Abstract
Rapidly growing mycobacteria (RGM) are human pathogens that are relatively easily identified by acid-fast staining but are proving difficult to treat in the clinic. In this study, we performed susceptibility testing of 40 international reference RGM species against 20 antimicrobial agents using the cation-adjusted Mueller-Hinton (CAMH) broth microdilution based on the minimum inhibitory concentration (MIC) assay recommended by the guidelines of the Clinical and Laboratory Standards Institute (CLSI). The results demonstrated that RGM organisms were resistant to the majority of first-line antituberculous agents but not to second-line fluoroquinolones or aminoglycosides. Three drugs (amikacin, tigecycline and linezolid) displayed potent antimycobacterial activity against all tested strains. Capreomycin, levofloxacin and moxifloxacin emerged as promising candidates for the treatment of RGM infections, and cefoxitin and meropenem were active against most strains. Mycobacterium chelonae (M. chelonae), M. abscessus, M. bolletii, M. fortuitum, M. boenickei, M. conceptionense, M. pseudoshottsii, M. septicum and M. setense were the most resistant RGM species. These results provide significant insight into the treatment of RGM species and will assist optimization of clinical criteria.
Keywords: Drug susceptibility, reference strains, rapidly growing mycobacteria (RGM), antimicrobial, minimum inhibitory concentration (MIC)
Introduction
All members of the genus Mycobacterium with the exception of Mycobacterium tuberculosis and M. leprae are considered nontuberculous mycobacteria (NTM) [1]. More than 160 species of NTM are documented in the Genus Mycobacterium database (http://www.bacterio.cict.fr/m/mycobacterium.html; accessed on 22 August 2013). These species are ubiquitously distributed in the environment in fresh and salt water, soil and biofilms [2]. Among NTM, rapidly growing mycobacteria (RGM) have recently gained increasing attention because they are associated with specific diseases and are characterized by extensive resistance to antimicrobial drugs [1]. RGMs are diverse and include Mycobacterium abscessus [3], M. chelonae, M. fortuitum [1], M. immunogenum, and M. smegmatis [1] groups. These strains can be cultured rapidly in as little as seven days [4,5]. To date, more than 50 species of RGM have been identified [6], many of which cause a broad spectrum of human such as pulmonary infections that resemble tuberculosis, infections in skin, soft tissue and the bloodstream, and osteomyelitis [1,7,8]. Among disease-causing RGM, M. abscessus is the most important respiratory pathogen and accounts for approximately 80% of RGM respiratory disease isolates [1].
Treatment of infections caused by RGM remains difficult [9] not least due to the difficulty of selecting the appropriate antimicrobial therapy [5]. In this study, we used the cation-adjusted Mueller-Hinton (CAMH) broth microdilution method to measure the minimum inhibitory concentration (MIC) of 20 antibiotic agents against 40 international reference RGM species in vitro to identify effective drugs for each species. The results may prove useful for clinical diagnosis and treatment of RGM, and contribute to international criteria for drug susceptibility patterns.
Materials and methods
Strains
40 international reference RGM strains were purchased from the German Collection of Microorganisms and Cell Cultures (Deutsche Sammlung Von Mikroorganismen and Zellkulturen, DSMZ) and the American Type Culture Collection (ATCC). Mycobacterium chelonae and M. pseudoshottsii were cultured at 32°C and 25°C, respectively, and all other strains were incubated at 37°C.
Antimicrobial agents
20 antimicrobial agents including rifampicin (RFP), isoniazid (INH), ethambutol (EMB), streptomycin (SM), amikacin (AM), kanamycin (KN), capreomycin (CPM), tobramycin (TOB), ofloxacin (OF), ciprofloxacin (CIP), levofloxacin (LEV), moxifloxacin (MOX), linezolid (LN), clarithromycin (CLR), sulfamethoxazole (SMZ), cefoxitin (FOX), minocycline (MIN), doxycycline (DOX), tigecycline (TIG) and meropenem (MEM) were purchased from Sigma-Aldrich (St. Louis, USA). Alamar Blue was obtained from Bio-Rad Corporation (California, USA). All antimicrobial solutions were prepared the day before the experiment and stored at -70°C.
