Abstract
Background: Acinetobacter baumannii (A. baumannii), especially the multidrug resistant A. baumannii (MDR-AB) is becoming a common opportunistic pathogen in hospital, and constitutes significant public health threats. This study aimed at investigating the relationship between drug resistance with expression of class A-D β-lactamase genes, mutation in membrane porin and over-expression of efflux pump genes among A. baumannii isolated from Zhengjiang, China. Methods: Antibiotic susceptibility assays were performed using Kirby-Bauer disc diffusion method. PCR was used to detect β-lactamase genes and carO, oprD, adeR, adeS. Real-time PCR was used to assess the mRNA expression level of efflux pump gene adeB. The software of DNAMAN was applied to assemble oprD and carO sequences, and the sequences were compared with those retrieved from GenBank (http://www.ncbi.nlm.nih.gov/). Results: 27 isolates (61.4%) in this study were MDR-AB, in which five β-lactamases including TEM, CTX-M-2, ADC, OXA-23 and OXA-51 were found, and the positive rate was 96.3% (26), 14.8% (4), 92.6% (25), 88.9% (24) and 92.6% (25), respectively. In addition, the expression level of adeB mRNA was significantly increased in MDR-AB, it might due to adeR mutation. Some mutations were also found in carO and oprD. Conclusion: MDR-AB showed high relationship with β-lactamase, mutation in membrane porin and overexpression of adeB, which may directly relates to the mutation in regulating gene adeR.
Keywords: A. baumannii, multidrug resistant, β-lactamase, membrane porin, efflux pump
Introduction
A. baumannii is a kind of nosocomial opportunistic pathogen, which can cause series of diseases, including pneumonia, skin and soft tissue infections, urinary tract infections and bacteraemia in immunocompromised patients [1], especially those in the intensive care units. Moreover, MDR-AB have emerged swiftly all over the world [2-5], bringing a knotty problem which is becoming much more difficult to cure in patients who have been infected. Thus, this pathogen possesses huge potential threat to human health, and its resistant mechanisms urgently need to be understood.
Although the mechanism of resistance in A. baumannii has largely been reported in recent years, which are associated with β-lactam antibiotics resistance includes producing varies enzymes, mutation in membrane porin, changing in conformation of PBP and over-expression of efflux pumps. In this study, we aim to characterize the frequency of class A~D β-lactamases, mutation in the membrane porin encoding genes and efflux pump relative gene among A. baumannii which were isolated in Zhenjiang, China.
Material and methods
Bacterial strains
44 strains of A. baumannii used in this study were isolated from inpatients with infectious diseases in the Affiliated Hospital of Jiangsu University and the First People’s Hospital of Zhengjiang during November 2012 to May 2013. Sputum constituted the most specimens in this study, which was about 90%. Isolates were identified by VITEK 2 compact automatic bacteria identification system (French). The bacteria were incubated on blood culture media and preserved at -80°C until they were used.
Antibiotic susceptibility testing
Antibiotic susceptibility assays were performed using Kirby-Bauer disc diffusion method. In brief, over night bacterial cultures were diluted to 0.5 with normal saline in McFar land, and then sub-cultured in 5 mL Mueller-Hinton broth (Bosai Biotechnology Co. Ltd). The measured inhibition zones were judged according to the CLSL 2014 standard. E. coli (ATCC25922) and P. aeruginosa (ATCC27853) were used as control strains. The following antibiotics were tested: Imipenem, Gentamicin, Amikacin, Ceftazidime, Ceftriaxone, Cefepime, Ciprofloxacin, Ampicillin/sulbactam, Sulfamethoxazole, Cefoperazone/sulbact, Piperacillin/tazobactam, Minocycline (Oxoid company, England).
PCR analysis
PCR was used to detect the β-lactamase genes carO, oprD, adeR and adeS. Primers for carO, oprD, adeR and adeS were synthesized by Wuxi Clone Gen-Tech Institute in China, others were purchased from Shanghai Hanyubiotech Co., Ltd. Among these, the primers for adeR and adeS were used to amplify the full-length fragment of efflux pump regulatory genes adeR and adeS. The PCR kits were provided by Takara Company. All of the reagents used in this experiment were used following the manufacturer’s instructions. Primers used are listed in Table 1.
Table 1.
