Abstract
MicroRNAs (miRNAs) are a class of short non-coding, single stranded RNAs, which perform post-transcriptional regulatory functions as tumor suppressors or oncogenes. Single nucleotide polymorphisms (SNPs) in miRs genes are currently being identified for contributing to cancer risk, prognosis and survival, however, an association between miR-149 rs2292832 T/C SNP and cancer risk is uncertain. Therefore, we performed an updated meta-analysis of all currently publications to clarify this relationship. From PubMed and Chinese language (WanFang) databases, we located articles published up to June 1, 2015, obtaining 21 case-control studies from 20 different articles containing 8913 cases and 9944 controls based on search criteria for cancer susceptibility related to the miR-149 rs2292832 T/C SNP. Odds ratios (OR) and 95% confidence intervals (CI) revealed association strengths. There had no association between this SNP and whole cancer risk. At the same time, in several subgroups, also no association was found in ethnicity, sex and smoking status. Nevertheless, poorly significant association was detected in cancer type (Digestive cancer: OR = 0.90, 95% CI = 0.81-1.00, Pheterogeneity = 0.142 for CT vs. TT) and source of control (population-based: OR = 1.15, 95% CI = 1.00-1.32, Pheterogeneity = 0.427 for CC vs. CT+TT) subgroups. The miR-149 rs2292832 T/C SNP may poorly decrease digestive cancer risk. Studies with larger samples and gene-environment interactions are warranted to understand the role of miR-149 polymorphisms, especially rs2292832 T/C SNP, in whole cancer risk.
Keywords: miR-149, sex, smoking, cancer, meta-analysis, polymorphism
Introduction
MicroRNAs are a family of short (approximately 22 nucleotides), non-coding, endogenous RNA molecules that regulate gene expression at the post-transcriptional level by suppressing translation of protein coding genes by destabilizing or cleaving target mRNAs [1]. Critical roles of miRNAs have been demonstrated in various key biological processes including growth of organism [2], hormone secretion [3], inflammatory reaction [4] and especially tumorigenesis [5,6]. Many evidences indicate that miRNAs are involved in malignant transformation of human cells [7] and miRNA profiling is significantly altered in human cancer [8-10]. Deregulation of miRNA expression seems to generate an important signaling cascade, since most of them have a large number of potential mRNA targets.
Sequence variants in miRNA genes are described as mechanisms that can contribute to their deregulation [11]. A mutation or a single nucleotide polymorphism (SNP) at a miRNA region might affect the transcription of miRNA primary transcripts, their processing to mature miRNA or miRNA target interactions [8,12]. Although the role of miRNA genetic variants in cancer susceptibility is largely unknown, the importance of miRNA SNPs has been implicated in many cancers. It is well known that common SNPs in miRNAs and SNPs within their targets (miRNA-binding SNPs) may affect miRNA target expression and functions and thus may contribute to cancer risk [13].
So far, many studies have investigated the association between the miR-149 rs2292832 T/C SNP and cancer risk. But the results were not conclusive and consistent. Furthermore, several meta-analysis have reported that there was no significant association between this SNP and cancer susceptibility, after that, a number of larger studies have been published, and some novel findings may be found in our current study. So we performed this updated meta-analysis of 20 published studies [14-33] to derive a more powerful estimation of the association between the miR-149 rs2292832 T/C SNP and cancer risk.
Materials and methods
Identification and eligibility of relevant studies
Searches were conducted in PubMed and in Chinese language (CNKI and WanFang) databases using key words ‘mir’, ‘cancer’ or ‘tumor’ or ‘carcinoma’, and ‘polymorphism’ or ‘variant’. No restrictions were placed on language or publication year and the last search was updated on June 1, 2015. A total of 494 articles were retrieved using the abovementioned terms and 20 articles contained the inclusion criteria. References of the retrieved and review articles were also screened by hand.
Inclusion criteria and exclusion criteria
Studies that were included in our analysis had to meet all of the following criteria: (1) the correlation between cancer risk and the miR-149 rs2292832 T/C SNP; (2) case-control study and (3) sufficient genotype (TT, TC and CC) numbers for cases and controls. The following exclusion criteria were used: (1) lack of a control population; (2) lack of available genotype frequency data; and (3) duplicated study.
Data extraction
Two of the authors extracted all data independently according to the selection criteria. The following items were collected: last name of first author, year of publication, country of origin, ethnicity, cancer type, the total and number of each genotype frequency in case/control groups, ‘source of control’, Hardy-Weinberg equilibrium (HWE) of controls, age range in case/control group, and genotyping method. Subgroup analysis, stratified by cancer type, was performed, especially including digestive cancer and none-digestive cancer. Ethnicity was categorized as Caucasian, Asian, Chinese and none Chinese. The ‘source of control’ subgroup analysis was performed on two groups and was classified as population-based (PB) or hospital-based (HB). Smoking (smoker or non-smoker) status and subject sex (male or female) were also included in our meta-analysis.