Drug susceptibility test
Strains were incubated on 7H10 agar and the drug susceptibility test was performed using the CAMH broth microdilution method in accordance with the standard operation procedures of the CLSI [10]. All experiments were repeated at least twice in 96-well microtitre plates. MICs of each antimicrobial agent for each strain are reported as the average of the two tests. Bacteria were adjusted using saline to a density of 0.5 McFarland standard units (1 × 107 colony forming units (CFU)/ml), and 50 μL of bacterial suspension were transferred to 10 ml of CAMH broth and vortexed thoroughly. Antimicrobial drugs were successively diluted two-fold in 100 μL CAMH broth and mixed with 100 μL of bacterial suspension to give the following final drug concentrations: rifampicin, isoniazid, ethambutol, streptomycin, tobramycin, sulfamethoxazole, cefoxitin, tigecycline and meropenem were 0.25-256 μg/mL; amikacin, kanamycin, capreomycin, ofloxacin, ciprofloxacin, levofloxacin, moxifloxacin, clarithromycin, doxycycline and minocycline were 0.03-32 μg/mL; linezolid was 0.06-64 μg/mL. Two negative controls (drug free, CAMH + bacteria; bacteria free, CAMH only) were included to define the appropriate time to add the Alamar Blue (drug free) and to quantify the level of interference from the CAMH media (bacteria free). 96-well microtitre plates were sealed in individual plastic bags and incubated at the appropriate temperatures in controlled incubators. Plates were checked after 72 h and the indicator (20 µL of Alamar Blue and 50 µL of sterile 5% Tween-80) turned pink (provided the drug-free growth control was sufficient). MICs were usually measured on day 3 or 4 and tests were repeated if growth in the drug-free control was insufficient on day 5. MIC breakpoints of antibiotics indicating susceptibility, moderate susceptibility and resistance were interpreted according to the approved guidelines established by the CLSI [10] and the World Health Organization (WHO) [11] (Table 1).
Table 1.
MIC (μg/ml) breakpoints of 20 selected antimicrobial agents
| Drug | MIC breakpoint | ||
|---|---|---|---|
|
|
|||
| Susceptibility | Moderate susceptibility | Resistance | |
| Rifampicin | — | — | ≥ 1 |
| Isoniazid | — | — | ≥ 1 |
| Ethambutol | — | — | ≥ 4 |
| Streptomycin | — | — | ≥ 5 |
| Amikacin | ≤ 16 | 32 | ≥ 64 |
| Kanamycin | — | — | ≥ 4 |
| Capreomycin | — | — | ≥ 2.5 |
| Tobramycin | ≤ 2 | 4 | ≥ 8 |
| Ofloxacin | — | — | ≥ 2 |
| Ciprofloxacin | ≤ 1 | 2 | ≥ 4 |
| Levofloxacin | ≤ 2 | 4 | ≥ 8 |
| Moxifloxacin | ≤ 1 | 2 | ≥ 4 |
| Linezolid | ≤ 8 | 16 | ≥ 32 |
| Sulfamethoxazole | ≤ 38 | — | ≥ 76 |
| Minocycline | ≤ 1 | 2-4 | ≥ 8 |
| Clarithromycin | ≤ 2 | 4 | ≥ 8 |
| Doxycycline | ≤ 1 | 2-4 | ≥ 8 |
| Tigecycline | ≤ 1 | 2-4 | ≥ 8 |
| Cefoxitin | ≤ 16 | 32-64 | ≥ 128 |
| Meropenem | ≤ 4 | 8-16 | ≥ 32 |
Note 1: + = resistant; - = susceptible. Note 2: M. stands for Mycobacterium.
Statistical analysis
Final MIC values for each antimicrobial agent are reported as means ± standard deviation as calculated using SPSS 17.0 software.
