Abstract
Chromosomal rearrangements and fusion genes play important roles in tumor development and progression. Four high-frequency prostate cancer (CaP) specific fusion genes, SDK1:AMACR, RAD50:PDLIM4, CTAGE5:KHDRBS3 and USP9Y:TTTY15 have been reported in Chinese CaP samples through a transcriptome sequencing study. We previously reported that USP9Y:TTTY15 is a transcription-mediated chimeric RNA, which is expressed in both tumor and non-malignant samples, and here we attempted to confirm the existence of the other three fusion genes SDK1:AMACR, RAD50:PDLIM and CTAGE5:KHDRBS3. We detected SDK1:AMACR fusion transcript in 23 of 100 Chinese CaP samples, but did not detect RAD50:PDLIM4 and CTAGE5:KHDRBS3 transcripts in any of those samples. SDK1:AMACR fusion transcript is Chinese CaP specific, which was neither detected in non-malignant prostate tissues adjacent to cancer from Chinese patient nor in CaP samples from UK patients. However, we did not detect genomic rearrangement of SDK1 gene by fluorescence in situ hybridization analysis, indicating that SDK1:AMACR is also a transcription-mediated chimeric RNA. Quantitative analysis demonstrated that high level AMACR expression was associated with SDK1:AMACR fusion status (P=0.004), suggesting that SDK1:AMACR fusion transcript may promote prostate carcinogenesis through increasing AMACR expression. However, the fusion status was not significantly correlated with any poor disease progression clinical features. The identification of the SDK1:AMACR fusion transcript in CaP cases from China but not from UK further supports our previous observation that different genetic alterations contribute to CaP in China and Western countries, although many genetic changes are also shared. Further studies are required to establish if CaPs with SDK1:AMACR represent a distinct subtype.
Keywords: Chinese CaP, chromosomal rearrangements, SDK1:AMACR, chimeric RNA
Introduction
Prostate cancer (CaP) is the most commonly diagnosed tumor and the second leading cause of deaths due to cancer in the Western male population [1]. The serum prostate-specific antigen (PSA) is helpful for early detection and monitoring disease progression of CaP, therefore improves the survival in some circumstances [2,3]. However, PSA level is specific for prostate tissue, but it is not specific for CaP. Benign prostatic hyperplasia (BPH), prostatitis and the effect of some medications can also lead to an elevated PSA, which weaken its specificity as a diagnostic marker for CaP. PSA screening remains controversial, since it results in over-diagnosis and over-treatment [4-6]. Extensive searches have been carried out to develop more CaP specific molecular markers, in particular those predicting cancer aggressiveness [7,8].
There is compelling evidence to consider genomic rearrangements as an initial event in tumorigenesis [9,10]. Fusion genes, initially identified in hematological malignancy and soft tissue sarcomas, have been used for tumor diagnosis, sub-classification, prognosis, recurrence monitoring and as therapeutic targets, such as BCR:ABL fusion gene in chronic myeloid leukemia [10]. It has been demonstrated that fusion genes play a critical role in CaP development and progression [9] and TMPRSS2:ERG, which was frequently detected in CaP, is the most commonly reported fusion gene to date in human malignancies [10-13].
CaP has an obvious difference in the incidence and mortality among different populations [14]. Little is known of the genetic mechanisms contributing to this discrepancy in prostatic cancer prevalence. We previously identified certain genetic difference between Chinese and Western CaPs [15-17]. Recently, it was reported that four new high-frequency fusion genes, USP9Y:TTTY15 (19/54), RAD50:PDLIM4 (15/54), CTAGE5:KHDRBS3 (20/54), SDK1:AMACR (13/54) were identified by transcriptome sequencing analysis of CaP cases from China [18]. Once confirmed, this finding may not only further highlight the genetic difference between Chinese and Western CaPs, but also provide new insight of prostate carcinogenesis and new population-based treatment strategy.
We previously reported that USP9Y:TTTY15 is a transcription-mediated chimeric RNA, which is expressed in both tumor and non-tumor samples [19]. In this study, we attempted to confirm the existence of fusion genes SDK1:AMACR, RAD50:PDLIM and CTAGE5:KHDRBS3 in a separate cohort of Chinese CaP samples. We detected high frequency of SDK1:AMACR fusion transcript in our CaP samples, which was specific for Chinese cancers and associated with high AMACR expression. However, SDK1:AMACR fusion was not associated with disease severity. We did not detect RAD50:PDLIM4 and CTAGE5:KHDRBS3 transcripts in any of our CaP samples.
Materials and methods
Samples
A total of 100 pairs of fresh cancer and their matched normal prostate tissue samples from Chinese CaP patients were collected from the First Affiliated Hospital, Zhejiang University Medical College, Hangzhou, China and were preserved in -80°C. The tissue morphology and Gleason grade of cancer lesions were confirmed by two pathologists. Detailed clinical pathological information for these samples is summarized in Table 1. Diagnostic PSA in two patients and the age information of one cases are missing. In addition, tissue microarrays (TMAs) containing 85 of the samples were made for fluorescence in situ hybridization (FISH). This study is approved by the ethical committee of First Affiliated Hospital, Zhejiang University Medical College. Twenty eight fresh frozen tissue samples of radical prostatectomy CaP from Barts Health patients were taken with informed patient content under Orchid Tissue Bank, ethically approved by East London and City Committee. CaP cell lines LNCaP, VCaP, 22RV1, DU145 and PC3 were also used for SDK1:AMACR fusion transcript analysis.