Statistical analysis
Odds ratios (OR) with 95% confidence intervals (CI) were used to measure the strength of the association between the miR-149 rs2292832 T/C SNP. The statistical significance of the summary OR was determined with the Z-test. A heterogeneity assumption was evaluated among studies using a Chi-square-based Q-test. A P value of > 0.10 for the Q-test indicated a lack of heterogeneity among the studies. If significant heterogeneity was detected, the random-effects model (DerSimonian-Laird method) was used. Otherwise, the fixed-effects model (Mantel-Haenszel method) was chosen [34,35].
We investigated the relationship between genetic variants of the miR-149 rs2292832 site and cancer risk by recessive genetic model (CC vs. CT+TT), comparison of heterozygotes (CT vs. TT) and the dominant genetic model (CC+CT vs. TT). A sensitivity analysis was performed by omitting studies, one after another, to assess the stability of results. The departure of the miR-149 rs2292832 SNP from expected frequencies under HWE was assessed in controls using the Pearson Chi-square test (P < 0.05 was considered significant). Publication bias was identified using Egger’s linear regression method and a funnel plot. A P-value < 0.05 in Egger’s linear regression indicated the presence of potential publication bias [36]. All statistical tests for this meta-analysis were performed with Stata software (version 11.0; StataCorp LP, College Station, TX).
Genotyping methods
Methods for genotyping for the miR-149 rs2292832 SNP was conducted in the retrieved literature using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP); matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS); high-resolution melting analysis (HRMA), TaqMan, SNaPshot and Sequenom®.
Results
Study characteristics
A total of 493 articles were collected from the PubMed database via a literature search using different combinations of key terms. As shown in Figure 1, 472 articles were excluded (79 were systematic review and meta-analysis, 12 were review, 381 were irrespective articles), furthermore, just only one article were found in WanFang database. The distribution of genotypes among controls was not consistent with HWE in both Lv et al. [37] and Chu et al. [38], which were also excluded, finally, 20 different articles about 21 case-control studies were included in our meta-analysis. There were 8913 cases and 9944 controls, including several kinds of cancer types. Study characteristics from published studies on the relationship between the miR-149 rs2292832 SNP and cancer risk are summarized in Table 1. Four different articles included the genotype detail and smoking status, and also four articles included information regarding sex. The cancer type of included studies contains ten articles about digestive cancer. In most of the studies, cases were histologically diagnosed, and controls were cancer-free.
Figure 1.
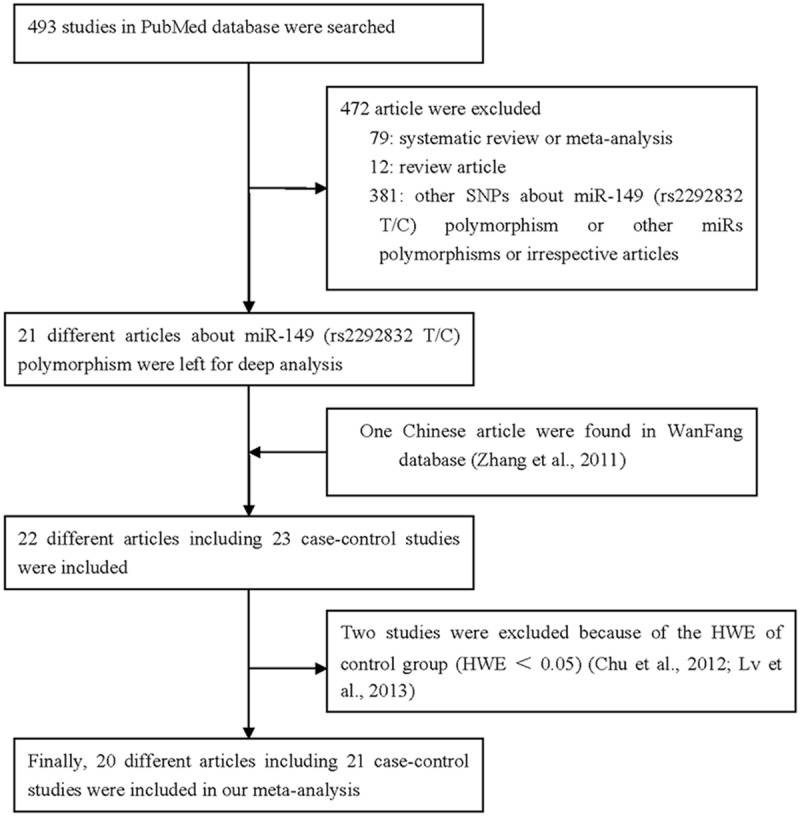
Flowchart illustrating the search strategy used to identify association studies of the miR-149 rs2292832 SNP and overall cancer risk for the meta-analysis.
Table 1.