Primers used in PCR assay
| Target gene | Primer sequence | Size (bp) | |
|---|---|---|---|
| Membraneporin | carO | P1: ATGAAAGTATTACGTGTTTTAGTGACAAC; | 729 |
| P2: TTACCAGTAGAATTCNACACCAACT | |||
| cprD | P1: ATGCTAAAAGCACAAAAACTTACATTAGCA; | 1320 | |
| P2: TTAGAATAATTTCACAGGAATATCTAAGAA | |||
| Class A β-lactamases | TEM | P1: AGGAAGAGTATGATTCAACA; | 535 |
| P2: CTCGTCGTTTGGTATGGC | |||
| SHV | P1: TGCGCAAGCTGCTGACCAGC; | 305 | |
| P2: TTAGCGYTGCCAGTGCTCGA | |||
| CTX-M-1 group | P1: ATGGTTAAAAAATCACTGCGYCAGTTC; | 876 | |
| P2: TCACAAACCGTYGGTGACGATTTTAGCCGC | |||
| CTX-M-2 group | P1: ATGATGACGCAGAGCATTCGCCGCTCA; | 876 | |
| P2: TCAGAAACCGTGGGTTACGATTTTCGC | |||
| CTX-M-9 group | P1: ATGGTGACAAAGAGAGTGCAACGG; | 876 | |
| P2: TTACAGCCCTTCGGCGATGATTCTCGC | |||
| PER | P1: AGTCAGCGGCTTAGATA; | 978 | |
| P2: CGTATGAAAAGGACAATC | |||
| GES | P1: ATGCGCTTCATTCACGCAC; | 846 | |
| P2: CTATTTGTCCGTGCTCAGG | |||
| VEB | P1: GCGGTAATTTAACCAGA; | 961 | |
| P2: GCCTATGAGCCAGTGTT | |||
| CARB | P1: AAAGCAGATCTTGTGACCTATTC; | 588 | |
| P2: TCAGCGCGACTGTGATGTATAAAC | |||
| Class B β-lactamases | IMP | P1: CGGCCKCAGGAGMGKCTTT; | 587 |
| P2: AACCAGTTTTGCYTTACYAT | |||
| VIM | P1: ATTCCGGTCGGMGAGGTCCG; | 633 | |
| P2: GAGCAAGTCTAGACCGCCCG | |||
| SIM | P1: ACAAGGGATTCGGCATCGTT; | 355 | |
| P2: TTATCTTGAGTGTGTCCTGG | |||
| Class C β-lactamases | DHA group | P1: AACTTTCACAGGTGTGCTGGGT; | 405 |
| P2: CCGTACGCATACTGGCTTTGC | |||
| ADC Group | P1: GGTATGGCYGTGGGBGTYATTC; | 739 | |
| P2: CTAAGASTTGGTCRAARGGT | |||
| Class D β-lactamases | OXA-1 group | P1: CTGTTGTTTGGGTTTCGCAAG; | 440 |
| P2: CTTGGCTTTTATGCTTGATG | |||
| OXA-2 group | P1: CAGGCGCYGTTCGYGATGAGTT; | 233 | |
| P2: GCCYTCTATCCAGTAATCGCC | |||
| OXA-10 group | P1: GTCTTTCRAGTACGGCATTA; | 822 | |
| P2: GATTTTCTTAGCGGCAACTTA | |||
| OXA-20 group | P1: TTGATAATCCGATTTCTAGCAC; | 801 | |
| P2: CTAGTTGGGTGGCAAAGCAT | |||
| OXA-23 group | P1: ATGAATAAATATTTTACTTGCTATGTG; | 822 | |
| P2: TTAAATAATATTCAGCTGTTTTAATGA | |||
| OXA-24 group | P1: CAAGAGCTTGCAAGACGGACT; | 420 | |
| P2: TCCAAGATTTTCTAGCRACTTATA | |||
| OXA-51 group | P1: ATGAACATTAAAGCACTCTTACTT; | 825 | |
| P2: CTATAAAATACCTAATTGTTCTAA | |||
| OXA-58 group | P1: TCGATCAGAATGTTCAAGCGC; | 530 | |
| P2: ACGATTCTCCCCTCTGCGC | |||
| Efflux pump regulatory gene | adeR | P1: GCTTGAGCGACTTCTTTTGAAT; | 1134 |
| P2: CTAATCCAGCCTTTTTCAATCG | |||
| adeS | P1: CCCCTAGCTGTAAAAGATGACG; | 1354 | |
| P2: ACCAATGGGGTCAAATACACA | |||
Sequencing and analyzing for carO and oprD
(www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html) was used to analyze the sequences of carO, oprD, adeR and adeS. Chromas were used to read each sequence, and then BLAST online to analysis the results. Software of DNAMAN was used to assemble oprD, adeR and adeS sequences. The sequences obtained in this experiment were compared with those present in the GenBank database from strain SDF (http://blast.ncbi.nlm.nih.gov). After BLAST online, translating the sequences of carO and oprD into amino acid, and then compared with amino acid sequence of strain SDF. Sequence from strain SDF was downloaded from www.biocyc.org. The three-dimensional structure of OprD was created according to the amino acid sequence of OprD using the molecular visualization tools Swiss-Pdb Viewer 3.7.