Table 1.
Clinicopathological details of the 100 Chinese CaP cases
| Values | |
|---|---|
| Age (year) | Mean (SD) |
| 66.45 (7.37) | |
| Clinical stage | |
| T1 | 2 |
| T2 | 39 |
| T3 | 56 |
| T4 | 3 |
| Gleason score | |
| 6 | 18 |
| 7 | 56 |
| 8 | 17 |
| 9 | 9 |
| Total PSA (ng/ml) at diagnosis | Mean (SD) |
| 22.28 (22.15) |
SD: standard deviation; PSA: prostate specific antigen.
RT-PCR and real time quantitative RT-PCR using SYBR green technology
Total RNA was extracted using Trizol (Invitrogen). Two micrograms of total RNA was used to synthesize cDNA in a 25 µl reaction mixture using Reverse Transcriptase M-MLV (R Nase H) and random primer as previously described [20]. The primers and annealing temperatures for the RT-PCR are listed in Table 2. The RT-PCR amplified product was detected by running 1.2% agarose gel.
Table 2.
All the primers used for RT-PCR
| Primers | Sequence | Annealing temperature | Length of product |
|---|---|---|---|
| RAD50:PDLIM4-F | ACTAAGTGAATGCGAGAAACACAA | 59°C or 62°C | 100-250 bp |
| RAD50:PDLIM4-R | ACAGACAGTGTGAGGTGATCGT | ||
| SDK1:AMACR-F | ACCTGGTCATTTCCAACATCAG | 54°C or 57°C | 150-400 bp |
| SDK1:AMACR-R | CAAAGCCAAATAGTTGATATCGTG | ||
| CTAGE5:KHDRBS5-F | TGCTGAAAATGAAGCCACTG | 59°C | 400 bp |
| CTAGE5:KHDRBS5-R | GGACTGGTGGAGATTGGCTA | ||
| CTAGE5:KHDRBS5-F | GTGGATGCAAGAGGCCCATTCT | 59°C or 62°C | 256 bp |
| CTAGE5:KHDRBS5-R | TAGACGCCCTTTGCTGTCCTC |
The amplification program for quantitative RT-PCR consisted of an initial denaturation step at 95°C for 30 s followed by incubations at 95°C for 5 s, 60°C for 30 s, and 72°C for 13 s for 50 cycles. All the reactions were performed in triplicate and all gene expression values were normalized using the housekeeping gene GAPDH and calculated using the comparative Ct method (ΔΔCt method). The transcript-specific primers used in this study were: AMACR-F: GATTTGGCCAGTCAGGAAGC, AMACR-R: GAACACCTGACAAAGCCAAATAGTT, GAPDH-F: AAGGTGAAGGTCGGAGTCAA and GAPDH-R: AATGAAGGGGTCATTGATGG.
The forward primer for AMACR is located in exon 2 and the reverse primer spans the junction of exon 2 and exon3 of AMACR.
Sequencing analysis
The expected band for each RT-PCR amplified product was gel-excised (GEL Extraction kit) and cloned into PMD 18-T vector (Takara, Dalian, China) with TA cloning technique following manufactory’s instructions. Briefly, the molar ratio of vector DNA and insert DNA ranged from 1:2~10 and the ligation reaction was performed at 16°C for 50 min. Then the plasmid was transformed into competent E. coli cells, which were cultured in LB growth medium containing Amp. We picked up monoclones for sequencing analysis using M13+ primers and the ABI3730XL sequencing machine.
Fluorescence in situ hybridization (FISH) analysis
Interphase FISH analysis of TMA samples was performed as previously described [21]. Using BAC probes, we applied the FISH signal break-apart approach on CaP samples to detect the genomic rearrangements of RAD50 (5’ BAC clone RP11-410H12 in red and 3’ BAC clone RP11-525D24 in green) and SDK1 (5’ BAC cloneRP11-133L20 in red and 3’ BAC clone RP11-644L16 in green), which are required to generate the genomic fusion of RAD50:PDLIM4 and SDK1:AMACR, respectively. If a fusion event occurs, it will either split apart the gene (red and green signals locate separately) or cause the deletion of the 3’ genome region (loss of the green signal). A schematic presentation ofthe FISH signals for potential SDK1 genomic rearrangements are shown in Figure 1.
Figure 1.
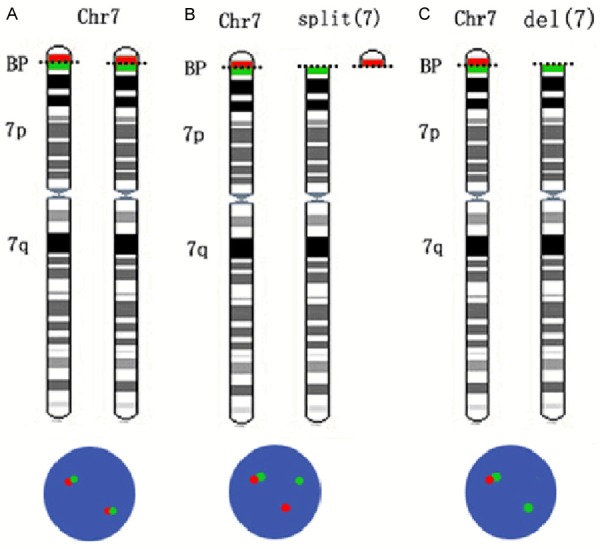
Schematic presentation of the detection of SDK1 rearrangements. A. A cell with normal SDK1 shows two pairs of green and red signals. B. Red and green signals split apart when a break at SDK1 occurs with both chromosome fragments remaining in the cell. C. Loss of green signal when the 3’ region of SDK1 is deleted. Chr: chromosome; BP: breakpoint.