Results
MICs were determined for 40 international reference RGM strains against 20 selected antimicrobial agents (Table 2). Among the four first-line antituberculous agents, 38/40 strains (95%) were highly resistant to isoniazid, and 32/40 strains (80%) presented resistance to rifampicin. A higher percentage of strains were susceptible to ethambutol (22/40, 55%) and streptomycin (18/40, 45%). Fluoroquinolones, which include ofloxacin (32/40, 80%), ciprofloxacin (37/40, 92.50%), levofloxacin (39/40, 97.50%) and moxifloxacin (39/40, 97.50%) exhibited powerful in vitro activity against most RGM strains tested. However, M. chelonae was resistant to all four fluoroquinolone agents. Aminoglycosides including amikacin (40/40, 100%), kanamycin (36/40, 90%), capreomycin (39/40, 97.50%) and tobramycin (31/40, 77.50%) were also effective in vitro, and all strains demonstrated complete susceptibility to tigecycline (40/40, 100%) and linezolid (40/40, 100%).
Table 2.
MICs (μg/mL) from antimicrobial susceptibility testing of 40 international reference rapidly growing mycobacteria
| Sp. (international code) | INH | RFP | EMB | SM | AM | KN | CPM | TOB | OF | CIP | LEV | MOX | LN | SMZ | MIN | DOX | TIG | CLR | FOX | MEM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M. chelonae (ATCC35752) | > 256 | > 256 | 128 | 32 | 4 | 16 | 16 | 2 | 32 | 4 | 16 | 4 | 8 | 64 | > 32 | > 32 | 1 | 0.06 | 128 | > 256 |
| M. abscessus (ATCC19977) | > 256 | 64 | 32 | 16 | 1 | 2 | 0.5 | 8 | 4 | 2 | 1 | 1 | 4 | 32 | 8 | 32 | 4 | < 0.03 | 32 | 64 |
| M. bolletii (DSM45149) | > 256 | 32 | 64 | 16 | 0.5 | 0.5 | 1 | 8 | 16 | 4 | 4 | 1 | 2 | 32 | > 32 | 32 | 0.06 | < 0.03 | 32 | 0.25 |
| M. massiliense (DSM45103) | > 256 | 32 | 64 | 2 | 0.13 | 2 | 0.03 | 32 | 0.06 | 0.03 | 0.03 | 0.03 | 4 | 64 | 8 | 0.5 | 0.06 | 0.03 | 64 | 16 |
| M. fortuitum (DSM44220) | 64 | 128 | 256 | 32 | 0.25 | 4 | < 0.03 | 16 | 0.13 | < 0.03 | < 0.03 | < 0.03 | 8 | 2 | 8 | 0.03 | 0. 5 | 1 | 32 | 4 |
| M. senegalense (ATCC35796) | > 256 | 8 | 64 | 4 | 0.5 | 0.25 | 0.13 | 4 | 4 | 4 | 2 | 0.13 | 1 | 16 | 0.25 | 0.06 | 0.03 | 0.13 | 4 | 2 |
| M. boenickei (DSM44677) | 32 | 128 | 4 | 8 | 0.5 | 4 | 0.06 | 4 | 0.5 | 0.25 | 0.25 | 0.06 | 8 | 4 | > 32 | 16 | 0.03 | 16 | 16 | 0.5 |
| M. goodii (DSM44492) | > 256 | 32 | 2 | 32 | 4 | 16 | 0.06 | 32 | 0.5 | 0.5 | 0.25 | 0.06 | 1 | 8 | 2 | 0.5 | 0.13 | > 32 | 8 | 0.25 |
| M. wolinskyi (DSM44493) | > 256 | 64 | 128 | 16 | 0.5 | 2 | 0.13 | 32 | 0.5 | 0.5 | 0.5 | 0.25 | 0.5 | 4 | 1 | 0.13 | 0.25 | > 32 | 32 | 1 |
| M. aichiense (ATCC27280) | > 256 | < 0.25 | 0.5 | < 0.25 | 0.06 | 0.03 | < 0.03 | 0.5 | < 0.03 | < 0.03 | < 0.03 | < 0.03 | 0.5 | 2 | 0.25 | < 0.03 | 0.03 | < 0.03 | 0.5 | 0.25 |
| M. aurum (ATCC23366) | > 256 | 4 | 1 | < 0.25 | 0.25 | 0.06 | < 0.03 | 4 | 0.06 | 0.03 | < 0.03 | < 0.03 | 0.5 | 4 | 0.25 | 0.03 | 0.13 | 0.06 | 16 | 16 |
