Abstract
Background: Interleukin-10 (IL-10) is a multifunctional cytokine with both immunosuppressive and anti-angiogenic properties and it plays an important role in the pathogenesis of cancer. A number of studies have examined the association between its promoter -1082/-819/-592 polymorphism and risk of lung cancer. However, the results are inconsistent and inconclusive. The aim of this study was to explore whether the IL-10 gene polymorphism contribute to the susceptibility of lung cancer. Method: We searched in PubMed, EMBASE, Cochrane Library as well as Chinese databases including China National Knowledge Infrastructure (CNKI) and Wan Fang database for all the relevant studies up to May 15, 2015. The data were extracted by two independent authors. The pooled odds ratios (ORs) with 95% confidence intervals (CIs) under co-dominant model, dominant model and recessive model were estimated. Results: A total of 8 studies involving 2033 cases and 3100 controls were included in the meta-analysis. The results revealed that the IL-10 -592C/A polymorphism was related to lung cancer susceptibility under all models (C allele vs. A allele: OR=1.195, 95% CI=1.075-1.329; CC vs. AA: OR=1.651, 95%=1.290-2.113; CA vs. AA: OR=1.229, 95%=1.029-1.468; CA+AA vs. CC: OR=0.832, 95%=0.704-0.984; CC+CA vs. AA: OR=1.301, 95%=1.100-1.538) and IL-10 -819C/T polymorphism was associated with lung cancer susceptibility under three models (C allele vs. T allele: OR=1.441, 95% CI=1.228-1.691; CC vs. TT: OR=2.444, 95%=1.732-3.449; CC+CT vs. TT: OR=1.496, 95%=1.172-1.908). For IL-10 -1082G/A, there was no significant association between its polymorphism and lung cancer risk. Conclusions: This meta-analysis demonstrated that two polymorphisms (-592C/A and -819C/T) in the promoter region of IL-10 gene were significantly associated with the risk of lung cancer in general population, while -1082G/A polymorphism did not affect susceptibility to lung cancer.
Keywords: IL-10, polymorphism, lung cancer, systematic review, meta-analysis
Introduction
Lung cancer has become the most frequently occurring cancer worldwide, which accounts for 13% of the total cancer cases and 18% of total deaths each year [1]. Yet even now, the potential mechanism of lung carcinogenesis is still unclear. Tobacco smoking is considered to be the leading cause of lung cancer, but only about 15% of heavy tobacco smokers ultimately develop lung cancer [2]. It is established that lung cancer is a complex multifactorial disease resulting from the interactions between various genetic and environmental factors3. Chronic inflammation has long been deemed to be a crucial factor in the development of carcinogenesis [4]. Previous studies have suggested that many pro-inflammatory cytokines were involved in the pathogenesis of lung cancer [5,6]. However, the molecular mechanisms underlying this relationship are not well explained.
Interleukin-10 (IL-10), mainly produced by many kinds of immune cells [7], is an anti-inflammatory cytokine involved in down-regulating cytotoxic and cell-mediated inflammatory responses [8]. And recent study has demonstrated that IL-10 has anti-angiogenic and immunosuppressive properties, which played an important role in the pathogenesis of cancer, including lung cancer [9].
The human IL-10 gene is located on chromosome 1q32 [10]. Promoter region polymorphisms appear to be correlated with variations in transcription. Three single nucleotide polymorphisms (SNPs), positioned at -1082G/A, -819C/T and -592C/A promoter regions, have been linked to effective production of IL-10. These polymorphisms have been investigated as potential susceptibility factors for lung cancer. Unfortunately, effects of IL-10 promoter polymorphisms on lung cancer genotypes in different studies are inconsistent and inconclusive [11-21]. Most studies used an inadequate sample sizes and lacked statistical power to obtain reliable conclusions. Therefore, we extensively reviewed literatures and conducted a meta-analysis to provide more credible evidence by systematically summarizing existed data.
Methods
Search strategy
An electronic search was performed in PubMed, EMBASE, Cochrane Library as well as Chinese databases including China National Knowledge Infrastructure (CNKI) and WanFang database for all the relevant studies utilizing the following search terms: “interleukin-10” or “IL-10”, “polymorphism” or “SNPs”, “lung cancer” or “lung carcinoma” (the latest research was updated to May 15, 2015). When more than one of the same patient population was included in several publications, only the most recent or complete study was used. Search and literature retrieval were completed independently by two investigators. Disagreements of the search result were settled by discussion among all the authors. No limitation was placed on publication status or language.
