed has promoter activity, the 2 kb DNA was cloned into the pGL4 reporter plasmid. Experimental results indicated that RUNX1 knockdown enhanced the luciferase activity driven by the potential promoter of miR-101 in 95D or H1299 cells (Figure 5E). These results strongly support that RUNX1 physically binds to miR-101 promoter to transcriptionally inactivate miR-101 expression in lung cancer cells.
Figure 5.
RUNX1 inhibits miR-101 expression in lung cancer cells. (A) Relative mean expression of miR-101 was detected by real-time RT-PCR in cells with RUNX1 knockdown. (B) A schematic representation of RUNX1 binding sites in the 2 kb putative miR-101 promoter upstream of the first base of the miR-101 precursor start site and the first base of the 2 kb set as 1. (C and D) ChIP-qPCR for the RUNX1 binding to the miR-101 promoter in (C) selected lung cancer cell lines and (D) RUNX1 knockdown cell lines by transfection of siRNAs. (E) Luciferase reporter assay for the luciferase activity driven by miR-101 promoter in H1299 or 95D cells transfected with RUNX1 siRNAs. vs related control, *P<0.01.
Discussion
Despite the impressive progress made in lung cancer diagnosis and therapy, the dismal 5-year survival rate for patients with lung cancer has not substantially changed. Thus, the molecular characterization of lung cancer progression and the identification of biomarkers for the selection of patients for targeted therapy could improve such dismal outcomes. Although miR-101 silencing occurs in lung cancer [11,14], the pathological contributions and molecular mechanisms remain elusive. In our present study, we confirmed miR-101 silencing in lung cancer, especially in metastatic lung cancer, and identified RUNX1 as a bona fide target of miR-101. Moreover, we demonstrated RUNX1 feedback negatively transcriptionally regulated miR-101, suggesting the crucial role of miR-101/RUNX1 regulatory loop in lung cancer progression.
miRNAs are the endogenous inhibitors of target genes expression and key regulators of most cellular processes. The miRNAs expression patterns are found to be associated with the cancer types and stages, thereby being explored as biomarkers for cancer diagnosis and prognosis [4,5]. Specifically, miR-101 is reported to be downregulated in multiple cancer types and miR-101 overexpressed impairs cancer cell expansion [8-14]. In agreement with this, our data showed that restored miR-101 expression sensitized lung cancer cells to chemotherapy and inhibited them invasion. Although most studies have reported miR-101 as a tumor suppressor, Sachdeva et al. reported that miR-101 was able to promote the estrogen-independent growth of breast cancer cells, and the suppression of membrane-associated guanylate kinase inverted 2 (MAGI-2) by miR-101 decreased the activity of PTEN, leading to Akt activation [15]. This indicates that miR-101 might also have an oncogenic potential which might favor tumor growth under certain conditions and implying that more novel targets should be identified to fully clarify the detailed roles and mechanisms of miR-101 in cancers, including lung cancer. Here, we demonstrated miR-101 levels inversely correlated with RUNX1 levels in lung cancer, and restored miR-101 downregulated RUNX1 expression. Overexpressed RUNX1 prevented the effects of miR-101 on chemotherapy sensitization and invasion inhibition in lung cancer cells, strongly suggesting the crucial contribution of miR-101 deregulation to lung cancer pathogenesis via regulating RUNX1 expression.
RUNX1 (Runt-related transcription factor 1) is a member of the RUNX transcription factor family (including RUNX1, RUNX2 and RUNX3) and plays an essential role in the cell lineage determination, development of normal hematopoiesis and even stem cell proliferation [16,17]. All RUNX1 proteins form a complex with the cofactor CBFB which enhances the DNA-binding ability of RUNX proteins and protects them from proteasomal degradation to increase their function [18]. RUNX1 has been reported to play an important role in various types of cancer by activating or repressing the transcription of key regulators of growth, survival and differentiation [19]. RUNX1 can act as a tumor suppressor or an oncogene in different cells and tissues through the regulation of cancer-related genes. RUNX1 has been regarded as a beneficial tumor suppressor for myeloid leukemogenesis and inactivating RUNX1 have frequently been found in patients with myelodysplastic syndrome and cytogenetically normal acute myeloid leukemia [20]. RUNX1-mediated hematopoiesis has been the subject of intense investigation. RUNX1 is involved in lineage commitment during myeloid, B-cell and T-cell differentiation. In general, loss of RUNX1 function leads to impaired differentiation and subsequent leukemia development [16,21]. On the other hand, several recent studies described the survival role of RUNX1 in sustaining leukemia cell growth [22-24], suggesting the context-and dosage-dependent roles of RUNX1 in various types of hematopoietic neoplasms. Although