Basic information for the included studies of the association between miR-149 rs2292832 T/C polymorphism and cancer risk
| First author Year | Country Ethnicity | Source of Control | Cancer Type | Case/Control | Methods | HWE | Age in Cases/Controls (Mean ± SD) |
|---|---|---|---|---|---|---|---|
| Zhang (2012) | China/Asian | PB | BC | 245/229 | PCR-RFLP | 0.213 | 54.66±11.18/54.51±11.41 |
| Hu (2009) | China/Asian | PB | BC | 1009/1093 | PCR-RFLP | 0.160 | 51.60±11.08/51.77±11.19 |
| Min (2012) | South Korea/Asian | HB | CC | 446/502 | PCR-RFLP | 0.948 | 61.89±12.35/61.74±12.11 |
| Zhang (2012) | China/Asian | PB | CC | 443/435 | PCR-RFLP | 0.431 | 62.39±10.81/62.24±10.62 |
| Vinci (2013) | Italy/Caucasian | HB | CC | 160/178 | TaqMan | 0.912 | NA |
| Dikeakos (2014) | Greece/Caucasian | HB | GC | 163/480 | PCR-RFLP | 0.450 | 60.58±9.67/61.15±13.12 |
| Ahn (2012) | South Korea/Asian | HB | GC | 461/447 | PCR-RFLP | 0.977 | 58.08±12.35/58.36±12.32 |
| Zhang (2012) | China/Asian | PB | GC | 274/269 | PCR-RFLP | 0.699 | 62.39± 10.81/62.24±10.62 |
| Hu (2013) | China/Asian | HB | Glioma | 680/690 | SNaPshot | 0.732 | 53.15±12.86/52.98±12.16 |
| Liu (2010) | USA/Caucasian | HB | HNSCC | 1109/1130 | PCR-RFLP | 0.271 | NA |
| Tu (2012) | China-Taiwan/Asian | HB | HNSCC | 273/122 | PCR-RFLP | 0.269 | 53.0±11.0/53.5±13.4 |
| Wang (2014) | China/Asian | HB | HC | 152/304 | PCR-RFLP | 0.623 | 53.5±9.4/53.0±11.5 |
| Liu (2014) | China/Asian | HB | HC | 327/327 | Sequenom® | 0.054 | 56.2±11.3/55.8±10.7 |
| Kou (2014) | China/Asian | HB | HC | 270/532 | PCR-RFLP | 0.877 | 55.8±10.6/52.6±11.2 |
| Kim (2012) | South Korea/Asian | HB | HC | 159/201 | PCR-RFLP | 0.345 | 56.06±11.02/53.58±11.17 |
| Zhang (2011) | China/Asian | PB | LC | 232/231 | PCR-RFLP | 0.123 | 64.4±10.0/63.9±9.9 |
| Tian (2009) | China/Asian | PB | LC | 1058/1035 | PCR-RFLP | 0.855 | 59.78±10.04/59.66±9.83 |
| Vinci (2011) | Italy/Caucasian | HB | LC | 101/129 | HRMA | 0.966 | NA |
| Huang (2013) | China/Asian | HB | NC | 158/242 | PCR-RFLP | 0.724 | 46.6±11.1/47.2±10.5 |
| Wei (2014) | China/Asian | PB | PTC | 838/1006 | MALDI-TOF-MS | 0.731 | 46.30±11.02/47.2±12.29 |
| Du (2014) | China/Asian | HB | RCC | 355/362 | TaqMan | 0.464 | 56.9±12.2/56.7±10.7 |
Annotation: BC, breast cancer; CC, colorectal cancer; GC, gastric cancer; HNSCC, head and neck squamous cell carcinoma; HC, hepatocellular carcinoma; LC, lung cancer; NC, nasopharyngeal carcinoma; PTC, papillary thyroid cancer; RCC, renal cell cancer; HB, hospital-based; PH, population-based; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; MALDI-TOF-MS, matrix-assisted laser desorption/ionization time of flight mass spectrometry; HRMA, high-resolution melting analysis. ®: registered trademark.
Quantitative synthesis
There were no association between miR-149 rs2292832 SNP and whole cancer susceptibility (Heterozygote comparison: OR = 0.97, 95% CI = 0.92-1.04, P heterogeneity = 0.152, dominant model: OR = 0.98, 95% CI = 0.90-1.07, P heterogeneity = 0.007 and recessive model: OR = 1.08, 95% CI = 0.95-1.24, P heterogeneity = 0.007) (Table 2).
Table 2.