Inclusion and exclusion criteria
Studies were included in this meta-analysis if they met the following criteria: (1). the study assessed the association between lung cancer and IL-10 polymorphisms (-1082G/A, -819C/T and -592C/A); (2). using case-control or cohort design; (3). data in the studies were adequate to calculate odds ratio (OR) and 95% confidence interval (95% CI); (4). the genotype was tested in controls to ensure its fitting with the Hardy-Weinberg equilibrium (HWE). The major reasons for exclusion from our studies were: (1). family-based or sibling-based association studies; (2). the study without control group; (3). literature with insufficient data for evaluating OR and 95% CI; (4). reviews and abstracts. For overlapping studies, only the one with the largest sample numbers was included.
Data extraction
Two investigators independently extracted the data from all eligible studies according to the inclusion and exclusion criteria. We verified accuracy of data by comparing collection forms from each investigator. Any discrepancy was resolved by discussion. The following information was collected from each study: name of the first author, year of publication, country of origin, ethnicity of research population, experimental method, sample sizes of cases and controls, genotype distributions of cases and controls, and the Hardy-Weinberg equilibrium (HWE) results in controls.
Statistical analysis
We measure the strength of the association between IL-10 SNPs (-1082G/A, -819C/T and -592C/A) and lung cancer by pooled OR with its corresponding 95% CI, which were estimated for allelic comparison, homozygote comparison, heterozygote comparison, dominant model and recessive model respectively. The P value of the pooled OR was considered significant if less than 0.05, which was examined by Z test. Heterogeneity across studies was determined by Chi-square test based Q statistic test and I [2] statistic and the presence of heterogeneity was confirmed if the result was P<0.05 and I2≥50%. In the condition of existence of heterogeneity, a random-effect model was utilized [22]; otherwise the fixed-effect model was employed to pool the results [23]. Additionally, in order to evaluate the stability of results, a sensitivity analysis was conducted. Both Begg’s test and Egger’s test were performed to test whether publication bias existed or not. All the analyses that have been mentioned were completed by STATA v.12.0.
Results
Characteristics of included studies
As shown in the Figure 1, 112 articles were found according to the search strategy and then 32 duplicates are removed. Meanwhile, 43 records without full-text were excluded. After reading the abstracts and full-texts of the remaining 37 articles, 29 records not conforming to the inclusion criteria were further excluded. Eventually, 8 studies involving a total of 2033 cases and 3100 controls met the criteria were included in the meta-analysis of which 9 articles were in English and other 1 in Chinese. The characteristics of included articles were showed in Table 1 and all of them were case-control study.
Figure 1.
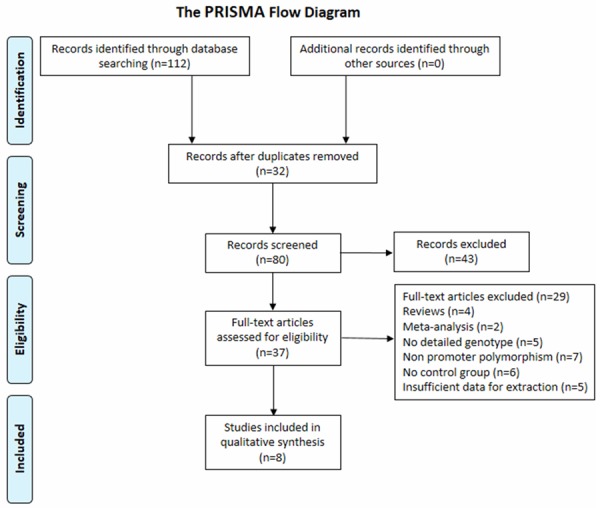
Flow chart of study selection.
Table 1.
Characteristics of studies included in the systematic review and meta-analysis
| Study | Year | Country | Ethnicity | Genotyping method | Cases | Controls | Genotype frequency in cases | Genotype frequency in controls | PHWE | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -592C/A | CC | CA | AA | CC | CA | AA | ||||||||
| Shih, C. M. | 2005 | Taiwan | Asian | PCR-RFLP | 154 | 205 | 18 | 70 | 66 | 13 | 76 | 116 | 0.9 | |
| Colakogullari, M. | 2008 | Turkey | Asian | PCR-SSP | 44 | 59 | 19 | 23 | 2 | 27 | 25 | 7 | 0.74 | |
| Vogel, U. | 2008 | Denmark | European | PCR | 403 | 744 | 241 | 149 | 13 | 452 | 250 | 42 | 0.34 | |