Statistical analysis
Chi-square test was performed when a cut off value was used to categorize the cases into groups. Unpaired t test were applied when the expression level of AMACR was compared between the positive and negative SDK1:AMACR samples. All statistical analysis is performed using SPSS 16.0 for Windows (SPSS, Chicago, IL, USA) with two tailed tests. A P value of less than 0.05 was considered statistically significant.
Results
Confirmation of the recurrent SDK1:AMACR fusion transcripts in Chinese CaP samples
Using the previously reported PCR primers, we attempted to detect the SDK1:AMACR fusion in 100 Chinese CaP samples. Based on the TMs of the primers, we tried three annealing temperatures at 54°C, 57°C and 60°C in 16 cancer samples and found a PCR product at around 250 bp in one sample using the lowest annealing temperatures of 54°C. Sequencing analysis confirmed a 241 bp fusion product of SDK1 (breakpoint after exon 22) with AMACR (breakpoint before exon 2) (Figure 2). This generated an in frame fusion with a predicted protein containing the most of SDK1 and AMACR. We analyzed the other 84 cancer samples using the annealing temperatures of 54°C and found 22 samples with a product at around 241 bp, which was confirmed as SDK1:AMACR (exon 22 to exon 2) fusion products by sequencing analysis in all the cases. In many positive samples, additional bands were also obtained but sequencing analysis of these bands (cut from the gel and then cloned) showed un-specific PCR products. Using an annealing temperatures of 57°C to PCR amplify those samples, in addition to the 241 bp specific band found in the 22 fusion transcript positive samples, we identified in sample 47C a 514 bp product (Figure 2). Sequencing analysis confirmed that the 514 bp product was a variation of the SDK1:AMACR fusion gene, which include part of the intron 2 of AMACR in the middle of the fusion (Figure 2C).
Figure 2.
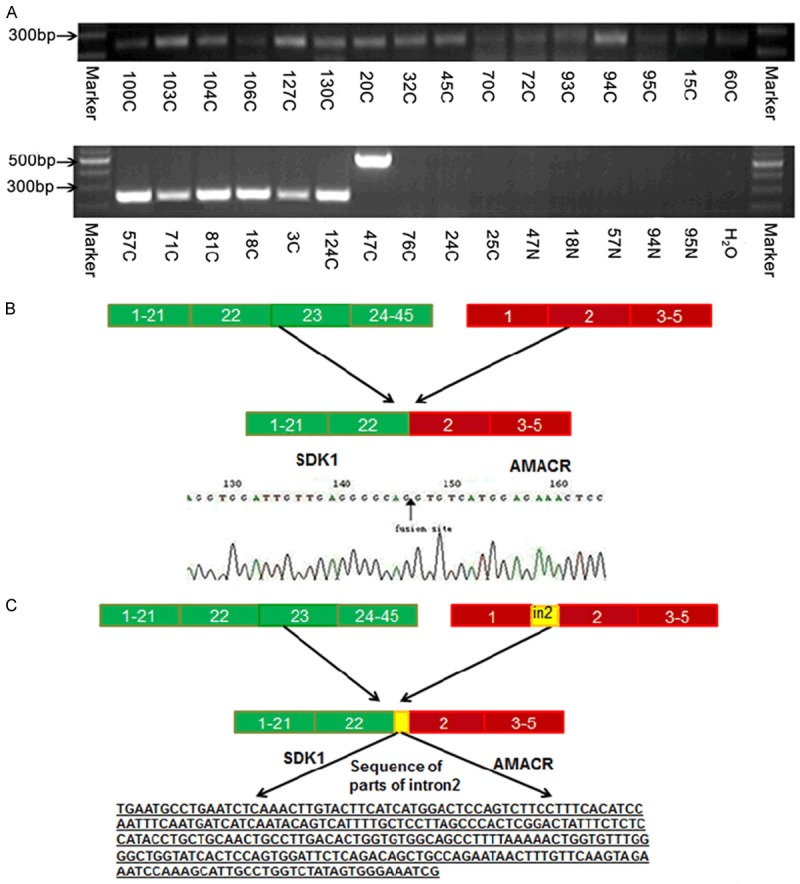
Detection of the SDK1:AMACR fusion transcript in CaP. (A) Examples of the RT-PCR products of SDK1:AMACR fusion transcript at 241 bp in CaP and a variation of 514 bp band (case 47C) along with case-matched adjacent normal tissue samples and a negative control without input cDNA template. (B) Schematic presentation of the alignment of SDK1 on chromosome 7 and AMACR on chromosome 5 and the fusion transcripts based on the sequencing data of the fusion at the end of AMACR exon 22 and the beginning of AMACR exon 2. (C) Schematic presentation of the alignment of the 514 bp fusion sequence for SDK1 and AMACR. In the middle of the fusion sequence presented in (B), there is additional sequence from AMACR intron 2 (underline). The position of the nucleotide fusion site is indicated by the blue arrows.