Total and stratified subgroup analysis for miR-149 rs2292832 T/C polymorphism and cancer risk
| Variables | Na | Cases/Controls | Heterozygote comparison | Dominant genetic model | Recessive genetic model | |||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||
| OR (95% CI) | P b | OR (95% CI) | P b | OR (95% CI) | P b | |||
| Total | 21 | 8913/9944 | 0.97 (0.92-1.04) | 0.152 | 0.98 (0.90-1.07) | 0.007 | 1.08 (0.95-1.24) | 0.007 |
| Ethnicity | ||||||||
| Asian | 17 | 7380/8027 | 0.96 (0.90-1.03) | 0.117 | 0.95 (0.87-1.05) | 0.008 | 1.04 (0.90-1.20) | 0.021 |
| Caucasian | 4 | 1533/1917 | 1.04 (0.90-1.20) | 0.435 | 1.06 (0.92-1.22) | 0.183 | 1.45 (0.90-2.34) | 0.031 |
| Chinese | 14 | 6314/6877 | 0.99 (0.92-1.06) | 0.141 | 0.98 (0.88-1.09) | 0.009 | 1.04 (0.88-1.23) | 0.007 |
| None-Chinese | 7 | 2599/3067 | 0.94 (0.84-1.06) | 0.273 | 0.97 (0.87-1.08) | 0.109 | 1.11 (0.93-1.33) | 0.133 |
| Cancer type | ||||||||
| BC | 2 | 1254/1322 | 0.92 (0.78-1.08) | 0.106 | 0.92 (0.79-1.07) | 0.101 | 0.97 (0.75-1.26) | 0.737 |
| CC | 3 | 1049/1115 | 0.85 (0.71-1.02) | 0.992 | 0.89 (0.75-1.06) | 0.937 | 1.14 (0.87-1.50) | 0.570 |
| GC | 3 | 898/1196 | 0.93 (0.67-1.29) | 0.064 | 1.00 (0.71-1.41) | 0.030 | 1.28 (0.96-1.71) | 0.223 |
| OC | 4 | 2031/2300 | 1.06 (0.93-1.20) | 0.181 | 1.07 (0.87-1.32) | 0.055 | 1.15 (0.85-1.56) | 0.073 |
| HNSCC | 2 | 1382/1252 | 1.00 (0.85-1.18) | 0.933 | 0.96 (0.82-1.12) | 0.405 | 0.61 (0.27-1.42) | 0.019 |
| HC | 4 | 908/1364 | 0.91 (0.68-1.23) | 0.058 | 0.92 (0.63-1.33) | 0.004 | 0.96 (0.59-1.54) | 0.017 |
| LC | 3 | 1391/1395 | 1.05 (0.90-1.23) | 0.928 | 1.08 (0.93-1.26) | 0.755 | 1.19 (0.94-1.51) | 0.304 |
| Digestive cancer | 10 | 2855/3675 | 0.90 (0.81-1.00) | 0.142 | 0.94 (0.80-1.10) | 0.013 | 1.14 (0.93-1.40) | 0.083 |
| None-digestive cancer | 11 | 6058/6269 | 1.01 (0.94-1.09) | 0.465 | 1.02 (0.95-1.10) | 0.110 | 1.04 (0.86-1.26) | 0.012 |
| Source of control | ||||||||
| HB | 14 | 4814/5646 | 0.98 (0.90-1.06) | 0.156 | 0.99 (0.87-1.12) | 0.008 | 1.04 (0.85-1.24) | 0.003 |
| PB | 7 | 4099/4298 | 0.97 (0.89-1.07) | 0.211 | 1.00 (0.92-1.09) | 0.124 | 1.15 (1.00-1.32) | 0.427 |
| Sex | ||||||||
| Female | 4 | 516/531 | 1.05 (0.80-1.37) | 0.869 | 1.07 (0.84-1.36) | 0.821 | 1.14 (0.74-1.77) | 0.327 |
| Male | 4 | 894/857 | 0.84 (0.52-1.35) | 0.012 | 0.91 (0.66-1.25) | 0.037 | 0.99 (0.72-1.36) | 0.570 |
| Smoking status | ||||||||
| Smoker | 4 | 487/468 | 1.01 (0.52-1.97) | 0.015 | 1.07 (0.67-1.72) | 0.019 | 0.87 (0.54-1.42) | 0.182 |
| Non-smoker | 4 | 255/519 | 0.90 (0.61-1.33) | 0.171 | 0.94 (0.69-1.28) | 0.429 | 1.42 (0.83-2.42) | 0.485 |
Number of comparisons;
P value of Q-test for heterogeneity test;
BC, breast cancer; OC, other cancer; CC, colorectal cancer; GC, gastric cancer; HNSCC; head and neck squamous cell carcinoma; HC, hepatocellular carcinoma; LC, lung cancer; NC, nasopharyngeal carcinoma; PTC, papillary thyroid cancer; RCC, renal cell cancer HB, hospital-based; PH, population-based.
When studies were stratified according to ethnicity, no association was also found in Asian, Caucasian, Chinese or None-Chinese. In the subgroup for sex and smoking status, the same result was detected (Table 2). When studies were stratified by cancer type, a significant association was identified between miR-149 rs2292832 SNP and digestive cancer (Heterozygote comparison: OR = 0.90, 95% CI = 0.81-1.00, P heterogeneity = 0.142, Figure 2), though no relationship was found for breast cancer, colorectal cancer, gastric cancer, head and neck squamous cell carcinoma, hepatocellular carcinoma and lung cancer (Table 1). Interestingly, in the subgroup of source of control, increased association was found in PB (recessive model: OR = 1.15, 95% CI = 1.00-1.32, P heterogeneity = 0.427, Figure 3), rather than HB (Table 2).
Figure 2.