| Liang, H. G. | 2011 | China | Asian | PCR-RFLP | 116 | 120 | 11 | 36 | 69 | 7 | 44 | 69 | 0.99 | |
| Hsia, T. C. | 2014 | Taiwan | Asian | PCR-RFLP | 358 | 716 | 40 | 145 | 173 | 71 | 277 | 368 | 0.08 | |
| Zhang, Y. M. | 2015 | China | Asian | PCR-RFLP | 330 | 336 | 110 | 156 | 64 | 75 | 176 | 85 | 0.37 | |
| -819C/T | CC | TC | TT | CC | TC | TT | ||||||||
| Seifart, C. | 2005 | German | European | PCR-RFLP | 77 | 242 | 42 | 31 | 4 | 140 | 88 | 14 | 0.97 | |
| Shih, C. M. | 2005 | Taiwan | Asian | PCR-RFLP | 154 | 205 | 30 | 58 | 66 | 15 | 86 | 104 | 0.63 | |
| Colakogullari, M. | 2008 | Turkey | Asian | PCR-SSP | 44 | 59 | 19 | 23 | 2 | 26 | 26 | 7 | 0.9 | |
| Zhang, Y. M. | 2015 | China | Asian | PCR-RFLP | 330 | 336 | 87 | 135 | 108 | 47 | 144 | 145 | 0.25 | |
| -1082G/A | AA | AG | GG | AA | AG | GG | ||||||||
| Seifart, C. | 2005 | German | European | PCR-RFLP | 115 | 243 | 30 | 54 | 31 | 86 | 115 | 42 | 0.74 | |
| Colakogullari, M. | 2008 | Turkey | Asian | PCR-SSP | 44 | 59 | 11 | 30 | 3 | 33 | 21 | 5 | 0.53 | |
| Hart, K. | 2011 | Norway | European | TaqMan | 436 | 435 | 120 | 207 | 109 | 104 | 226 | 105 | 0.41 | |
| Hsia, T. C. | 2014 | Taiwan | Asian | PCR-RFLP | 358 | 716 | 273 | 69 | 16 | 561 | 130 | 25 | 0.07 | |
PHWE, p value for Hardy-Weinberg Equilibrium test in controls.
Quantitative data synthesis
A summary of this meta-analysis results concerning the relationships between IL-10 polymorphism and lung cancer is provided in Table 2. Six studies determined IL-10 -592C/A polymorphism were included in pooling. Total sample sizes for lung cancer and control groups were 1405 and 2180. Overall, the results of pooling all studies showed that IL-10 -592C/A polymorphism exhibited the obvious association with lung cancer susceptibility in general population under allelic, homozygous, heterozygous, recessive and dominant model (C allele vs. A allele: OR=1.195, 95% CI=1.075-1.329, P=0.001; CC vs. AA: OR=1.651, 95%=1.290-2.113, P<0.001; CA vs. AA: OR=1.229, 95%=1.029-1.468, P=0.023; CA+AA vs. CC: OR=0.832, 95%=0.704-0.984, P=0.032; CC+CA vs. AA: OR=1.301, 95%=1.100-1.538, P=0.002) (Figure 2). No significant heterogeneity was found in all the five models. The fixed-effect model was therefore chosen to synthesize the data.
Table 2.
The meta-analysis results of association between IL-10 polymorphisms and lung cancer
| SNPs | Contrast model | OR (95% CI) | P | Test for heterogeneity | Publication bias (Egger‘s test) | ||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| I2 (%) | P | t | P | ||||
| -592C/A | C vs. A | 1.195 (1.075-1.329) | 0.001 | 35.3 | 0.172 | 0.25 | 0.813 |
| CC vs. AA | 1.651 (1.290-2.113) | 0.000 | 0.0 | 0.562 | 1.01 | 0.372 | |
| CA vs. AA | 1.229 (1.029-1.468) | 0.023 | 30.7 | 0.205 | 1.89 | 0.132 | |
| CA+AA vs. CC | 0.832 (0.704-0.984) | 0.032 | 51.2 | 0.069 | -0.83 | 0.452 | |
| CC+CA vs. AA | 1.301 (1.100-1.538) | 0.002 | 25.6 | 0.242 | 1.69 | 0.167 | |
| -819C/T | C vs. T | 1.441 (1.228-1.691) | 0.000 | 49.7 | 0.114 | -1.83 | 0.210 |
| CC vs. TT | 2.444 (1.732-3.449) | 0.000 | 0.0 | 0.446 | -0.47 | 0.683 | |
| CT vs. TT | 1.220 (0.939-1.586) | 0.137 | 0.0 | 0.663 | 1.25 | 0.337 | |
| CT+TT vs. CC* | 0.638 (0.354-1.151) | 0.136 | 77.0 | 0.005 | 0.50 | 0.667 | |
| CC+CT vs. TT | 1.496 (1.172-1.908) | 0.001 | 0.0 | 0.785 | 0.51 | 0.661 | |
| -1082G/A | A vs. G* | 0.815 (0.626-1.061) | 0.128 | 67.6 | 0.026 | -3.05 | 0.093 |
| AA vs. GG | 0.849 (0.641-1.126) | 0.256 | 48.0 | 0.124 | -1.30 | 0.322 | |
| AG vs. GG | 0.843 (0.651-1.091) | 0.193 | 0.0 | 0.426 | 1.07 | 0.397 | |
| AG+GG vs. AA* | 1.340 (0.855-2.100) | 0.201 | 77.6 | 0.004 | 1.78 | 0.145 | |
| AA+AG vs. GG | 0.839 (0.658-1.070) | 0.158 | 4.7 | 0.369 | -0.17 | 0.881 | |
Random-effect model was used.