| M. chubuense (ATCC27278) | > 256 | 1 | 0.5 | 0.5 | 0.25 | 0.03 | < 0.03 | 2 | 0.03 | < 0.03 | < 0.03 | < 0.03 | 0.25 | 1 | 0.25 | 0.13 | 0.13 | < 0.03 | 128 | 16 |
| M. duvalii (ATCC43910) | > 256 | 0.5 | 0.25 | 0.5 | 0.25 | 0.5 | < 0.03 | 2 | 0.06 | < 0.03 | 0.06 | < 0.03 | 0.5 | 8 | 0.5 | 0.03 | 0.06 | 0.5 | 8 | 2 |
| M. flavescens (ATCC14474) | > 256 | 32 | 0.5 | 1 | 0.13 | 0.13 | < 0.03 | 2 | 0.13 | < 0.03 | 0.06 | < 0.03 | 0.5 | 0.5 | 0.5 | 0.06 | 0.03 | 8 | 8 | 64 |
| M. gilvum (ATCC43909) | > 256 | 0.25 | 0.25 | 1 | 0.25 | 0.25 | 0.03 | 1 | 0.13 | < 0.03 | 0.03 | 0.03 | 0.25 | 4 | 0.25 | 0.03 | 0.06 | 0.03 | 4 | 2 |
| M. neoaurum (ATCC25795) | > 256 | 1 | 8 | 0.5 | 0.5 | 0.06 | < 0.03 | 0.25 | 0.06 | 0.06 | 0.03 | < 0.03 | 0.5 | 0.5 | 0.03 | 0.06 | 0.03 | 1 | 8 | 0.5 |
| M. obuense (ATCC27023) | > 256 | 2 | 64 | < 0.25 | 0.13 | 0.13 | < 0.03 | 1 | < 0.03 | < 0.03 | < 0.03 | < 0.03 | 0.5 | 32 | 0.25 | < 0.03 | 0.06 | < 0.03 | < 0.25 | 0.5 |
| M. parafortuitum (ATCC19686) | 0.5 | 2 | 2 | 0.5 | 0.5 | 0.5 | < 0.03 | 0.5 | 0.06 | 0.06 | 0.06 | < 0.03 | 0.5 | 2 | 0.25 | 0.03 | 0.06 | 1 | 4 | 0.25 |
| M. rhodesiae (ATCC27024) | 8 | < 0.25 | < 0.25 | < 0.25 | 0.13 | < 0.03 | < 0.03 | 0.25 | 0.06 | < 0.03 | 0.03 | < 0.03 | 0.25 | 1 | 0.25 | 0.06 | 0.03 | < 0.03 | < 0.25 | 1 |
| M. tokaiense (ATCC27282) | 4 | < 0.25 | 0.5 | 0.5 | 0.25 | 0.25 | < 0.03 | 0.5 | 0.06 | 0.03 | 0.03 | < 0.03 | 0.5 | 8 | 0.25 | 0.13 | 0.03 | 0.13 | 128 | 0.25 |
| M. porcinum (ATCC33776) | 16 | 64 | 64 | 16 | 0.5 | 1 | 0.13 | 2 | 1 | 0.25 | 0.5 | 0.06 | 8 | 4 | 16 | 16 | 0.06 | 2 | 8 | 4 |
| M. pulveris (ATCC35154) | > 256 | 64 | 4 | 2 | 0.5 | 0.13 | 0.03 | 2 | > 32 | 0.5 | 1 | 0.13 | 2 | 256 | 0.25 | 0.06 | 0.03 | 2 | 256 | 1 |
| M. austroafricanum (ATCC33464) | 16 | 1 | 0.5 | < 0.25 | 0.25 | 0.06 | 0.03 | 2 | 0.25 | 0.13 | 0.06 | 0.03 | 0.25 | 256 | 0.25 | 0.03 | 0.13 | <0.03 | 8 | 0.5 |
| M. chitae (ATCC19627) | > 256 | 8 | < 0.25 | 0.5 | 0.13 | 0.03 | < 0.03 | 0.5 | 0.03 | < 0.03 | < 0.03 | < 0.03 | 0.5 | 0.5 | 2 | 0.03 | 0.06 | 4 | 4 | 4 |
| M. aubagnense (DSM45150) | > 256 | 2 | < 0.25 | 1 | 0.06 | 0.5 | < 0.03 | 1 | 0.25 | 0.13 | 0.06 | 0.06 | 0.25 | 1 | 16 | 2 | < 0.03 | < 0.03 | 2 | 0.25 |
| M. brisbanense (DSM44680) | > 256 | 128 | > 256 | 16 | 0.13 | 2 | 0.13 | 8 | 0.5 | 0.25 | 0.25 | 0.06 | 0.5 | 4 | 16 | 4 | 0.13 | 0.13 | 64 | 4 |
| M. brumae (DSM44177) | > 256 | 128 | 64 | 32 | 0.06 | < 0.03 | < 0.03 | 0.5 | < 0.03 | < 0.03 | < 0.03 | < 0.03 | 1 | 2 | 0.25 | < 0.03 | 0.25 | < 0.03 | 64 | 32 |
| M. canariasense (DSM44828) | 16 | 4 | 1 | 0.5 | 0.5 | 0.5 | 0.06 | 0.25 | 0.5 | 0.25 | 0.25 | 0.03 | 0.5 | 0.5 | 0.25 | 0.13 | 0.13 | 0.13 | 4 | 4 |
| M. conceptionense (DSM45102) | > 256 | 128 | 256 | 32 | 0.5 | 1 | 0.06 | 4 | 0.5 | 0.25 | 0.25 | 0.06 | 16 | 128 | 0.5 | 0.13 | < 0.03 | 0.13 | 128 | 0.25 |