We also investigate whether SDK1:AMACR fusion exists at genomic level in 18 SDK1:AMACR fusion transcript positive sampled by dual-color FISH analysis, but we did not found evidence of the truncation of SDK1 in any of the 18 CaP cases analyzed (Figure 3A).
Figure 3.
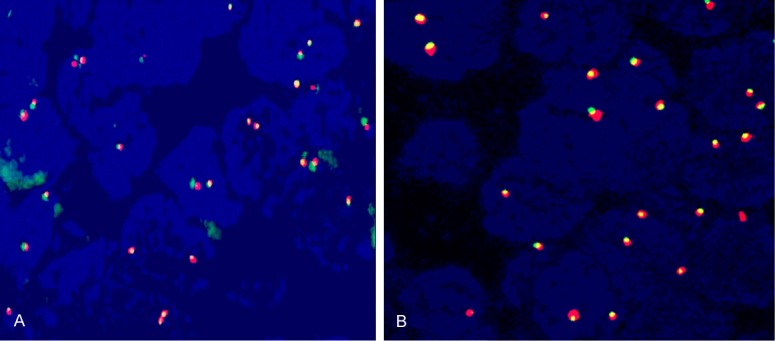
Interphase FISH on formalin-fixed, paraffin-embedded tissue samples to analyze (A) SDK1 and (B) RAD50 genomic status. The overlap of red and green indicates that no genomic rearrangement occurred to SDK1 and RAD50 genomic regions.
Correlation of SDK1:AMACR fusion transcripts with clinicopathological parameters in Chinese CaP cases
We further analyzed the correlation of the existence of SDK1:AMACR fusion transcripts with clinical data in those 100 Chinese CaP cases (23 fusion positive and 77 fusion negative), where the clinical data were available. There was no statistically significant correlation of SDK1:AMACR fusion with clinicopathological parameters (Table 3).
Table 3.
Correlation between clinicopathological parameters and expression of SDK1:AMACR fusion transcript
| Variable | SDK1:AMACR (-) | SDK1:AMACR (+) | P value |
|---|---|---|---|
| Age ≤70 | 52 | 14 | P=0.554 |
| Age >70 | 25 | 9 | |
| PSA ≤10 | 24 | 10 | P=0.292 |
| PSA >10 | 52 | 13 | |
| GS ≤7 | 54 | 20 | P=0.106 |
| GS >7 | 23 | 3 | |
| Clinical stage ≤3 | 67 | 20 | P=0.994 |
| Clinical stage >3 | 10 | 3 | |
| Meta negative | 71 | 21 | P=0.889 |
| Meta positive | 6 | 2 |
Correlation of AMACR expression levels with SDK1:AMACR status in Chinese CaP cases
We further analyzed the expression difference of the AMACR by quantitative RT-PCR between SDK1:AMACR fusion positive (n=18) and negative (n=25) samples. The fusion positive samples expressed a significantly higher AMACR than the negative cases (P=0.004). While in 12 SDK1:AMACR positive cases, AMACR expressed at a level closed to the median expression detected in the SDK1:AMACR negative cases, in six SDK1:AMACR positive cases, >2 fold higher expression of AMACR than any of the expression level of the fusion negative samples was detected (Figure 4). The patients with both SDK1:AMACR fusion and relatively higher expression of AMACR did not have a history of using anti-androgen or taking other therapeutic drugs.
Figure 4.
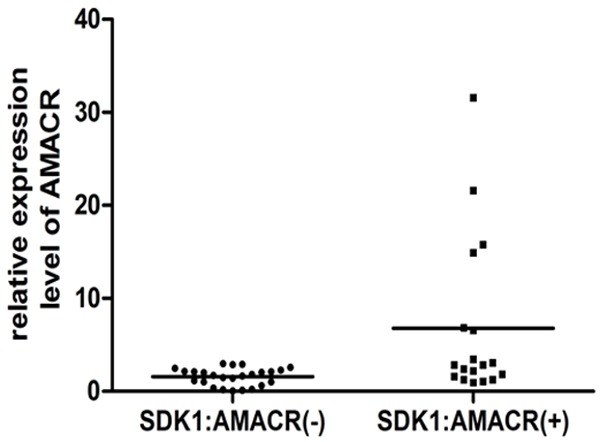
The expression level of AMACR in the fusion positive and fusion negative Chinese CaP samples. The expression level of AMACR in all samples was relative to a fusion transcript negative samples (101C), which was set to 1.
Lack of RAD50:PDLIM4 and CTAGE5:KHDRBS3 fusions in our CaP samples
While we confirmed the SDK1:AMACR fusion transcripts, we did not detect any RAD50:PDLIM4 and CTAGE5:KHDRBS3 fusion transcripts using a considerable number of our Chinese CaP samples. For CTAGE5:KHDRBS3, we initially tried to detect the fusion product in fourteen cancer samples used the previously described primer pair [22] and a range of annealing temperatures at 51°C, 53°C, 55°C, 57°C and 59°C. We found many PCR product bands with the most dominant one at around 300 bp in all cases even at the highest annealing temperatures of 59°C. When 51°C and 53°C were used for annealing temperatures, there was a band at the expected fusion product size of 400 (Figure 5A). Sequencing analysis of this PCR product (cut from the gel) from all the samples showed unspecific PCR products. We then designed another pair of PCR primer with expected PCR products of 256 bp (Table 2), which is nested in the above primer pair. Using the annealing temperatures of 59°C and 62°C, we analyzed 32 cancer samples and found an expected size of PCR product in 14 samples (Figure 5B). However, none of the PCR products were fusion transcripts by sequencing analysis. Finally we applied nested PCR to amplify the fusion transcript. Although we detected PCR products at around expected size of 256 bp in 16 and 13 out of 51 samples with the annealing temperature of 59°C and 62°C respectively, sequencing analysis of all those PCR products did not detected the expected fusion product.