Figure 2.
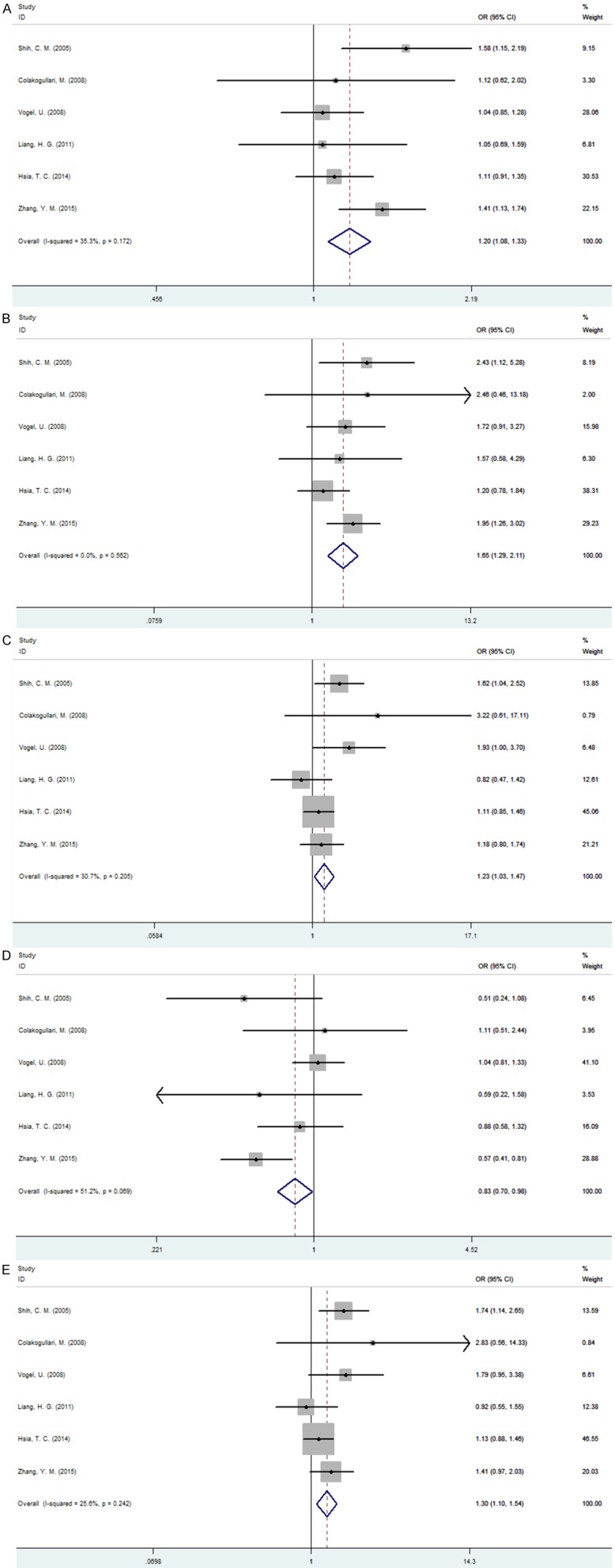
Forest plot of association between IL-10-5922C/A polymorphism and lung cancer. A. C allele vs. A allele; B. CC vs. AA; C. CA vs. AA; D. CA+AA VS. CC; E. CC+CA vs. AA.
Four studies involving a total of 605 cases and 842 controls that identified the association between IL-10 -819C/T polymorphism and lung cancer risk were included in this meta-analysis. The results suggested that IL-10 -819C/T polymorphism was associated with lung cancer susceptibility under allelic, homozygous and dominant model (C allele vs. T allele: OR=1.441, 95% CI=1.228-1.691, P<0.001; CC vs. TT: OR=2.444, 95%=1.732-3.449, P<0.001; CC+CT vs. TT: OR=1.496, 95%=1.172-1.908, P=0.001). However, under heterozygous and recessive model, we observed no association between IL-10 -819C/T polymorphism and lung cancer risk (CT vs. TT: OR=1.220, 95% CI=0.939-1.586, P=0.137; CT+TT vs. CC: OR=0.638, 95%=0.354-1.151, P=0.136) (Figure 3). Meanwhile, significant heterogeneity was detected under the recessive model and a random-effect model was utilized to pool the data.
Figure 3.