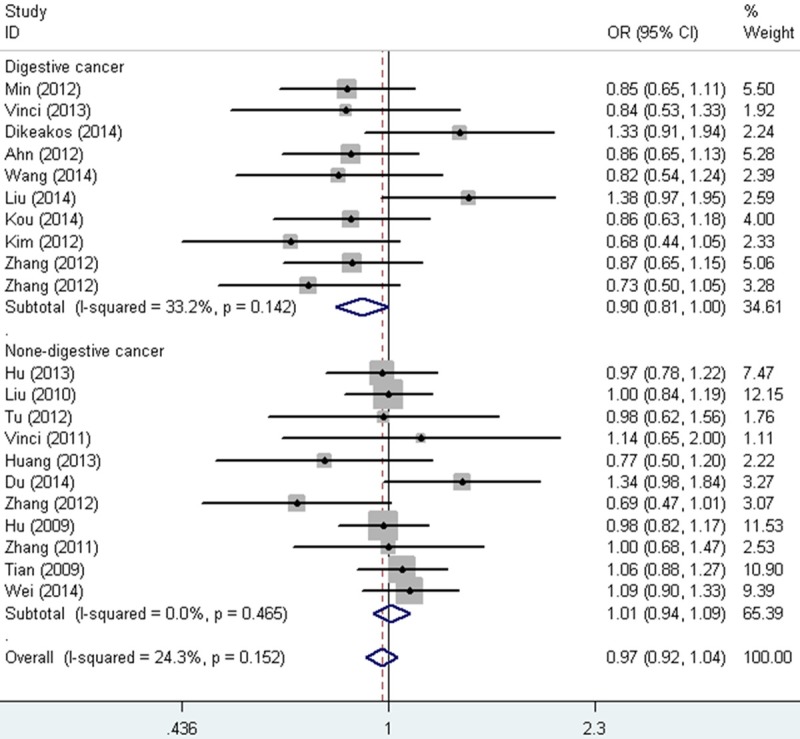
Forest plot of digestive risk associated with the miR-149 rs2292832 SNP (CT vs. TT). The squares and horizontal lines correspond to the study-specific OR and 95% CI. The area of the squares reflects the weight (inverse of the variance). The diamond represents the summary OR and 95% CI.
Figure 3.
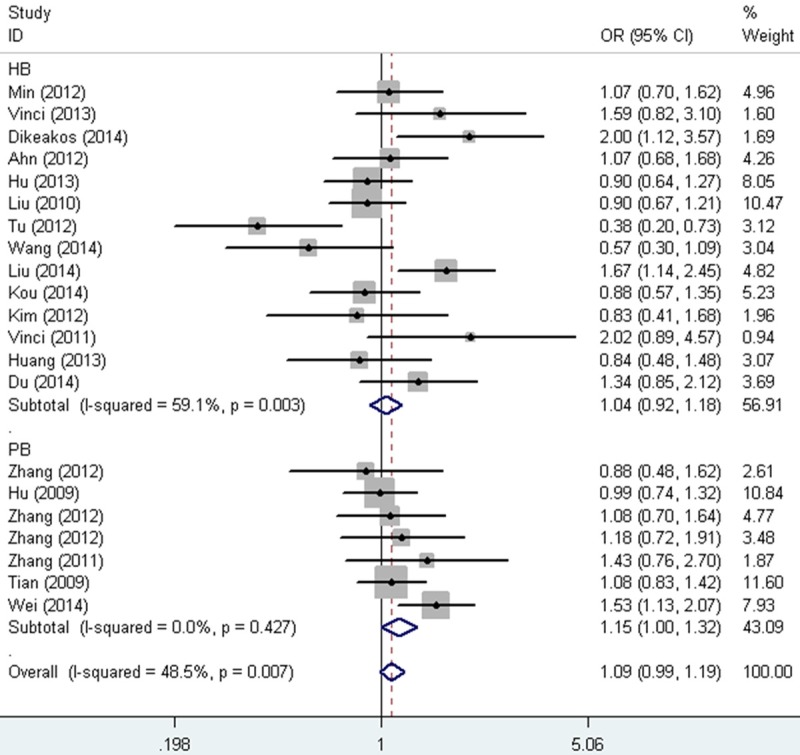
Forest plot of cancer risk associated with the miR-149 rs2292832 SNP (CC vs. CT+TT) by source of control. The squares and horizontal lines correspond to the study-specific OR and 95% CI. The area of the squares reflects the weight (inverse of the variance). The diamond represents the summary OR and 95% CI.
Sensitivity analysis and bias diagnosis
We used a sensitivity analysis to determine whether modifying the meta-analysis inclusion criteria affected the final results. No other single study influenced the summary OR qualitatively (Figure 4). Egger’s test was performed to assess publication bias and to provide statistical evidence of funnel plot symmetry, and data did not reveal evidence of publication bias (date not shown).
Figure 4.