| M. confluentis (DSM44017) | > 256 | 8 | 32 | 4 | 0.13 | 0.03 | < 0.03 | 0.25 | 0.03 | < 0.03 | < 0.03 | < 0.03 | 2 | 1 | 1 | 0.06 | 0.25 | 2 | 4 | 2 |
| M. fluoranthenivorans (DSM44556) | > 256 | 0.5 | 0.5 | < 0.25 | 0.06 | 0.13 | < 0.03 | 1 | 0.06 | < 0.03 | 0.03 | < 0.03 | 0.25 | 64 | 0.25 | 0.06 | 0.25 | < 0.03 | 2 | 16 |
| M. moriokaense (DSM44221) | > 256 | 8 | 16 | 8 | 2 | 0.25 | 0.25 | 8 | > 32 | 0.5 | 0.5 | 0.03 | 2 | 64 | 0.25 | 0.03 | 0.03 | 0.5 | 16 | 2 |
| M. mucogenicum (DSM44625) | 0.5 | 32 | 16 | 8 | 1 | 2 | 0.5 | 4 | 2 | 1 | 1 | 0.25 | 2 | 16 | 0.25 | 0.06 | < 0.03 | 0.06 | 16 | 0.25 |
| M. poriferae (DSM44585) | > 256 | 128 | 128 | 16 | 4 | 1 | 0.13 | 8 | 1 | 2 | 2 | 0.13 | 1 | 4 | 0.25 | 0.25 | 0.03 | < 0.03 | 32 | 2 |
| M. pseudoshottsii (DSM45108) | > 256 | 256 | 128 | 32 | 2 | 0.5 | 1 | 4 | 8 | 2 | 4 | 1 | 8 | 4 | 0.25 | 32 | < 0.03 | 0.03 | > 256 | 128 |
| M. psychrotolerans (DSM44697) | 4 | < 0.25 | < 0.25 | < 0.25 | 0.06 | < 0.03 | < 0.03 | 4 | < 0.03 | < 0.03 | < 0.03 | < 0.03 | 0.03 | 0.25 | 1 | 0.13 | 0.03 | < 0.03 | < 0.25 | 2 |
| M. septicum (DSM44393) | 256 | 16 | 4 | 16 | 0.13 | 0.5 | < 0.03 | 1 | 0.25 | 0.06 | 0.13 | 0.06 | 1 | 1 | 32 | 8 | 1 | 1 | 128 | 16 |
| M. setense (DSM45070) | > 256 | 64 | 16 | 8 | 0.5 | 1 | < 0.03 | 4 | 0.25 | 0.06 | 0.13 | 0.03 | 4 | > 256 | > 32 | > 32 | 0.13 | 4 | 16 | 0.25 |
| M. vanbaalenii (DSM7251) | > 256 | 1 | 0.5 | < 0.25 | 0.13 | 0.13 | 0.25 | 1 | 0.25 | 0.13 | 0.13 | 0.03 | 0.25 | 8 | 16 | < 0.03 | 0.25 | < 0.03 | 4 | 64 |
| M. murale (DSM44340) | > 256 | < 0.25 | 0.25 | 0.5 | 0.13 | 1 | < 0.03 | 1 | 0.03 | < 0.03 | < 0.03 | < 0.03 | 0.5 | 2 | 0.25 | 0.13 | 0.13 | 0.06 | 16 | 32 |
| Standard Deviation | 74.64 | 59.64 | 67.62 | 11.78 | 1.05 | 3.62 | 3.54 | 8.36 | 6.1 | 1.3 | 3 | 0.85 | 3.38 | 59.94 | 7.09 | 9.62 | 0.69 | 3.48 | 56.2 | 25.94 |
Note 1: INH: isoniazid, RFP: rifampicin, EMB: ethambutol, SM: streptomycin, AM: amikacin, KN: kanamycin, CPM: capreomycin, TOB: tobramycin, OF: ofloxacin, CIP: ciprofloxacin, LEV: levofloxacin, MOX: moxifloxacin, LN: linezolid, SMZ: sulfamethoxazole, MIN: minocycline, CLR: clarithromycin, DOX: doxycycline, TIG: tigecycline, FOX: cefoxitin, MEM: meropenem. Note 2: Numbers in bold type and underlined indicate sensitive strains; numbers in bold type only indicate moderately sensitive strains. Note 3: M. stands for Mycobacterium.
Clarithromycin (36/40, 90%) showed excellent activity against all RGM organisms except M. boenickei, M. goodie and M. wolinskyi. Minocycline (29/40, 72.50%) and doxycycline (32/40, 80%) also displayed significant in vitro antimycobacterial activity but M. chelonae, M. abscessus, M. boenickei, M. bolletii, M. septicum and M. setense showed resistance. Cefoxitin (33/40, 82.50%) and meropenem (33/40, 82.50%) exhibited considerable activity against all RGM strains excluding M. chelonae and M. pseudoshottsii, while sulfamethoxazole (32/40, 80%) inhibited all strains apart from M. chelonae, M. pulveris, M. austroafricanum, M. conceptionense and M. setense.