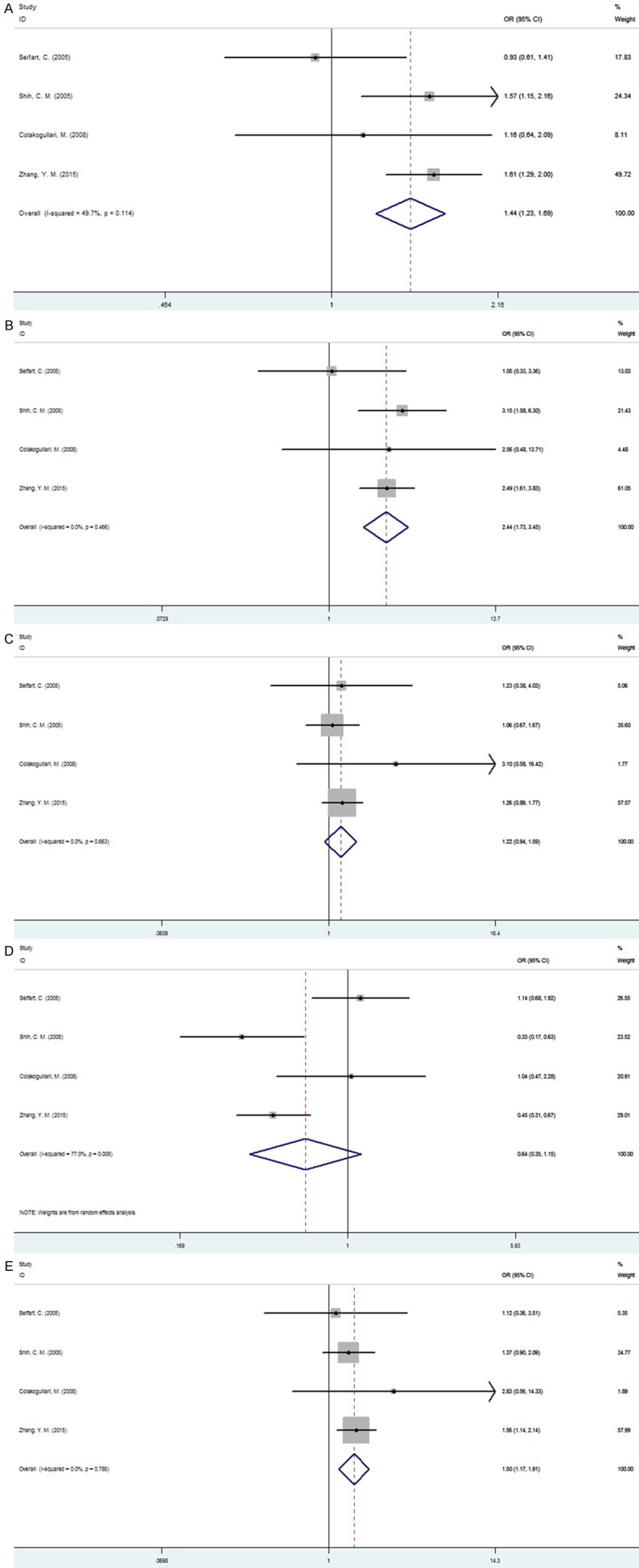
Forest plot of association between IL-10-8192C/T polymorphism and lung cancer. A. C allele vs. T allele; B. CC vs. TT; C. CT vs. TT; D. CT+TT VS. CC; E. CC+CT vs. TT.
For the IL-10 -1082G/A polymorphism, 4 studies (953 cases and 1453 controls) were pooled to perform the meta-analysis and the results indicated there was no significant between IL-10 -1082G/A polymorphism and lung cancer susceptibility under all the five models (A allele vs. G allele: OR=0.815, 95% CI=0.626-1.061, P=0.128; AA vs. GG: OR=0.849, 95%=0.641-1.126, P=256; AG vs. GG: OR=0.843, 95%=0.651-1.091, P=0.193; AG+GG vs. AA: OR=1.340, 95%=0.855-2.100, P=0.201; AA+AG vs. GG: OR=0.839, 95%=0.658-1.070, P=0.158) (Figure 4). Under allelic and recessive model, because of significant between-study heterogeneity for the association, we used random-effect model to complete the synthesis of data.
Figure 4.