Figure 5.
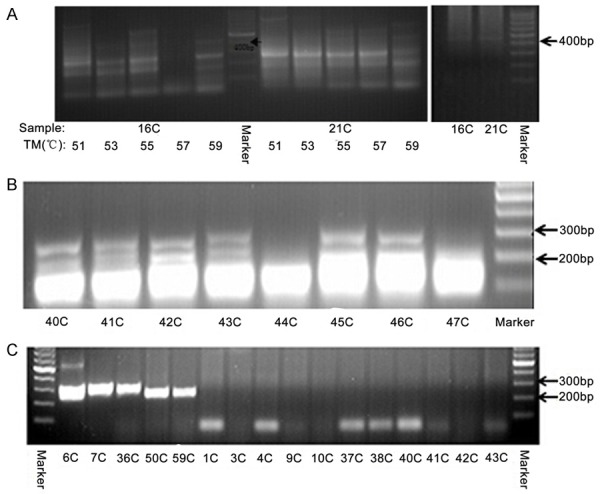
Examples of the RT-PCR products using primers for CTAGE5:KHDRBS3 and RAD50:PDLIM4 fusion transcripts. A. Examples of the RT-PCR products around the expected size (about 400 bp) for CTAGE5:KHDRBS3 fusion transcript. B. Examples of the RT-PCR products with the expected size around 256 bp (in samples 40C, 41C, 42C, 43C, 45C and 46C) for CTAGE5:KHDRBS3 fusion transcript. C. Examples of the RT-PCR products at the expected size between 100-250 bp in five samples, 6C, 7C, 36C, 50C and 59C for RAD50:PDLIM4 fusion transcript.
For the RAD50:PDLIM4 fusion, we performed the RT-PCR only with the previously reported primers [22]. The initial test using different annealing temperatures at 55°C, 57°C and 59°C in three cancer samples yielded multiple bands at all conditions, with 59°C annealing temperature better than the other two. We then tried annealing temperatures at 59°C and 62°C in eight more cancer samples and found one case with PCR product around the expected 225 bp size at both PCR conditions but with more specific PCR band at the annealing temperature at 62°C. Using the later PCR condition, we analyzed 42 more CaP samples and found the PCR product at 225 bp in four more samples (Figure 5C). However, sequencing analysis of the five PCR products did not identify any fusion products.
To investigate RAD50:PDLIM4 fusion at genomic level, we also determined the existence of any truncations of RAD50 as a result of the fusion event by dual-color FISH analysis. We did not found evidence of the truncation of RAD50 in any of the 85 CaP cases analyzed (Figure 3B).
Discussion
With the recent investigation of fusion genes in solid tumors, fusion gene has now been recognized as a common mechanism in the development and/or progression of many human tumors [10,23]. CaP is the carcinoma where fusion gene is most commonly found. However, apart from TMPRSS2:ERG, all the other fusion genes have been only found at a low frequency and TMPRSS2:ERG was found at a much lower frequency in CaP samples in East Asian population compared to the Western countries [15,24]. The reporting of the four high frequency fusions in Chinese CaP [22] poses the potential to reveal the specific mechanisms, genetic pathways and unique therapeutic approaches for cancer in this population. However, in our previous study we identified that one of the four fusion transcripts, USP9Y:TTTY15, is a transcription-mediated chimeric RNA, which was detected in both tumor and non-malignant samples as well as non-prostate tissues [19]. In this independent study, we confirmed that SDK1:AMACR is frequently and specifically expressed in Chinese CaP samples, but we did not detect the other two reported fusions with different PCR conditions on a considerable large number of Chinese CaP samples. Our FISH data also demonstrated that there was no genomic rearrangement of RAD50, which is required to generate the RAD50:PDLIM4 genomic fusion. Therefore RAD50:PDLIM4 and CTAGE5:KHDRBS3 genomic fusions are unlikely to exist in Chinese CaP and even those fusion transcripts may be rare.
The SDK1:AMACR fusion transcripts identified in 23% samples in our study is comparable with the 24% (13/54) positive cases reported by Ren [18]. SDK1 is regulated by androgen through the androgen-responsive Serum Response Factor (SRF) [25,26]. AMACR was initially identified as a CaP over expressed gene through a microarray analysis of subtracted cDNA library of CaP and non-malignant tissues [27] and it is commonly over expressed in CaP [28]. Over expression of AMACR has been considered as a biomarker for CaP [29-33]. AMACR expression is not regulated by androgen, but it depends on tumor differentiation [34,35]. Therefore, similar to TMPRSS2:ERG fusion, SDK1:AMACR fusion may also lead to AMACR over expression in those Chinese CaPs, driving by SDK1 promoter under androgen control. By quantitative detection of the expression level of AMACR in CaP samples, we found that the SDK1:AMACR fusion transcripts was associated with over expression of AMACR. However, our FISH data rules out the genomic fusion of these two genes. Therefore, SDK1:AMACR, although specific to Chinese CaP, it is also a transcription-mediated chimeric RNA. The transcription of this fusion RNA may use a novel mechanism, transcription-mediated chimeric RNA, for AMACR over expression in CaP cells. Many mechanisms have been used to generate transcription-mediated chimeric RNA, but the biological significance and controls to use those processes generating those fusion transcripts are not known yet [36-38].