Analysis of the mRNA level of efflux pump gene AdeB
RT-PCR was used to detect the mRNA level of efflux pump gene adeB. Primer of 16SrRNA was synthesized by Shanghai Hanyubiotech Co., Ltd and Primer of adeB was from Hinggins et al [6]. The RT-PCR kits were provided by Takara Company. All of the reagents were used following the manufacturer’s instructions. Primers used in this study were listed in Table 2. Relative quantification was used to analyze the data according to the formula 2-ΔCt (ΔCt = Ctunknown-Cthousekeeping gene).
Table 2.
Primer sequence of efflux pump genes and 16SrRNA
| Target gene | Primer sequence | Size (bp) |
|---|---|---|
| adeB | P1: GGATTATGGCGACAGAAGGA | 104 bp |
| P2: AATACTGCCGCCAATACCAG | ||
| 16SrRNA | P1: GTAGCGGTGAAATGCGTAGA | 85 bp |
| P2: CTTTCGTACCTCAGCGTCAG |
Statistical analysis
Data were shown as the mean ± S.E.M. The software SPSS was used to perform the statistical analysis of the data with homogeneity of variance test and non-parametric test. P<0.05 was considered to be statistically significant.
Results
Characterization of the isolates
MDR-AB refers to resistance to more than 3 of the 5 classes of antibiotics resistance including aminoglycoside, beta lactamase inhibitor penicillinum, cephalosporins, carbon alkene and fluoroquinolone. In this study, 27 strains of A. baumanii were multidrug-resistant in 44 isolates, and their antibiotic susceptibility patterns were shown in Table 3.
Table 3.
The results of antibiotic susceptibility patterns of A. baumannii isolates
| Antibiotic | R (%) | I (%) | S (%) |
|---|---|---|---|
| Imipenem | 27 (61.4) | 2 (4.5) | 15 (34.1) |
| Gentamicin | 28 (63.6) | 1 (2.3) | 15 (34.1) |
| Amikacin | 29 (65.9) | 0 (0.0) | 15 (34.1) |
| Ceftazidime | 28 (63.6) | 0 (0.0) | 16 (36.4) |
| Ceftriaxone | 31 (70.5) | 1 (2.3) | 12 (27.2) |
| Cefepime | 30 (68.2) | 3 (6.8) | 11 (25.0) |
| Ciprofloxacin | 28 (63.6) | 0 (0.0) | 16 (36.4) |
| Ampicillin/sulbactam | 12 (27.2) | 10 (22.7) | 22 (50.1) |
| Sulfamethoxazole | 29 (65.9) | 2 (4.5) | 13 (29.6) |
| Cefoperazone/sulbact | 20 (45.5) | 3 (6.8) | 21 (47.7) |
| Piperacillin/tazobactam | 27 (61.4) | 3 (6.8) | 14 (31.8) |
| Minocycline | 18 (40.9) | 6 (13.6) | 20 (45.5) |
The break point of Kirby-Bauer disc diffusion in A. baumannii to Cefoperazone/sulbact was accepted according to the standard (S: ≥21 mm; I: 16 mm-20 mm; R: ≤15 mm) [7]. S = susceptible; I = intermediate; R = resistant.
TEM, CTX-M-2, ADC, OXA-23, OXA-51 were expressed inMDR-AB
Among the β-lactamase producing MDR-AB isolates, only TEM, CTX-M-2, ADC, OXA-23 and OXA-51 were found in this study. It was therefore suggested that the multidrug resistance of the isolates might be associated with these β-lactamase genes (Table 4).
Table 4.