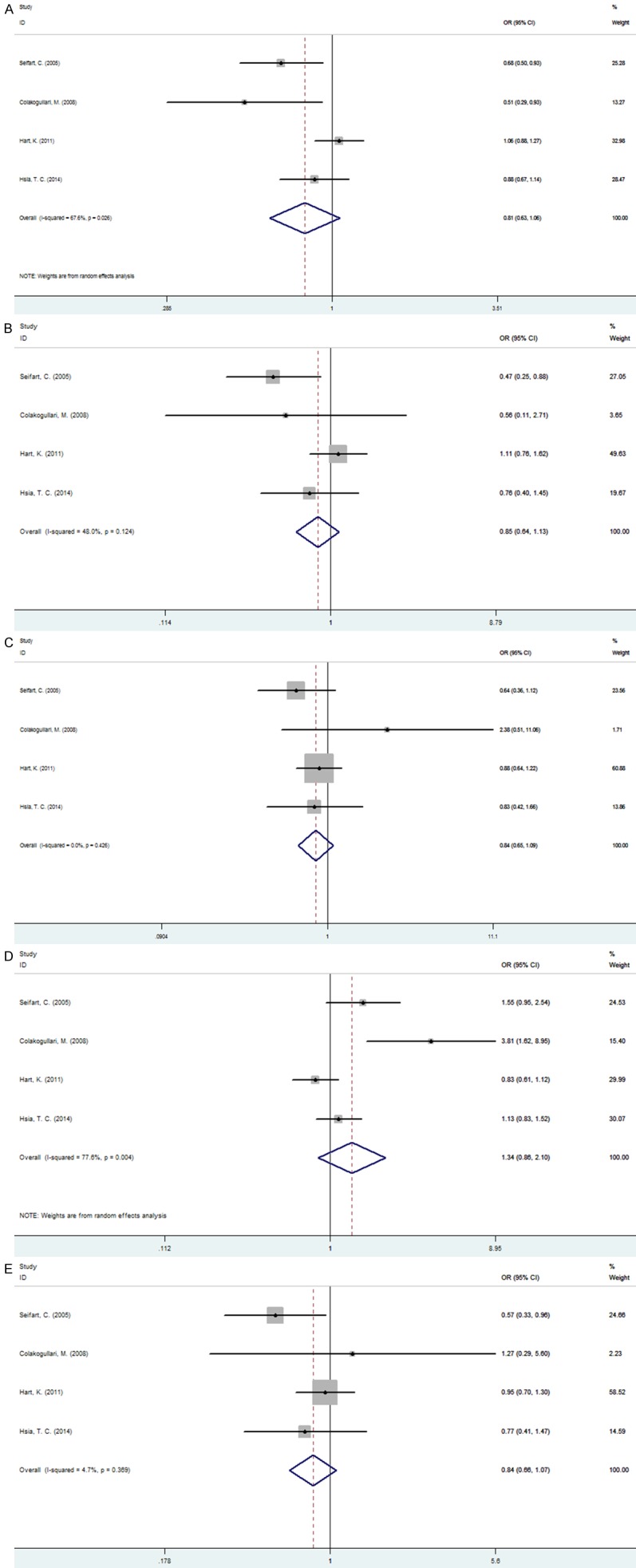
Forest plot of association between IL-10-1082G/A polymorphism and lung cancer. A. A allele vs. G allele; B. AA vs. GG; C. AG vs. GG; D. AG+GG VS. AA; E. AA+AG vs. GG.
Since there was above-mentioned significant heterogeneity for association between IL-10 polymorphism and risk of lung cancer, we performed a subgroup analysis to explore source of heterogeneity. We introduced variables including year of publication, ethnicity of research population, experimental method, sample sizes of cases and controls. Nevertheless, these variables cannot explain the source of heterogeneity.
Publication bias and sensitivity analysis
Begg’s funnel plot and Egger’s test were performed to assess the publication bias of included studies. The shapes of funnel plot were symmetrical, which have not implied the existence of publication bias. All the Egger’s test results of three SNPs were demonstrated in Table 2, all P values were greater than 0.05 and thus there was no obvious publication bias in the meta-analysis.
Sensitivity analysis was performed to evaluate the stability of the result. Each data set was omitted individually to investigate the impact of a single study on the pooled ORs. The exclusion of any single study did not alter the overall conclusion, indicating that results were reliable.
Discussion
To date, convincing evidence indicate that the outcome of lung cancer is modulated by the environment and host genetic components. Many investigations have confirmed that cytokines appear to play the critical roles in the development of lung cancer. Polymorphisms in several cytokine genes have been described and demonstrated to influence gene transcription, leading to inter individual variations in cytokine [24].
IL-10 is a potent immunomodulatory molecule that inhibits the synthesis of pro-inflammatory cytokines, and upregulates of B cell production and differentiation [25]. IL-10 production has also been implicated in the development of various types of cancers, including lung cancer, and may also protect tumors by inhibiting cytotoxic T lymphocyte (CTL)-mediated tumor-specific cell lysis [12], suggesting that IL-10 gene may play an important role in the pathogenesis of lung cancer. Furthermore, Polymorphisms in the promoter of the IL-10 gene, consisting of three SNPs (-1082G/A, -819C/T and -592C/A), have been reported to influence its production capacity [26] and to be associated with the risk of different cancer types. Although a number of studies have investigated the association between the IL-10 promoter polymorphism and susceptibility to lung cancer, the results were controversial and unconvincing. Therefore, it is necessary to use a quantitative approach for combining the results of these studies, and for estimating and explaining their diversity.