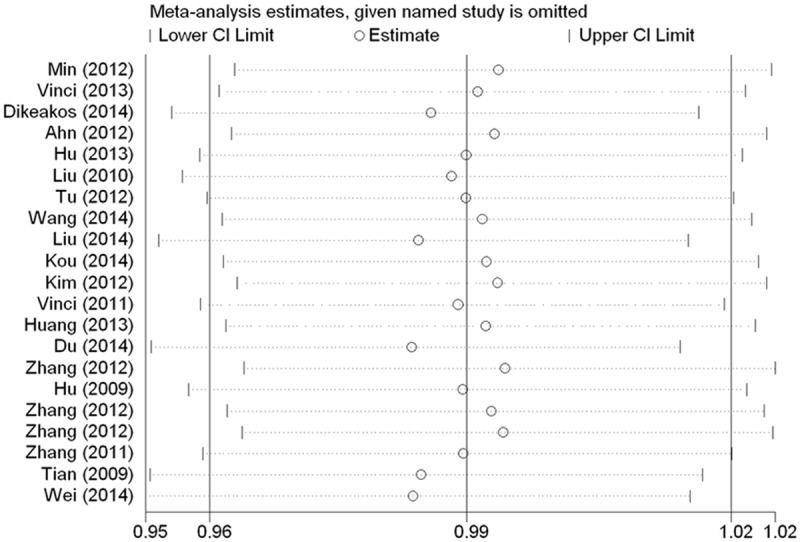
Sensitivity analysis between the miR-149 rs2292832 SNP polymorphism and whole cancer risk.
Discussion
Recent studies have shown that miRNAs may play an important role in human carcinogenesis, and SNPs located either in the pre-miRNAs or within miRNA-binding sites are likely to affect the expression and function of the miRNA targets [39-41] and thus may contribute to the susceptibility to cancer.
Concerning miR-149, it is a pro-apoptotic miRNA that represses the expression of Akt1 and E2F1. Silencing of Akt1 and E2F1 induces apoptosis in human cancer cell lines [42,43], moreover, miR-149 is known as a tumor suppressor with the function of inhibiting cell growth and invasion by binding to the target gene specificity protein 1 [44]. The miR-149 rs2292832 SNP may alter the expression of mature miRNAs or their binding activities to target mRNA, thereby influencing cancer risk through variable mechanisms.
To date, numerous genetic association studies have been conducted to examine the relationship between the miR-149 rs2292832 SNP and the risk of several cancers, including breast cancer, colorectal cancer, gastric cancer, head and neck squamous cell carcinoma, hepatocellular carcinoma and lung cancer. Conflicting results were obtained. Limited sample sizes may account for the inconclusive result. A meta-analysis provides a means for effectively increasing the size of the sample by pooling data from individual correlation studies, thus enhancing the statistical power of the analysis to estimate genetic effects [45]. So we applied this method to demonstrate statistically significant genetic associations.
We used the meta-analysis, containing 8913 cases and 9944 controls, to conclude a convincing association. In the present study, we found that individuals carried CT genotype may have a decreased association with digestive cancer than TT genotype. Recently, two meta-analysis, including Zhang et al. [46] and Xu et al., [47] did not find any association between the miR-149 rs2292832 SNP and cancer susceptibility. However, it is necessary to conduct large sample studies with homogeneous cancer patients and different ethnic backgrounds which may be a way to maximize study efficacy and overcome the limitations of individual studies.
Limitations in the present meta-analysis include the suboptimal number of published studies for a comprehensive analysis, especially in terms of linking smoking status, sex and other cancer types. Secondly, interactions between different polymorphic loci of the same miRNA may modulate cancer risk, which should be included in future research and analysis. In addition, our meta-analysis was based on unadjusted estimates. A more precise analysis should be conducted if individual data are available to adjust for other covariates including age, sex, family history, environmental factors, cancer stage, and lifestyle. Finally, controls may not have been truly healthy individuals.
In summary, in the present meta-analysis, a poorly decreased association was found between the miR-149 rs2292832 SNP and digestive cancer risk. To further confirm the results, larger scale case-control studies with different ethnic groups and multiple cancer types are needed.
Acknowledgements
This work was supported by a grant from The National Natural Science Funds (No. 81101938). Shenyang Science and Technology Project (F14-231-1-56) and Liaoning Province Natural Science Funds (2015020516; 2015020430).
Disclosure of conflict of interest
None.