The prevalence of β-lactamase genes in MDR-AB
| β-lactamase | Positive | Negative |
|---|---|---|
| TEM | 96.3% (26) | 3.7% (1) |
| CTX-M-2 | 85.2% (23) | 14.8% (4) |
| ADC | 92.6% (25) | 7.4% (2) |
| OXA-23 | 88.9% (24) | 11.1% (3) |
| OXA-51 | 92.6% (25) | 7.4% (2) |
Point mutation of CarO in the isolates
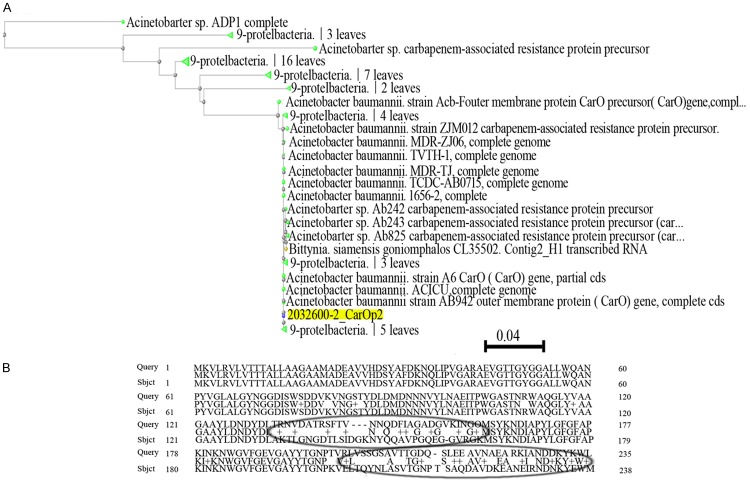
Sequencing results showed that the carO was obtained in 25 of the 27 MDR-AB isolates, which was different from strain SDF. After translating it into amino acid sequence, there were some point mutations were found. Apart from some sense mutation, they were also deficient in 4 amino acids locus at positions 144, 145, 146 and 213 in the isolates, and the amino acid distance was showed in Figure 1.
Figure 1.
Homologic analysis of CarO in A. baumannii isolates. A. The amino acid distance based on CarO of A. baumannii isolates. B. The sense mutation of amino acid of CarO in MDR-AB strains compared with strain SDF.
Point mutation of OprD in the isolates
Like carO analysis, 29 of the 44 strains expressed oprD, and the sequences of OprD from 27 strains of MDR-AB were also showed absolutely the same in this study. Their sense mutations were T→98→A and D→278→G, and the amino acid distance was showed in Figure 2 and the three-dimensional structure of OprD was showed in Figure 3.
Figure 2.
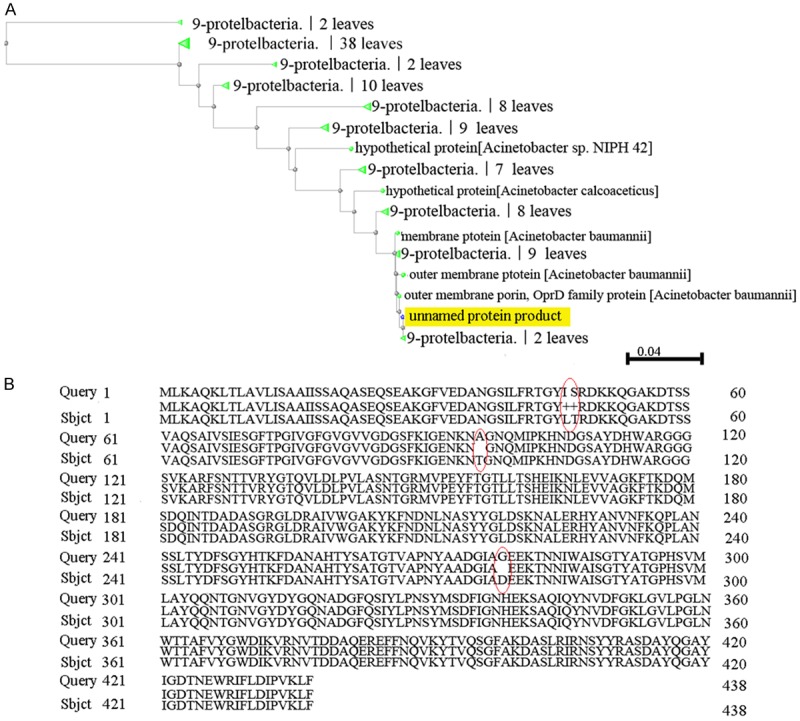
The variability analysis of OprD in A. baumannii isolates. A. The amino acid distance based on CarO of A. baumannii isolates. B. The structure of amino acids of OprD from 29 MDR-AB strains was obviously different from strain SDF, there were somesense mutations on amino acid of OprD were found in A. baumannii isolates.
Figure 3.