Susceptibility rates were determined for the 20 selected antimicrobial agents against the 40 international reference RGMs (Table 3), and the most susceptible species were M. aichiense, M. duvalii, M. gilvum, M. parafortuitum, M. rhodesiae and M. psychrotolerans. The most resistant species was M. chelonae.
Table 3.
Susceptibility of 40 international reference rapidly growing mycobacterial strains towards 20 selected antimicrobial agents
| Sp. (international code) | INH | RFP | EMB | SM | AM | KN | CPM | TOB | OF | CIP | LEV | MOX | LN | SMZ | MIN | DOX | TIG | CLR | FOX | MEM | Susceptibility rate (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M. chelonae (ATCC35752) | + | + | + | + | - | + | + | - | + | + | + | + | - | + | + | + | - | - | + | + | 25 |
| M. abscessus (ATCC19977) | + | + | + | + | - | - | - | + | + | - | - | - | - | - | + | + | - | - | - | + | 55 |
| M. bolletii (DSM45149) | + | + | + | + | - | - | - | + | + | + | - | - | - | - | + | + | - | - | - | - | 55 |
| M. massiliense (DSM45103) | + | + | + | - | - | - | - | + | - | - | - | - | - | + | + | - | - | - | - | - | 70 |
| M. fortuitum (DSM44220) | + | + | + | + | - | + | - | + | - | - | - | - | - | - | - | - | - | - | - | - | 70 |
| M. senegalense (ATCC35796) | + | + | + | - | - | - | - | - | + | + | - | - | - | - | - | - | - | - | + | - | 75 |
| M. boenickei (DSM44677) | + | + | + | + | - | + | - | - | - | - | - | - | - | - | + | + | - | + | - | - | 60 |
| M. goodii (DSM44492) | + | + | - | + | - | + | - | + | - | - | - | - | - | - | - | - | - | + | - | - | 70 |
| M. wolinskyi (DSM44493) | + | + | + | + | - | - | - | + | - | - | - | - | - | - | - | - | - | + | - | - | 70 |
| M. aichiense (ATCC27280) | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 95 |
| M. aurum (ATCC23366) | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 90 |
| M. chubuense (ATCC27278) | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | - | 85 |
| M. duvalii (ATCC43910) | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 95 |
| M. flavescens (ATCC14474) | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | - | + | 80 |
| M. gilvum (ATCC43909) | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 95 |
| M. neoaurum (ATCC25795) | + | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 85 |
| M. obuense (ATCC27023) | + | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 85 |
| M. parafortuitum (ATCC19686) | - | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 95 |
| M. rhodesiae (ATCC27024) | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 95 |
| M. tokaiense (ATCC27282) | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | - | 90 |
| M. porcinum (ATCC33776) | + | + | + | + | - | - | - | - | - | - | - | - | - | - | + | + | - | - | - | - | 70 |