Although the above data, both from the literature and our experiments, suggest that AMACR over expression and SDK1:AMACR contribute to prostate carcinogenesis, the fusion status was not correlated with any clinical parameters of CaP we analyzed. Further study of its clinical significant is required. In consistent with our finding of the lack of relationship between the SDK1:AMACR and PSA level, previous studies showed that, although AMACR expression was strongly associated with prostate, it was not associated with PSA level. AMACR over expression was detected in several human cancers and their precursor lesions, including CaP precursor, high-grade prostatic intraepithelial neoplasia [28], suggesting that AMACR may have a role in the early steps of cancer development. High level AMACR expression in normal prostate tissue is associated with increased risk of developing CaP [28]. However, the mechanism how AMACR promotes carcinogenesis is not clear yet. Methylacyl-CoA racemase (AMACR) is a mitochondrial and peroxisomal enzyme. It is essential in the metabolism of branched chain fatty acid and bile acid intermediates [39]. Red meat and dairy product, which are associated with branched chain fatty acid, are suggestive risk factor for CaP, indicating that AMACR may contribute to prostate carcinogenesis through branched chain fatty acid metabolism pathway.
In gastrointestinal stromal tumor, where AMACR is also over expressed, AMACR affects cell proliferation but not apoptosis [40]. AMACR amplification and overexpression in primary imatinib-naïve gastrointestinal stromal tumors is a driver of cell proliferation, indicating adverse prognosis [40]. In CaP, Zha S also found that AMACR knockdown by RNA interference inhibited the growth of cancer cells [35]. However, in gastrointestinal stromal tumor, AMACR over expression was associated with poor prognosis [40], but in CaP, it has been reported that decreased AMACR expression in localized cancer was associated with poor prognosis [41]. AMACR may be differentially associated with other factors involved in disease progression in different cancer cells, while have similar role in cell proliferation and cancer development for different tissue types.
As SDK1 is controlled by androgen and SDK1:AMACR fusion transcript was associated with increased AMACR expression, this transcription-mediated fusion transcript may, similar to the genomic fusion of these genes, also put AMACR expression under the control of SDK1 promoter, which is stimulated by androgen. Therefore, AMACR expression in those SDK1:AMACR fusion transcript positive cases is likely influenced by androgen stimulation or AR activity. As generation of this transcription-mediated fusion transcript needs a complicated transcript level control, it may not as efficient as the consequence of genomic fusion to stimulate the expression of the downstream gene by an androgen regulated promoter. As the product contains half of SDK1 and most of the sequence of AMACR, it is also possible that the fusion protein may gain novel oncogenic functions. While the role of SDK1:AMACR fusion products in CaP development and progression requires further clarification, it is equally important to investigate the mechanism how promoter on a different chromosome can be used to drive a gene expression and if the generation of SDK1:AMACR is influenced by androgen level or AR activity, which will help us to prevent this large proportion of SDK1:AMACR fusion transcript positive CaP in Chinese men.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Neal DE. PSA testing for prostate cancer improves survival--but can we do better? Lancet Oncol. 2010;11:702–703. doi: 10.1016/S1470-2045(10)70152-2. [DOI] [PubMed] [Google Scholar]
- 3.Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nat Rev Cancer. 2008;8:268–278. doi: 10.1038/nrc2351. [DOI] [PubMed] [Google Scholar]
- 4.Draisma G, Etzioni R, Tsodikov A, Mariotto A, Wever E, Gulati R, Feuer E, de Koning H. Lead time and overdiagnosis in prostate-specific antigen screening: importance of methods and context. J Natl Cancer Inst. 2009;101:374–383. doi: 10.1093/jnci/djp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tawadros T, Doerfler A, Treuthardt C, Praz V, Levi F, Zouhair A, Berthold D, Bauer J, Iselin C, Jichlinski P. [Prostate cancer screening: an update] . Rev Med Suisse. 2009;5:2438–2441. [PubMed] [Google Scholar]
- 6.Barry MJ. Screening for Prostate Cancer-The Controversy That Refuses to Die. N Engl J Med. 2009;360:1351–1354. doi: 10.1056/NEJMe0901166. [DOI] [PubMed] [Google Scholar]