This current meta-analysis of 8 case-control studies including 2033 cases and 3100 controls evaluated the association between IL-10 polymorphism and lung cancer risk. We found that -592C/A polymorphism was a risk factor for developing lung cancer between patients with lung cancer and control subjects. The results demonstrated that compared with homozygote (-592AA) genotype, both the variant (-592CC) genotype and the heterozygote (-592CA) genotype were significantly associated with lung cancer risk. In addition, the C allele carriers (CC+CA) had a 30% increased risk of lung cancer, as compared with those individuals with the AA homozygote. Overall, the -592C allele may contribute to the susceptibility of lung cancer. Similarly, -819C/T polymorphism was also significantly associated with lung cancer risk, with carriers of the C allele (CC+CT) having a 50 increased risk. However, compared with homozygote (-819TT) genotype, the heterozygote (-819CT) genotype was not associated with lung cancer risk, indicating that the statistical difference under the dominant model originated from the variant homozygote (-819CC) genotype which had a 140% increased risk of lung cancer. In terms of -1082G/A polymorphism, the results in pooling all studies presented no evidence to support the association between this polymorphism and susceptibility of lung cancer. Due to the limit of the number of studies, we should prudently consider the conclusions. The current consensus is that lung cancer is a multi-factorial disease that results from complex interactions between many environmental and genetic factors, and the interaction among some other SNPs might affect the relationship of each polymorphism included with the development of lung cancer. Hence, if we only consider suspected gene polymorphisms in lung cancer neglecting the role of environmental factors or other genes, we might fail to conclude a real association.
In addition, the existence of heterogeneity may potentially affect the interpretation of the results, which may be attributed to the year of publication, ethnicity of research population, experimental method, sample sizes of cases and controls, or the interaction with other risk factors. In our meta-analysis, there was significant heterogeneity for association between two IL-10 promoter polymorphisms (-819C/T and -1082G/A) and risk of lung cancer under certain genetic models. However, subgroup analysis in consideration of the potential confounders did not address the heterogeneity.
There are several limitations that should be considered when interpreting our results. Firstly, this meta-analysis was based on a relatively small number of studies, especially in the case of -819C/T and -1082G/A polymorphisms, which respectively included only four studies. Secondly, given that only published studies were included in this study, there may be publication bias, although our results of publication bias showed no significance. Thirdly, significant between study heterogeneity was detected in some comparisons, and as such, results may be distorted. Moreover, we included no prospective study to confirm the correlation between IL-10 promoter polymorphisms and risk of lung cancer, which can provide a higher credibility.
Conclusions
In conclusion, this meta-analysis demonstrated that two polymorphisms (-592C/A and -819C/T) in the promoter region of IL-10 gene were significantly associated with the risk of lung cancer in general population, while -1082G/A polymorphism did not affect susceptibility to lung cancer. However, taking above-mentioned shortcomings into consideration, more evidence of prospective, multi-centric and multi-populational trials are needed to further explore the association between the IL-10 promoter polymorphism and susceptibility to lung cancer.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Toh CK, Gao F, Lim WT, Leong SS, Fong KW, Yap SP, Hsu AA, Eng P, Koong HN, Thirugnanam A, Tan EH. Never-smokers with lung cancer: epidemiologic evidence of a distinct disease entity. J. Clin. Oncol. 2006;24:2245–2251. doi: 10.1200/JCO.2005.04.8033. [DOI] [PubMed] [Google Scholar]
- 3.Yokota J, Shiraishi K, Kohno T. Genetic basis for susceptibility to lung cancer: Recent progress and future directions. Adv Cancer Res. 2010;109:51–72. doi: 10.1016/B978-0-12-380890-5.00002-8. [DOI] [PubMed] [Google Scholar]
- 4.Seitz HK, Stickel F. Risk factors and mechanisms of hepatocarcinogenesis with special emphasis on alcohol and oxidative stress. Biol Chem. 2006;387:349–360. doi: 10.1515/BC.2006.047. [DOI] [PubMed] [Google Scholar]
- 5.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 6.Takizawa H, Tanaka M, Takami K, Ohtoshi T, Ito K, Satoh M, Okada Y, Yamasawa F, Umeda A. Increased expression of inflammatory mediators in small-airway epithelium from tobacco smokers. Am J Physiol Lung Cell Mol Physiol. 2000;278:L906–913. doi: 10.1152/ajplung.2000.278.5.L906. [DOI] [PubMed] [Google Scholar]