| M. pulveris (ATCC35154) | + | + | + | - | - | - | - | - | + | - | - | - | - | + | - | - | - | - | + | - | 70 |
| M. austroafricanum (ATCC33464) | + | + | - | - | - | - | - | - | - | - | - | - | - | + | + | - | - | - | - | - | 85 |
| M. chitae (ATCC19627) | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 90 |
| M. aubagnense (DSM45150) | + | + | - | - | - | - | - | - | - | - | - | - | - | - | + | - | - | - | - | - | 85 |
| M. brisbanense (DSM44680) | + | + | + | + | - | - | - | + | - | - | - | - | - | - | + | - | - | - | - | - | 70 |
| M. brumae (DSM44177) | + | + | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | 75 |
| M. canariasense (DSM44828) | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 90 |
| M. conceptionense (DSM45102) | + | + | + | + | - | - | - | - | - | - | - | - | - | + | - | - | - | - | + | - | 70 |
| M. confluentis (DSM44017) | + | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 85 |
| M. fluoranthenivorans (DSM44556) | + | - | - | - | - | - | - | - | - | - | - | - | - | + | - | - | - | - | - | - | 90 |
| M. moriokaense (DSM44221) | + | + | + | + | - | - | - | + | + | - | - | - | - | + | - | - | - | - | - | - | 65 |
| M. mucogenicum (DSM44625) | - | + | + | + | - | - | - | - | + | - | - | - | - | - | - | - | - | - | - | - | 80 |
| M. poriferae (DSM44585) | + | + | + | + | - | - | - | + | - | - | - | - | - | - | - | - | - | - | - | - | 75 |
| M. pseudoshottsii (DSM45108) | + | + | + | + | - | - | - | - | + | - | - | - | - | - | - | + | - | - | + | + | 60 |
| M. psychrotolerans (DSM44697) | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 95 |
| M. septicum (DSM44393) | + | + | + | + | - | - | - | - | - | - | - | - | - | - | + | + | - | - | + | - | 65 |
| M. setense (DSM45070) | + | + | + | + | - | - | - | - | - | - | - | - | - | + | + | + | - | - | - | - | 65 |
| M. vanbaalenii (DSM7251) | + | + | - | - | - | - | - | - | - | - | - | - | - | - | + | - | - | - | - | + | 80 |
| M. murale (DSM44340) | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | 90 |
Note 1: + = resistant; - = susceptible. Note 2: M. stands for Mycobacterium.
Antibacterial susceptibility distributions were determined for al 40 RGMs (Figure 1), and amikacin, linezolid and tigecycline scored 100% susceptibility, meaning all strains were susceptible to these agents. Capreomycin, levofloxacin and moxifloxacin were also among the highest scoring for susceptibility and are therefore good candidates for the treatment of RGM infections. M. abscessus and M. bolletii were susceptible to 11 of the tested agents, and M. massiliense was susceptible to 14 (Figure 2). M. fortuitum, M. senegalense and M. boenickei were susceptible or moderately susceptible to 12-15 drugs (Figure 3), and M. goodii and M. wolinskyi were susceptible to 14 of the compounds tested (Figure 4).