RUNX1 has been considered a hematopoietic gene, recent evidence has revealed a pivotal role for RUNX1 in solid tumors. RUNX1 is highly expressed in human breast epithelial cells. RUNX1 knockdown causes hyperproliferation and abnormal morphogenesis in MCF10A cell [25]. In prostate cancer, the RUNX1 promoter is bound by EZH2 and is negatively regulated by histone H3 lysine 27 (K27) trimethylation. Repression of RUNX1 is important for the growth promotion ability of EZH2 in AR-independent cells. In clinical prostate cancer samples, the RUNX1 expression level is negatively associated with EZH2 and that RUNX1 loss correlated with poor prognosis [26]. In contrast to the tumor suppressor role for RUNX1 in breast cancer, RUNX1 appears to have survival/growth-promoting roles in several epithelial cancers. RUNX1 is highly expressed in various epithelial tumors including skin and head/neck squamous cell carcinomas (SCC) [27]. Genetic deletion of RUNX1 in mouse inhibits tumor formation in a murine model of chemically induced skin SCC. RUNX1 also supports the tumorigenesis of oral SCC induced by oncogenic KrasG12D. In line with these observations, RUNX1 knockdown in human skin and head/neck SCC cells induces growth arrest. Mechanistically, p21 (CIP1/WAF1) is upregulated in RUNX1 depleted mouse keratinocytes and p21 depletion rescues cell proliferation of RUNX1-depleted cells in vitro [28]. Additionally, RUNX1 prevents expression of SOCS3 and SOCS4 transcription, which leads to the activation of STAT3 [27]. Thus, the combination of p21 repression and STAT3 activation appears to underlie RUNX1-mediated SCC formation. Recently, RUNX1 was shown to be upregulated in human epithelial ovarian carcinoma tissues and associated with ovarian carcinoma cell proliferation, migration and invasion [29]. In agreement, we demonstrated that RUNX1 was overexpressed in lung cancer and RUNX1 knockdown sensitized lung cancer cells to chemotherapy and inhibited invasion. As a master regulator for hematopoiesis and cancer, RUNX1 function is tightly controlled. Several regulatory mechanisms exist for fine-tuning RUNX1 activity. These include alternative splicing, transcriptional control, translational control and posttranslational modifications [18]. Recently, miR-302b was identified to target RUNX1 to suppress human epithelial ovarian cancer cell growth [30]. In our present study, we also demonstrated RUNX1 is a direct target of miR-101. Moreover, we found RUNX1 feedback transcriptionally inactivates miR-101 expression in lung cancer cells. But functional consequences of aberrant RUNX1 expression and details in RUNX1-mediated miR-101 dysregulation in lung cancer still need to be clarified experimentally in the further studies.
In summary, we here confirm the downregulation of miR-101 in lung cancer and demonstrate that miR-101 increases chemotherapeutic-sensitivity and inhibits invasion via targeting RUNX1, and RUNX1 feedback transcriptionally inactivates miR-101 expression. Our results provide new insight into the role of miR-101/RUNX1 regulatory loop in lung cancer progression and suggest potential molecular target for the treatment of lung cancer.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Kumar MS, Armenteros-Monterroso E, East P, Chakravorty P, Matthews N, Winslow MM, Downward J. HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature. 2014;505:212–7. doi: 10.1038/nature12785. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol. 2014;11:473–81. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 4.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 5.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–91. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Song Y, Wang Y, Luo J, Yu W. MicroRNA-148a suppresses epithelial-to-mesenchymal transition by targeting ROCK1 in non-small cell lung cancer cells. Mol Cell Biochem. 2013;380:277–82. doi: 10.1007/s11010-013-1682-y. [DOI] [PubMed] [Google Scholar]
- 7.Liu ZL, Wang H, Liu J, Wang ZX. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem. 2013;372:35–45. doi: 10.1007/s11010-012-1443-3. [DOI] [PubMed] [Google Scholar]
- 8.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, Brenner JC, Yu J, Kim JH, Han B, Tan P, Kumar-Sinha C, Lonigro RJ, Palanisamy N, Maher CA, Chinnaiyan AM. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–9. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konno Y, Dong P, Xiong Y, Suzuki F, Lu J, Cai M, Watari H, Mitamura T, Hosaka M, Hanley SJ, Kudo M, Sakuragi N. MicroRNA-101 targets EZH2, MCL-1 and FOS to suppress proliferation, invasion and stem cell-like phenotype of aggressive endometrial cancer cells. Oncotarget. 2014;5:6049–62. doi: 10.18632/oncotarget.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smits M, Nilsson J, Mir SE, van der Stoop PM, Hulleman E, Niers JM, de Witt Hamer PC, Marquez VE, Cloos J, Krichevsky AM, Noske DP, Tannous BA, Würdinger T. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget. 2010;1:710–20. doi: 10.18632/oncotarget.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang JG, Guo JF, Liu DL, Liu Q, Wang JJ. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J Thorac Oncol. 2011;6:671–8. doi: 10.1097/JTO.0b013e318208eb35. [DOI] [PubMed] [Google Scholar]