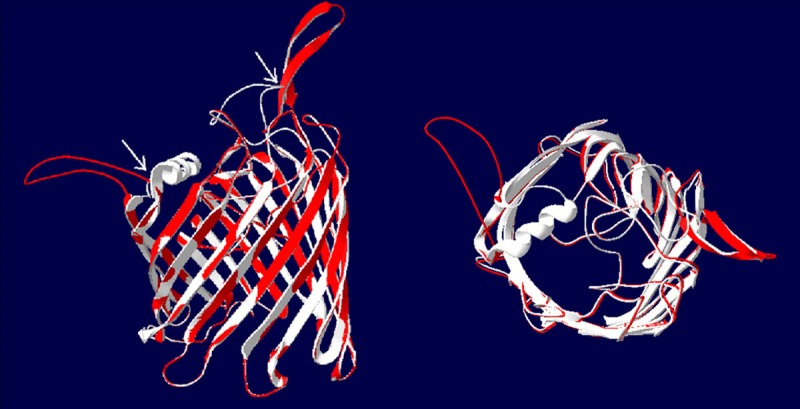
The three-dimensional structure of OprD. The PCR products of OprD genes from 29 MDR-AB strains were sequenced, and the homology analysis showed that there were mutations in MDR-AB strains compared with the SDF strain. It was significant difference in three dimensional structure of protein between MDR-AB and SDF strains by molecular modeling and comparison of overlapping molecular structure. The white region isthe structure of OprD and the red region is the structure of MDR-AB. Left: the frontage of β-barrel structure; Right: the side of β-barrel structure.
The mutation of adeR and adeS
It was reported that adeR and adeS, as main efflux pump regulator genes, involved in the regulation of expression of AdeABC efflux pump system genes [8]. In our study, 3 strains were selected randomly from the MDR-AB strains, and the full length of their adeR and adeS genes was amplified by PCR, followed by blasting online with strain SDF genes. The result showed that there was a single mutation A→G in 2639 of adeR, which cause an Lys219→Glu amino acid replacement, while nonsense mutation was found in adeS (Figure 4A, 4B).
Figure 4.
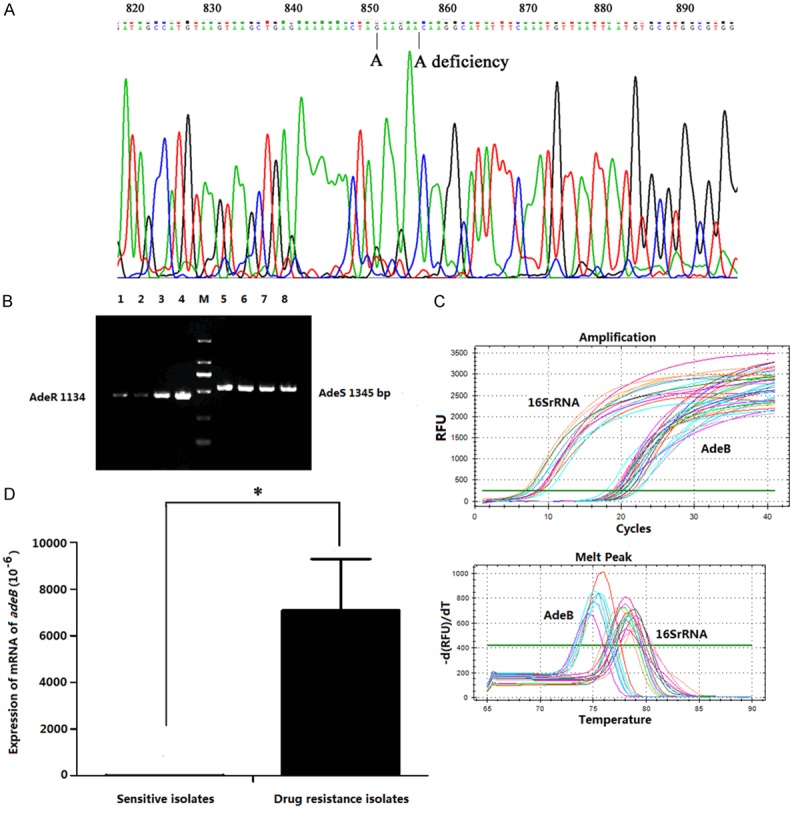
The expression and characteristics of efflux pump genes. PCR amplification and DNA sequence analysis were showed in (A and B), there was a single mutation A→G in 2639 of adeR, which causean Lys219→Glu amino acid replacement; the deficiency of base “A” in 2633 of adeR displayed a nonsense mutation, in addition, nonsense mutation was found in adeS (A and B). (C) The results of qRT-PCR for detecting mRNA levels of 16SrRNA and adeB. (D) Comparison of the levels of adeB mRNA expression between drug sensitive isolates and MDR-AB strains. *P<0.01.
In addition, the adeB gene could be detected in all MDR-AB strains. The mRNA expression level of adeB was also detected using qRT-PCR, and the result showed that the adeB mRNA in MDR-AB was significantly increased (Figure 4C, 4D).