- 7.Cuzick J, Thorat MA, Andriole G, Brawley OW, Brown PH, Culig Z, Eeles RA, Ford LG, Hamdy FC, Holmberg L, Ilic D, Key TJ, La Vecchia C, Lilja H, Marberger M, Meyskens FL, Minasian LM, Parker C, Parnes HL, Perner S, Rittenhouse H, Schalken J, Schmid HP, Schmitz-Drager BJ, Schroder FH, Stenzl A, Tombal B, Wilt TJ, Wolk A. Prevention and early detection of prostate cancer. Lancet Oncol. 2014;15:e484–492. doi: 10.1016/S1470-2045(14)70211-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Truong M, Yang B, Jarrard DF. Toward the detection of prostate cancer in urine: a critical analysis. J Urol. 2013;189:422–429. doi: 10.1016/j.juro.2012.04.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salagierski M, Schalken JA. Molecular diagnosis of prostate cancer: PCA3 and TMPRSS2:ERG gene fusion. J Urol. 2012;187:795–801. doi: 10.1016/j.juro.2011.10.133. [DOI] [PubMed] [Google Scholar]
- 10.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 11.Esgueva R, Perner S, J LaFargue C, Scheble V, Stephan C, Lein M, Fritzsche FR, Dietel M, Kristiansen G, Rubin MA. Prevalence of TMPRSS2-ERG and SLC45A3-ERG gene fusions in a large prostatectomy cohort. Mod Pathol. 2010;23:539–546. doi: 10.1038/modpathol.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD, Pienta KG, Kuefer R, Vessella R, Sun XW, Meyerson M, Lee C, Sellers WR, Chinnaiyan AM, Rubin MA. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 13.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 14.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 15.Mao X, Yu Y, Boyd LK, Ren G, Lin D, Chaplin T, Kudahetti SC, Stankiewicz E, Xue L, Beltran L, Gupta M, Oliver RT, Lemoine NR, Berney DM, Young BD, Lu YJ. Distinct genomic alterations in prostate cancers in Chinese and Western populations suggest alternative pathways of prostate carcinogenesis. Cancer Res. 2010;70:5207–5212. doi: 10.1158/0008-5472.CAN-09-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue L MX, Ren G, Stankiewicz E, Kudahetti SC, Lin D, Beltran L, Berney DM, Lu YJ. Chinese and Western prostate cancers show alternate pathogenetic pathways in association with ERG status. Am J Cancer Res. 2012;2:736–744. [PMC free article] [PubMed] [Google Scholar]
- 17.Ren G, Liu X, Mao X, Zhang Y, Stankiewicz E, Hylands L, Song R, Berney DM, Clark J, Cooper C, Lu YJ. Identification of frequent BRAF copy number gain and alterations of RAF genes in Chinese prostate cancer. Genes Chromosomes Cancer. 2012;51:1014–1023. doi: 10.1002/gcc.21984. [DOI] [PubMed] [Google Scholar]
- 18.Ren S, Peng Z, Mao JH, Yu Y, Yin C, Gao X, Cui Z, Zhang J, Yi K, Xu W, Chen C, Wang F, Guo X, Lu J, Yang J, Wei M, Tian Z, Guan Y, Tang L, Xu C, Wang L, Gao X, Tian W, Wang J, Yang H, Wang J, Sun Y. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res. 2012;22:806–821. doi: 10.1038/cr.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren G, Zhang Y, Mao X, Liu X, Mercer E, Marzec J, Ding D, Jiao Y, Qiu Q, Sun Y, Zhang B, Yeste-Velasco M, Chelala C, Berney D, Lu YJ. Transcription-mediated chimeric RNAs in prostate cancer: time to revisit old hypothesis? OMICS. 2014;18:615–624. doi: 10.1089/omi.2014.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noel EE, Yeste-Velasco M, Mao X, Perry J, Kudahetti SC, Li NF, Sharp S, Chaplin T, Xue L, McIntyre A, Shan L, Powles T, Oliver RT, Young BD, Shipley J, Berney DM, Joel SP, Lu YJ. The Association of CCND1 Overexpression and Cisplatin Resistance in Testicular Germ Cell Tumors and Other Cancers. Am J Pathol. 2010;176:2607–2615. doi: 10.2353/ajpath.2010.090780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren G, Liu X, Mao X, Zhang Y, Stankiewicz E, Hylands L, Song R, Berney DM, Clark J, Cooper C, Lu YJ. Identification of frequent BRAF copy number gain and alterations of RAF genes in chinese prostate cancer. Genes Chromosomes and Cancer. 2012;51:1014–1023. doi: 10.1002/gcc.21984. [DOI] [PubMed] [Google Scholar]
- 22.Ren S, Peng Z, Mao JH, Yu Y, Yin C, Gao X, Cui Z, Zhang J, Yi K, Xu W, Chen C, Wang F, Guo X, Lu J, Yang J, Wei M, Tian Z, Guan Y, Tang L, Xu C, Wang L, Tian W, Wang J, Yang H, Sun Y. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res. 2012;22:806–821. doi: 10.1038/cr.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards PA. Fusion genes and chromosome translocations in the common epithelial cancers. J Pathol. 2010;220:244–254. doi: 10.1002/path.2632. [DOI] [PubMed] [Google Scholar]
- 24.Boyd LK, Mao X, Lu YJ. The complexity of prostate cancer: genomic alterations and heterogeneity. Nat Rev Urol. 2012;9:652–664. doi: 10.1038/nrurol.2012.185. [DOI] [PubMed] [Google Scholar]