References
- 1.Ruvkun G. Molecular biology. Glimpses of a tiny RNA world. Science. 2001;294:797–799. doi: 10.1126/science.1066315. [DOI] [PubMed] [Google Scholar]
- 2.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 3.Baroukh N, Ravier MA, Loder MK, Hill EV, Bounacer A, Scharfmann R, Rutter GA, Van Obberghen E. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J Biol Chem. 2007;282:19575–19588. doi: 10.1074/jbc.M611841200. [DOI] [PubMed] [Google Scholar]
- 4.Carissimi C, Fulci V, Macino G. MicroRNAs: novel regulators of immunity. Autoimmun Rev. 2009;8:520–524. doi: 10.1016/j.autrev.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Carissimi C, Fulci V, Macino G. MicroRNAs: novel regulators of immunity. Autoimmun Rev. 2009;8:520–524. doi: 10.1016/j.autrev.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Ruvkun G. Clarifications on miRNA and cancer. Science. 2006;311:36–37. doi: 10.1126/science.311.5757.36d. [DOI] [PubMed] [Google Scholar]
- 7.Negrini M, Ferracin M, Sabbioni S, Croce CM. MicroRNAs in human cancer: from research to therapy. J Cell Sci. 2007;120:1833–1840. doi: 10.1242/jcs.03450. [DOI] [PubMed] [Google Scholar]
- 8.Wu M, Jolicoeur N, Li Z, Zhang L, Fortin Y, L’Abbe D, Yu Z, Shen SH. Genetic variations of microRNAs in human cancer and their effects on the expression of miRNAs. Carcinogenesis. 2008;29:1710–1716. doi: 10.1093/carcin/bgn073. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 10.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 11.Diederichs S, Haber DA. Sequence variations of microRNAs in human cancer: alterations in predicted secondary structure do not affect processing. Cancer Res. 2006;66:6097–6104. doi: 10.1158/0008-5472.CAN-06-0537. [DOI] [PubMed] [Google Scholar]
- 12.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 14.Hu Z, Liang J, Wang Z, Tian T, Zhou X, Chen J, Miao R, Wang Y, Wang X, Shen H. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat. 2009;30:79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 15.Vinci S, Gelmini S, Mancini I, Malentacchi F, Pazzagli M, Beltrami C, Pinzani P, Orlando C. Genetic and epigenetic factors in regulation of microRNA in colorectal cancers. Methods. 2013;59:138–146. doi: 10.1016/j.ymeth.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Li G, Wei S, Niu J, El-Naggar AK, Sturgis EM, Wei Q. Genetic variants in selected premicroRNA genes and the risk of squamous cell carcinoma of the head and neck. Cancer. 2010;116:4753–4760. doi: 10.1002/cncr.25323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du M, Lu D, Wang Q, Chu H, Tong N, Pan X, Qin C, Yin C, Wang M, Zhang Z. Genetic variations in microRNAs and the risk and survival of renal cell cancer. Carcinogenesis. 2014;35:1629–1635. doi: 10.1093/carcin/bgu082. [DOI] [PubMed] [Google Scholar]
- 18.Dikeakos P, Theodoropoulos G, Rizos S, Tzanakis N, Zografos G, Gazouli M. Association of the miR-146aC > G, miR-149T > C, and miR-196a2T > C polymorphisms with gastric cancer risk and survival in the Greek population. Mol Biol Rep. 2014;41:1075–1080. doi: 10.1007/s11033-013-2953-0. [DOI] [PubMed] [Google Scholar]
- 19.Zhang MW, Yu YX, Jin MJ, Pan YF, Jiang X, Li QL, Ma XY, Zhang SC, Chen K. [Association of miR-605 and miR-149 genetic polymorphisms with related risk factors of lung cancer susceptibility] . Zhejiang Da Xue Xue Bao Yi Xue Ban. 2011;40:265–71. doi: 10.3785/j.issn.1008-9292.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Zhang MW, Jin MJ, Yu YX, Zhang SC, Liu B, Jiang X, Pan YF, Li QI, Ma SY, Chen K. Associations of lifestyle-related factors, hsa-miR-149 and hsa-miR-605 gene polymorphisms with gastrointestinal cancer risk. Mol Carcinog. 2012;51:E21–E31. doi: 10.1002/mc.20863. [DOI] [PubMed] [Google Scholar]
- 21.Min KT, Kim JW, Jeon YJ, Jang MJ, Chong SY, Oh D, Kim NK. Association of the miR-146aC > G, 149C > T, 196a2C > T, and 499A > G polymorphisms with colorectal cancer in the Korean population. Mol Carcinog. 2012;51:E65–E73. doi: 10.1002/mc.21849. [DOI] [PubMed] [Google Scholar]
- 22.Zhang M, Jin M, Yu Y, Zhang S, Wu Y, Liu H, Liu H, Chen B, Li Q, Ma X, Chen K. Associations of miRNA polymorphisms and female physiological characteristics with breast cancer risk in Chinese population. Eur J Cancer Care (Engl) 2012;21:274–280. doi: 10.1111/j.1365-2354.2011.01308.x. [DOI] [PubMed] [Google Scholar]
- 23.Kim WH, Min KT, Jeon YJ, Kwon CI, Ko KH, Park PW, Hong SP, Rim KS, Kwon SW, Hwang SG, Kim NK. Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene. 2012;504:92–7. doi: 10.1016/j.gene.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 24.Ahn DH, Rah H, Choi YK, Jeon YJ, Min KT, Kwack K, Hong SP, Hwang SG, Kim NK. Association of the miR-146aC > G, miR-149T > C, miR-196a2T > C, and miR-499A > G polymorphisms with gastric cancer risk and survival in the Korean population. Mol Carcinog. 2013;52:E39–E51. doi: 10.1002/mc.21962. [DOI] [PubMed] [Google Scholar]