Discussion
The role of A. baumannii is strengthened by relatively high resistance to numerous antibiotics which is determined by both natural and acquired mechanisms. In multi-drug resistant strains of A. baumannii, the drugs of choice are the Carbapenems. Unfortunately, the development of resistance dose not elude even this family of antimicrobial agents, due to the production of carbapenemase or β-lactamases. In addition, resistance to a wide range of antibiotics can be caused by a single mutation in a gene. Emerging expression of β-lactamase and mutation in membrane porin undoubtedly become a significant mechanism of MDR-AB.
Expression of β-lactamase is often assumed to be the result of resistant to β-lactams antibiotics. According to amino acid sequence and conserved motifs, β-lactamases are defined to classes A, B, C, and D [9]. Class C β-lactams means AmpC enzymes, AmpC enzymes are cephalosporinase encoded by chromosomally in Acinetobacter spp and P. aeruginosa [10]. Chromosomal AmpC β-lactams have been identified in several works in A. baumannii such as Acinetobacter-derived cephalosporinase (ADC) [11,12]. The expression of AmpC enzymes was reported to be promoted by insertion such as IASba1 upstream the blaampC gene [11], and to be associated with cephalosporin resistance [13-15].
Our data has revealed that TEM, ADC, OXA-23 and OXA-51 expressed highly in A. baumannii isolates, which were more than 90%, specifically the TEM. Extended spectrum β-lactamases (ESBLs) identified in Acinetobacter spp were TEM, SHV, PER and CTX [10,16], and the most popular was also TEM. In this study, the positive rate of TEM was as high as 96.3%, except one strain which did not expressed TEM, all the others showed obvious stripe. TEM could be mediated not only by chromosome, but also plasmid and TEM-1 is reported to be associated with sulbactam resistance in A. baumannii [17]. CTX-M group was not found in our A. baumannii isolates, but it is known to be the most common type of class A β-lactamases strains in Turkey among ESBL-producing E. coli [18].
Carbapenem resistance in A. baumannii was reported to be due to the emergence and dissemination of OXA-type carbapenemase encoding genes [19], such as OXA-51-like gene [20]. Pagano et al revealed that upstream of OXA-23-like gene only in isolates resistant to carbapenems, whereas ISAba1 upstream of OXA-51-like gene was presented in both susceptible and resistant isolates, and ISAba1/blaOXA-51-like gene alone could not lead to resistance to carbapenems [21]. The positive rate of OXA-51 in this experiment was as high as 92.6% and OXA-23 was 88.9%, and the confirming fact was that resistance rate to imipenem was up to the 96.8% among MDR-AB strains. Recent study showed that IMP increased carbapenemase activities in multidrug-resistant Pseudomonas aeruginosa [22], but no metallo-β-lactamase gene was found in those isolates.
There are limited reports on mutations in the membrane porin of Acinetobacter baumannii. In our experiment, the sequencing results of CarO and OprD were used to compare with strain SDF, and we found some sense mutations in most of the A. baumannii isolates. It can be speculated that the channel would be changed subsequently after increasing mutations in membrane porin, and could resulted in antibiotics being not able to enter or the amount entered being decreased, which will result in resistance.
Furthermore, more and more evidences have shown the relationship between multiple drug resistance of A. baumannii and expression of efflux system [8,23-27]. As it is known, under antibiotic, point mutation of efflux pump regulatory gene induce the over-expression of efflux pump system, decreasing the concentration of antibiotics in A. baumannii. Our data showed that the expression of adeB had a close relationship with the mutation of adeR, it might expel the antibiotics from bacteria, and mediate drug resistance.
In conclusion, the mutual effect of various of drug resistance mechanisms lead to the overwhelming resistance to majority of antibiotics, and MDR-AB have populated all over the world, it not only can be spread within a country, but also disseminated from one country to another [28-30]. In this study, we found that those strains of MDR-AB have high relationship with some β-lactamase and mutations in carO and oprD, and mutation in adeR causing over-expression of adeB. Consequently, prevention the spread of β-lactamase producing isolates should be managed carefully, in order to prevent the spread of MDR-AB within hospitals.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31270947, 81370084 and 31470881), Natural Science Foundation of Jiangsu Province (BK2011472) and Postdoctoral Foundation of China (2014T70490 and 2013T60508).
Disclosure of conflict of interest
None.