- 7.Fillatreau S, Gray D, Anderton SM. Not always the bad guys: B cells as regulators of autoimmune pathology. Nat Rev Immunol. 2008;8:391–397. doi: 10.1038/nri2315. [DOI] [PubMed] [Google Scholar]
- 8.Wu MS, Huang SP, Chang YT, Shun CT, Chang MC, Lin MT, Wang HP, Lin JT. Tumor necrosis factor-alpha and interleukin-10 promoter polymorphisms in Epstein-Barr virus-associated gastric carcinoma. J Infect Dis. 2002;185:106–109. doi: 10.1086/324771. [DOI] [PubMed] [Google Scholar]
- 9.Chow MT, Moller A, Smyth MJ. Inflammation and immune surveillance in cancer. Semin Cancer Biol. 2012;22:23–32. doi: 10.1016/j.semcancer.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Jones EA, Flavell RA. Distal enhancer elements transcribe intergenic RNA in the IL-10 family gene cluster. J Immunol. 2005;175:7437–7446. doi: 10.4049/jimmunol.175.11.7437. [DOI] [PubMed] [Google Scholar]
- 11.Seifart C, Plagens A, Dempfle A, Clostermann U, Vogelmeier C, von Wichert P, Seifart U. TNFalpha, TNF-beta, IL-6, and IL-10 polymorphisms in patients with lung cancer. Dis Markers. 2005;21:157–165. doi: 10.1155/2005/707131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shih CM, Lee YL, Chiou HL, Hsu WF, Chen WE, Chou MC, Lin LY. The involvement of genetic polymorphism of IL-10 promoter in non-small cell lung cancer. Lung cancer. 2005;50:291–297. doi: 10.1016/j.lungcan.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Colakogullari M, Ulukaya E, Yilmaztepe Oral A, Aymak F, Basturk B, Ursavas A, Oral HB. The involvement of IL-10, IL-6, IFN-gamma, TNFalpha and TGF-beta gene polymorphisms among Turkish lung cancer patients. Cell Biochem Funct. 2008;26:283–290. doi: 10.1002/cbf.1419. [DOI] [PubMed] [Google Scholar]
- 14.Vogel U, Christensen J, Wallin H, Friis S, Nexø BA, Raaschou-Nielsen O, Overvad K, Tjønneland A. Polymorphisms in genes involved in the inflammatory response and interaction with NSAID use or smoking in relation to lung cancer risk in a prospective study. Mutat Res. 2008;639:89–100. doi: 10.1016/j.mrfmmm.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Hart K, Landvik NE, Lind H, Skaug V, Haugen A, Zienolddiny S. A combination of functional polymorphisms in the CASP8, MMP1, IL10 and SEPS1 genes affects risk of non-small cell lung cancer. Lung Cancer. 2011;71:123–129. doi: 10.1016/j.lungcan.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 16.Liang HG, Fu ZR, Qi Xm. Relationship between the genetic polymorphism in Interleukin-10-592C/A and susceptibility to non-small cell lung cancer. China Journal of Modern Medicine. 2011:189–193. [Google Scholar]
- 17.Wang YC, Sung WW, Wu TC, Wang L, Chien WP, Cheng YW, Chen CY, Shieh SH, Lee H. Interleukin-10 haplotype may predict survival and relapse in resected non-small cell lung cancer. PLoS One. 2012;7:e39525. doi: 10.1371/journal.pone.0039525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reyes-Gibby CC, Wang J, Spitz M, Wu X, Yennurajalingam S, Shete S. Genetic variations in interleukin-8 and interleukin-10 are associated with pain, depressed mood, and fatigue in lung cancer patients. J Pain Symptom Manage. 2013;46:161–172. doi: 10.1016/j.jpainsymman.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang YC, Sung WW, Wang L, Cheng YW, Chen CY, Wu TC, Shieh SH, Lee H. Different impact of IL10 haplotype on prognosis in lung squamous cell carcinoma and adenocarcinoma. Anticancer Res. 2013;33:2729–2735. [PubMed] [Google Scholar]
- 20.Hsia TC, Chang WS, Liang SJ, Chen WC, Tu CY, Chen HJ, Yang MD, Tsai CW, Hsu CM, Tsai CH, Bau DT. Interleukin-10 (IL-10) promoter genotypes are associated with lung cancer risk in Taiwan males and smokers. Anticancer Res. 2014;34:7039–7044. [PubMed] [Google Scholar]
- 21.Zhang YM, Mao YM, Sun YX. Genetic polymorphisms of IL-6 and IL-10 genes correlate with lung cancer in never-smoking Han population in China. Int J Clin Exp Med. 2015;8:1051–1058. [PMC free article] [PubMed] [Google Scholar]
- 22.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 23.Egger M, Smith GD, Phillips AN. Meta-analysis: principles and procedures. BMJ. 1997;315:1533–1537. doi: 10.1136/bmj.315.7121.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng WJ, He Q, Yang JX, Wang BX, Lu MM, Wang S, Wang J. Meta-analysis of association between cytokine gene polymorphisms and lung cancer risk. Mol Biol Rep. 2012;39:5187–5194. doi: 10.1007/s11033-011-1315-z. [DOI] [PubMed] [Google Scholar]
- 25.Taga K, Tosato G. IL-10 inhibits human T cell proliferation and IL-2 production. J Immunol. 1992;148:1143–1148. [PubMed] [Google Scholar]
- 26.Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]