- 25.Vinci S, Gelmini S, Pratesi N, Conti S, Malentacchi F, Simi L, Pazzagli M, Orlando C. Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clin Chem Lab Med. 2011;49:2073–2080. doi: 10.1515/CCLM.2011.708. [DOI] [PubMed] [Google Scholar]
- 26.Hu E, Wang D, Zhang X, Li J, Hu Y, Gong H, Liu E. Four common polymorphisms in microRNAs and the risk of adult glioma in a Chinese casecontrol study. J Mol Neurosci. 2013;51:933–940. doi: 10.1007/s12031-013-9980-0. [DOI] [PubMed] [Google Scholar]
- 27.Huang GL, Lu Y, Pu XX, He YX, Chen ML, Li YZ, Tang SY, Che H, He Z. Association study between miR-149 gene polymorphism and nasopharyngeal carcinoma. Biomed Rep. 2013;1:599–603. doi: 10.3892/br.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian T, Shu Y, Chen J, Hu Z, Xu L, Jin G, Liang J, Liu P, Zhou X, Miao R, Ma H, Chen Y, Shen H. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18:1183–1187. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 29.Wang XH, Wang FR, Tang YF, Zou HZ, Zhao YQ. Association of miR-149C > T and miR-499A > G polymorphisms with the risk of hepatocellular carcinoma in the Chinese population. Genet Mol Res. 2014;13:5048–5054. doi: 10.4238/2014.July.4.20. [DOI] [PubMed] [Google Scholar]
- 30.Liu MF, Chen WQ, He YZ, Gu YL. Role of miR-149C > T polymorphisms on the risk of hepatocellular carcinoma in a Chinese population. Genet Mol Res. 2014;13:7184–7189. doi: 10.4238/2014.September.5.4. [DOI] [PubMed] [Google Scholar]
- 31.Wei WJ, Lu ZW, Li DS, Wang Y, Zhu YX, Wang ZY, Wu Y, Wang YL, Ji QH. Association of the miR-149 Rs2292832 polymorphism with papillary thyroid cancer risk and clinicopathologic characteristics in a Chinese population. Int J Mol Sci. 2014;15:20968–20981. doi: 10.3390/ijms151120968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kou JT, Fan H, Han D, Li L, Li P, Zhu J, Ma J, Zhang ZH, He Q. Association between four common microRNA polymorphisms and the risk of hepatocellular carcinoma and HBV infection. Oncol Lett. 2014;8:1255–1260. doi: 10.3892/ol.2014.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tu HF, Liu CJ, Chang CL, Wang PW, Kao SY, Yang CC, Yu EH, Lin SC, Chang KW. The association between genetic polymorphism and the processing efficiency of miR-149 affects the prognosis of patients with head and neck squamous cell carcinoma. PLoS One. 2012;7:e51606. doi: 10.1371/journal.pone.0051606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 35.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 36.Hayashino Y, Noguchi Y, Fukui T. Systematic evaluation and comparison of statistical tests for publication bias. J Epidemiol. 2005;15:235–243. doi: 10.2188/jea.15.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lv M, Dong W, Li L, Zhang L, Su X, Wang L, Gao L, Zhang L. Association between genetic variants in pre-miRNA and colorectal cancer risk in a Chinese population. J Cancer Res Clin Oncol. 2013;139:1405–1410. doi: 10.1007/s00432-013-1456-7. [DOI] [PubMed] [Google Scholar]
- 38.Chu YH, Tzeng SL, Lin CW, Chien MH, Chen MK, Yang SF. Impacts of microRNA gene polymorphisms on the susceptibility of environmental factors leading to carcinogenesis in oral cancer. PLoS One. 2012;7:e39777. doi: 10.1371/journal.pone.0039777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Z, Li Z, Jolicoeur N, Zhang L, Fortin Y, Wang E, Wu M, Shen SH. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35:4535–4541. doi: 10.1093/nar/gkm480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibé B, Bouix J, Caiment F, Elsen JM, Eychenne F, Larzul C, Laville E, Meish F, Milenkovic D, Tobin J, Charlier C, Georges M. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 41.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin T, Ding Z, Li N, Xu J, Luo G, Liu J, Shen J. 2-Tellurium-bridged β-cyclodextrin, a thioredoxin reductase inhibitor, sensitizes human breast cancer cells to TRAIL-induced apoptosis through DR5 induction and NF-κB suppression. Carcinogenesis. 2011;32:154–167. doi: 10.1093/carcin/bgq234. [DOI] [PubMed] [Google Scholar]
- 43.Lin RJ, Lin YC, Yu AL. miR-149 induces apoptosis by inhibiting Akt1 and E2F1 in human cancer cells. Mol Carcinog. 2010;49:719–727. doi: 10.1002/mc.20647. [DOI] [PubMed] [Google Scholar]
- 44.Wang F, Ma YL, Zhang P, Shen TY, Shi CZ, Yang YZ, Moyer MP, Zhang HZ, Chen HQ, Liang Y, Qin HL. SP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancer. J Pathol. 2013;229:12–24. doi: 10.1002/path.4078. [DOI] [PubMed] [Google Scholar]
- 45.Munafò MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20:439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 46.Xu L, Zhou X, Qiu MT, Yin R, Wu YQ, Xu L. Lack of association between hsa-miR-149 rs2292832 polymorphism and cancer risk: a metaanalysis of 12 studies. PLoS One. 2013;8:e73762. doi: 10.1371/journal.pone.0073762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Liu YF, Gan Y. Lack of association between miR-149 C > T polymorphism and cancer susceptibility: a meta-analysis based on 4,677 cases and 4,830 controls. Mol Biol Rep. 2012;39:8749–8753. doi: 10.1007/s11033-012-1735-4. [DOI] [PubMed] [Google Scholar]