References
- 1.Richmond GE, Chua KL, Piddock LJ. Efflux in Acinetobacter baumannii can be determined by measuring accumulation of H33342 (bis-benzamide) J Antimicrob Chemother. 2013;68:1594–1600. doi: 10.1093/jac/dkt052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morfin-Otero R, Alcántar-Curiel MD, Rocha MJ, Alpuche-Aranda CM, Santos-Preciado JI, Gayosso-Vázquez C, Araiza-Navarro JR, Flores-Vaca M, Esparza-Ahumada S, González-Díaz E, Pérez-Gómez HR, Rodríguez-Noriega E. Acinetobacter baumannii Infections in a Tertiary Care Hospital in Mexico over the Past 13 Years. Chemotherapy. 2013;59:57–65. doi: 10.1159/000351098. [DOI] [PubMed] [Google Scholar]
- 3.Chen LK, Liu YL, Hu A, Chang KC, Lin NT, Lai MJ, Tseng CC. Potential of bacteriophage [greek small letter phi] AB2 as an environmental biocontrol agent for the control of multidrug-resistant Acinetobacter baumannii. BMC Microbiol. 2013;13:154. doi: 10.1186/1471-2180-13-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin MF, Liou ML, Tu CC, Yeh HW, Lan CY. Molecular Epidemiology of Integron-Associated Antimicrobial Gene Cassettes in the Clinical Isolates of Acinetobacter baumannii from Northern Taiwan. Ann Lab Med. 2013;33:242–247. doi: 10.3343/alm.2013.33.4.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peymani A, Farajnia S, Nahaei MR, Sohrabi N, Abbasi L, Ansarin K, Azhari F. Prevalence of class 1 integron among multidrug-resistant Acinetobacter baumannii in Tabriz, northwest of Iran. Pol J Microbiol. 2012;61:57–60. [PubMed] [Google Scholar]
- 6.Higgins PG, Wisplinghoff H, Stefanik D, Seifert H. Selection of topoisomerase mutations and overexpression of adeB mRNA transcripts during an outbreak of Acinetobacter baumannii. J Antimicrob Chemother. 2004;54:821–823. doi: 10.1093/jac/dkh427. [DOI] [PubMed] [Google Scholar]
- 7.Levin AS. Multiresistant Acinetobacter infections: A role for sulbactam combinations in overcoming an emerging worldwide problemc. Clin Microbiol Infect. 2002;8:144–153. doi: 10.1046/j.1469-0691.2002.00415.x. [DOI] [PubMed] [Google Scholar]
- 8.Marchand I, Damier-Piolle L, Courvalin P, Lambert T. Expression of the RND-type dfflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother. 2004;48:3298–3304. doi: 10.1128/AAC.48.9.3298-3304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush K. The ABCD’s of beta-lactamase nomenclature. J Infect Chemother. 2013;19:549–559. doi: 10.1007/s10156-013-0640-7. [DOI] [PubMed] [Google Scholar]
- 10.Zavascki AP, Carvalhaes CG, Picão RC, Gales AC. Multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: resistance mechanisms and implications for therapy. Expert Rev Anti Infect Ther. 2010;8:71–93. doi: 10.1586/eri.09.108. [DOI] [PubMed] [Google Scholar]
- 11.Hujer KM, Hamza NS, Hujer AM, Perez F, Helfand MS, Bethel CR, Thomson JM, Anderson VE, Barlow M, Rice LB, Tenover FC, Bonomo RA. Identification of a new allelic variant of the Acinetobacter baumannii cephalosporinase, ADC-7 beta-lactamase: defining a unique family of class C enzymes. Antimicrob Agents Chemother. 2005;49:2941–2948. doi: 10.1128/AAC.49.7.2941-2948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Héritier C, Poirel L, Nordmann P. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin Microbiol Infect. 2006;12:123–130. doi: 10.1111/j.1469-0691.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 13.Rezaee MA, Pajand O, Nahaei MR, Mahdian R, Aghazadeh M, Ghojazadeh M, Hojabri Z. Prevalence of Ambler class A beta-lactamases and ampC expression in cephalosporin-resistant isolates of Acinetobacter baumannii. Diagn Microbiol Infect Dis. 2013;76:330–334. doi: 10.1016/j.diagmicrobio.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Hamidian M, Hall RM. ISAba1 targets a specific position upstream of the intrinsic ampC gene of Acinetobacter baumannii leading to cephalosporin resistance. J Antimicrob Chemother. 2013;68:2682–2683. doi: 10.1093/jac/dkt233. [DOI] [PubMed] [Google Scholar]