- 25.Verone AR, Duncan K, Godoy A, Yadav N, Bakin A, Koochekpour S, Jin JP, Heemers HV. Androgen-responsive serum response factor target genes regulate prostate cancer cell migration. Carcinogenesis. 2013;34:1737–1746. doi: 10.1093/carcin/bgt126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heemers HV, Schmidt LJ, Sun Z, Regan KM, Anderson SK, Duncan K, Wang D, Liu S, Ballman KV, Tindall DJ. Identification of a clinically relevant androgen-dependent gene signature in prostate cancer. Cancer Res. 2011;71:1978–1988. doi: 10.1158/0008-5472.CAN-10-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu JC, Stolk JA, Zhang XQ, Silva SJ, Houghton RL, Matsumura M, Vedvick TS, Leslie KB, Badaro R, Reed SG. Identification of differentially expressed genes in human prostate cancer using subtraction and microarray. Cancer Res. 2000;60:1677–1682. [PubMed] [Google Scholar]
- 28.Zhou M, Chinnaiyan AM, Kleer CG, Lucas PC, Rubin MA. Alpha-methylacyl-CoA racemase: A novel tumor marker over-expressed in several human cancers and their precursor lesions. Am J Surg Pathol. 2002;26:926–931. doi: 10.1097/00000478-200207000-00012. [DOI] [PubMed] [Google Scholar]
- 29.Rubin MA, Zhou M, Dhanasekaran SM, Varambally S, Barrette TR, Sanda MG, Pienta KJ, Ghosh D, Chinnaiyan AM. alpha-methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. J Am Med Assoc. 2002;287:1662–1670. doi: 10.1001/jama.287.13.1662. [DOI] [PubMed] [Google Scholar]
- 30.Zehentner BK, Secrist H, Zhang X, Hayes DC, Ostenson R, Goodman G, Xu J, Kiviat M, Kiviat N, Persing DH, Houghton RL. Detection of alpha-methylacyl-coenzyme-A racemase transcripts in blood and urine samples of prostate cancer patients. Mol Diagn Ther. 2006;10:397–403. doi: 10.1007/BF03256217. [DOI] [PubMed] [Google Scholar]
- 31.Jiang Z, Woda BA, Rock KL, Xu YD, Savas L, Khan A, Pihan G, Cai F, Babcook JS, Rathanaswami P, Reed SG, Xu JC, Fanger GR. P504S: A new molecular marker for the detection of prostate carcinoma. Am J Surg Pathol. 2001;25:1397–1404. doi: 10.1097/00000478-200111000-00007. [DOI] [PubMed] [Google Scholar]
- 32.Luo J, Zha S, Gage WR, Dunn TA, Hicks JL, Bennett CJ, Ewing CN, Platz EA, Ferdinandusse S, Wanders RJ, Trent JM, Isaacs WB, De Marzo AM. alpha-Methylacyl-CoA racemase: A new molecular marker for prostate cancer. Cancer Res. 2002;62:2220–2226. [PubMed] [Google Scholar]
- 33.Kaic G, Tomasovic-Loncaric C. Alpha-methylacyl-CoA racemase (AMACR) in fine-needle aspiration specimens of prostate lesions. Diagn Cytopathol. 2009;37:803–808. doi: 10.1002/dc.21103. [DOI] [PubMed] [Google Scholar]
- 34.Kuefer R, Varambally S, Zhou M, Lucas PC, Loeffler M, Wolter H, Mattfeldt T, Hautmann RE, Gschwend JE, Barrette TR, Dunn RL, Chinnaiyan AM, Rubin MA. alpha-Methylacyl-CoA racemase: expression levels of this novel cancer biomarker depend on tumor differentiation. Am J Pathol. 2002;161:841–848. doi: 10.1016/s0002-9440(10)64244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zha S, Ferdinandusse S, Denis S, Wanders RJ, Ewing CM, Luo J, De Marzo AM, Isaacs WB. Alpha-methylacyl-CoA racemase as an androgen-independent growth modifier in prostate cancer. Cancer Res. 2003;63:7365–7376. [PubMed] [Google Scholar]
- 36.Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 37.Horiuchi T, Aigaki T. Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol Cell. 2006;98:135–140. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Zhao L, Jiang H, Wang W. Short homologous sequences are strongly associated with the generation of chimeric RNAs in eukaryotes. J Mol Evol. 2009;68:56–65. doi: 10.1007/s00239-008-9187-0. [DOI] [PubMed] [Google Scholar]
- 39.Amery L, Fransen M, De Nys K, Mannaerts GP, Van Veldhoven PP. Mitochondrial and peroxisomal targeting of 2-methylacyl-CoA racemase in humans. J Lipid Res. 2000;41:1752–1759. [PubMed] [Google Scholar]
- 40.Li CF, Chen LT, Lan J, Chou FF, Lin CY, Chen YY, Chen TJ, Li SH, Yu SC, Fang FM, Tai HC, Huang HY. AMACR amplification and overexpression in primary imatinib-naive gastrointestinal stromal tumors: a driver of cell proliferation indicating adverse prognosis. Oncotarget. 2014;5:11588–11603. doi: 10.18632/oncotarget.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubin MA, Bismar TA, Andren O, Mucci L, Kim R, Shen R, Ghosh D, Wei JT, Chinnaiyan AM, Adami HO, Kantoff PW, Johansson JE. Decreased alpha-methylacyl CoA racemase expression in localized prostate cancer is associated with an increased rate of biochemical recurrence and cancer-specific death. Cancer Epidemiol Biomarkers Prev. 2005;14:1424–1432. doi: 10.1158/1055-9965.EPI-04-0801. [DOI] [PubMed] [Google Scholar]