- 12.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–42. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 13.He XP, Shao Y, Li XL, Xu W, Chen GS, Sun HH, Xu HC, Xu X, Tang D, Zheng XF, Xue YP, Huang GC, Sun WH. Downregulation of miR-101 in gastric cancer correlates with cyclooxygenase-2 overexpression and tumor growth. FEBS J. 2012;279:4201–12. doi: 10.1111/febs.12013. [DOI] [PubMed] [Google Scholar]
- 14.Yan F, Shen N, Pang J, Xie D, Deng B, Molina JR, Yang P, Liu S. Restoration of miR-101 suppresses lung tumorigenesis through inhibition of DNMT3a-dependent DNA methylation. Cell Death Dis. 2014;5:e1413. doi: 10.1038/cddis.2014.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sachdeva M, Wu H, Ru P, Hwang L, Trieu V, Mo YY. MicroRNA-101-mediated Akt activation and estrogen-independent growth. Oncogene. 2011;30:822–31. doi: 10.1038/onc.2010.463. [DOI] [PubMed] [Google Scholar]
- 16.Link KA, Chou FS, Mulloy JC. Core binding factor at the crossroads: determining the fate of the HSC. J Cell Physiol. 2010;222:50–6. doi: 10.1002/jcp.21950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim W, Barron DA, San Martin R, Chan KS, Tran LL, Yang F, Ressler SJ, Rowley DR. RUNX1 is essential for mesenchymal stem cell proliferation and myofibroblast differentiation. Proc Natl Acad Sci U S A. 2014;111:16389–94. doi: 10.1073/pnas.1407097111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goyama S, Huang G, Kurokawa M, Mulloy JC. Posttranslational modifications of RUNX1 as potential anticancer targets. Oncogene. 2015;34:3483–92. doi: 10.1038/onc.2014.305. [DOI] [PubMed] [Google Scholar]
- 19.Ito Y. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res. 2008;99:33–76. doi: 10.1016/S0065-230X(07)99002-8. [DOI] [PubMed] [Google Scholar]
- 20.Nishimoto N, Arai S, Ichikawa M, Nakagawa M, Goyama S, Kumano K, Takahashi T, Kamikubo Y, Imai Y, Kurokawa M. Loss of AML1/Runx1 accelerates the development of MLL-ENL leukemia through down-regulation of p19ARF. Blood. 2011;118:2541–50. doi: 10.1182/blood-2010-10-315440. [DOI] [PubMed] [Google Scholar]
- 21.Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–87. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 22.Ben-Ami O, Friedman D, Leshkowitz D, Goldenberg D, Orlovsky K, Pencovich N, Lotem J, Tanay A, Groner Y. Addiction of t(8;21) and inv(16) acute myeloid leukemia to native RUNX1. Cell Rep. 2013;4:1131–43. doi: 10.1016/j.celrep.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 23.Goyama S, Schibler J, Cunningham L, Zhang Y, Rao Y, Nishimoto N, Nakagawa M, Olsson A, Wunderlich M, Link KA, Mizukawa B, Grimes HL, Kurokawa M, Liu PP, Huang G, Mulloy JC. Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J Clin Invest. 2013;123:3876–88. doi: 10.1172/JCI68557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilkinson AC, Ballabio E, Geng H, North P, Tapia M, Kerry J, Biswas D, Roeder RG, Allis CD, Melnick A, de Bruijn MF, Milne TA. RUNX1 is a key target in t(4;11) leukemias that contributes to gene activation through an AF4-MLL complex interaction. Cell Rep. 2013;3:116–27. doi: 10.1016/j.celrep.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Brugge JS, Janes KA. Intersection of FOXO-and RUNX1-mediated gene expression programs in single breast epithelial cells during morphogenesis and tumor progression. Proc Natl Acad Sci U S A. 2011;108:E803–12. doi: 10.1073/pnas.1103423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takayama K, Suzuki T, Tsutsumi S, Fujimura T, Urano T, Takahashi S, Homma Y, Aburatani H, Inoue S. RUNX1, an androgen- and EZH2-regulated gene, has differential roles in AR-dependent and -independent prostate cancer. Oncotarget. 2015;6:2263–76. doi: 10.18632/oncotarget.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheitz CJ, Lee TS, McDermitt DJ, Tumbar T. Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J. 2012;31:4124–39. doi: 10.1038/emboj.2012.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoi CS, Lee SE, Lu SY, McDermitt DJ, Osorio KM, Piskun CM, Peters RM, Paus R, Tumbar T. Runx1 directly promotes proliferation of hair follicle stem cells and epithelial tumor formation in mouse skin. Mol Cell Biol. 2010;30:2518–36. doi: 10.1128/MCB.01308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keita M, Bachvarova M, Morin C, Plante M, Gregoire J, Renaud MC, Sebastianelli A, Trinh XB, Bachvarov D. The RUNX1 transcription factor is expressed in serous epithelial ovarian carcinoma and contributes to cell proliferation, migration and invasion. Cell Cycle. 2013;12:972–86. doi: 10.4161/cc.23963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge T, Yin M, Yang M, Liu T, Lou G. MicroRNA-302b suppresses human epithelial ovarian cancer cell growth by targeting RUNX1. Cell Physiol Biochem. 2014;34:2209–20. doi: 10.1159/000369664. [DOI] [PubMed] [Google Scholar]