- 15.Hamidian M, Hancock D, Pand RM. Hall, Horizontal transfer of an ISAba125-activated ampC gene between Acinetobacter baumannii strains leading to cephalosporin resistance. J Antimicrob Chemother. 2013;68:244–245. doi: 10.1093/jac/dks345. [DOI] [PubMed] [Google Scholar]
- 16.Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007;5:939–51. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 17.Krizova L, Poirel L, Nordmann P, Nemec A. TEM-1 beta-lactamase as a source of resistance to sulbactam in clinical strains of Acinetobacter baumannii. J Antimicrob Chemother. 2013;68:2786–2791. doi: 10.1093/jac/dkt275. [DOI] [PubMed] [Google Scholar]
- 18.Copur Cicek A, Saral A, Ozad Duzgun A, Yasar E, Cizmeci Z, Ozlem Balci P, Sari F, Firat M, Altintop YA, Ak S, Caliskan A, Yildiz N, Sancaktar M, Esra Budak E, Erturk A, Birol Ozgumus O, Sandalli C. Nationwide study of Escherichia coli producing extended-spectrum beta-lactamases TEM, SHV and CTX-M in Turkey. J Antibiot (Tokyo) 2013;66:647–650. doi: 10.1038/ja.2013.72. [DOI] [PubMed] [Google Scholar]
- 19.Clímaco EC, Oliveira ML, Pitondo-Silva A, Oliveira MG, Medeiros M, Lincopan N, da Costa Darini AL. Clonal complexes 104, 109 and 113 playing a major role in the dissemination of OXA-Carbapenemase-Producing Acinetobacter baumannii in Southeast Brazil. Infect Genet Evol. 2013;19:127–133. doi: 10.1016/j.meegid.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 20.Zander E, Higgins PG, Fernández-González A, Seifert H. Detection of intrinsic blaOXA-51-like by multiplex PCR on its own is not reliable for the identification of Acinetobacter baumannii. Int J Med Microbiol. 2013;303:88–89. doi: 10.1016/j.ijmm.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Pagano M, Martins AF, Machado AB, Barin J, Barth AL. Carbapenem-susceptible Acinetobacter baumannii carrying the ISAba1 upstream blaOXA-51-like gene in Porto Alegre, southern Brazil. Epidemiol Infect. 2012;141:330–333. doi: 10.1017/S095026881200074X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tada T, Miyoshi-Akiyama T, Shimada K, Shimojima M, Kirikae T. IMP-43 and IMP-44 Metallo beta-Lactamases with Increased Carbapenemase Activities in Multidrug-Resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57:4427–4432. doi: 10.1128/AAC.00716-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magnet S, Courvalin P, Lambert T. Resistance-nodulation cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother. 2001;45:3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wieczorek P, Sacha P, Hauschild T, Zórawski M, Krawczyk M, Tryniszewska E. Multidrug resistant Acinetobacterbaumannii--the role of AdeABC (RND family) efflux pump in resistance to antibiotics. Folia Histochem Cytobiol. 2008;46:257–267. doi: 10.2478/v10042-008-0056-x. [DOI] [PubMed] [Google Scholar]
- 25.Ruzin A, Keeney D, Bradford PA. AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus-Acinetobacter baumannii complex. JAC. 2007;59:1001–1004. doi: 10.1093/jac/dkm058. [DOI] [PubMed] [Google Scholar]
- 26.Chu YW, Chau SL, Houang ET. Presence of active efflux systems AdeABC, AdeDE and AdeXYZ in different Acinetobacter genomic DNA groups. J Med Microbiol. 2006;55:477–478. doi: 10.1099/jmm.0.46433-0. [DOI] [PubMed] [Google Scholar]
- 27.Higgins PG, Wisplinghoff H, Stefanik D, Seifert H. Selection of topoisomerase mutations and overexpression of adeB mRNA transcripts during an outbreak of Acinetobacter baumannii. J Antimicrob Chemother. 2004;54:821–823. doi: 10.1093/jac/dkh427. [DOI] [PubMed] [Google Scholar]
- 28.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villalón P, Valdezate S, Cabezas T, Ortega M, Garrido N, Vindel A, Medina-Pascual MJ, Saez-Nieto JA. Endemic and epidemic Acinetobacter baumannii clones: a twelve-year study in a tertiary care hospital. BMC Microbiol. 2015;15:47. doi: 10.1186/s12866-015-0383-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dexter C, Murray GL, Paulsen IT, Peleg AY. Community-acquired Acinetobacter baumannii: clinical characteristics, epidemiology and pathogenesis. Expert Rev Anti Infect Ther. 2015;13:567–573. doi: 10.1586/14787210.2015.1025055. [DOI] [PubMed] [Google Scholar]