Figure 1.
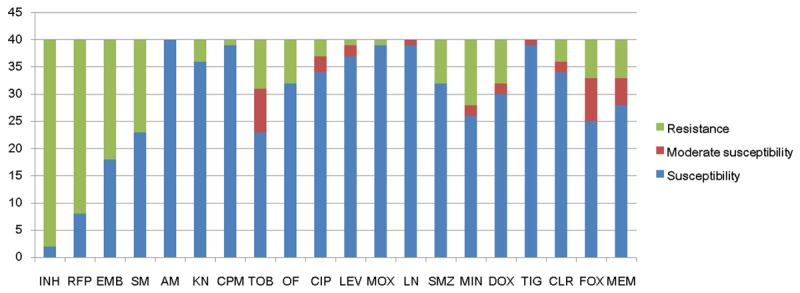
Susceptibility of 40 reference rapidly growing mycobacteria to 20 selected antimicrobial agents.
Figure 2.
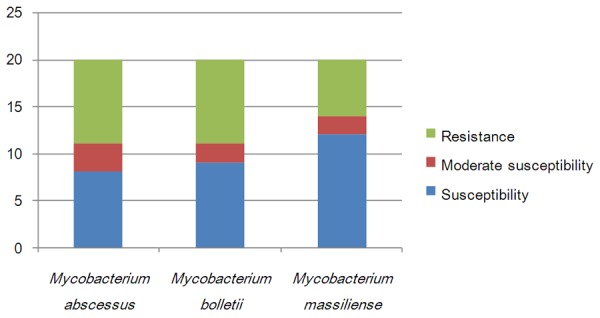
Susceptibility of the Mycobacterium abscessus group to 20 selected antibacterial agents.
Figure 3.
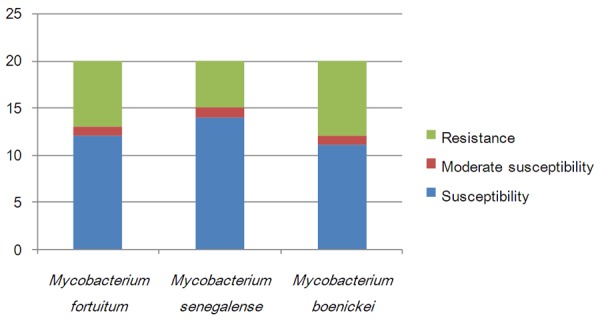
Susceptibility of the Mycobacterium fortuitum group to 20 selected antibacterial agents.
Figure 4.
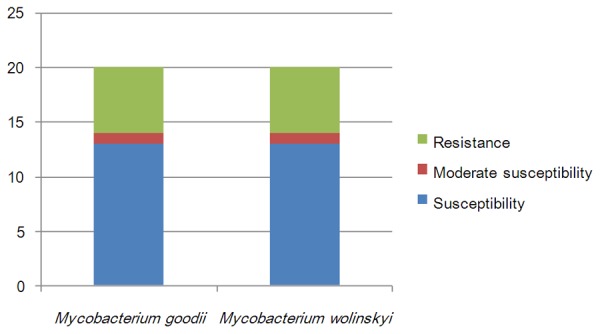
Susceptibility of the Mycobacterium smegmatis group to 20 selected antibiotics.
Discussion
In this study, the susceptibility patterns of 40 international reference RGM strains towards 20 selected antimicrobial agents were determined using the CAMH broth microdilution method. To our knowledge, this is the most extensive susceptibility analysis of RGM reference strains to date.
Antimicrobial sensitivity is known to vary across RGM species [1]. In the present study we found that the majority of RGM strains were resistant to the first-line antituberculous agents, with isoniazid displaying the highest MIC, consistent with previous reports [1,12-14]. Aminoglycosides and fluoroquinolones are the current second-line conventional antituberculous drugs, and amikacin exhibits excellent activity towards M. abscessus, M. chelonae and M. fortuitum, while tobramycin was effective against M. chelonae [1,13,15-17]. Our results showed that all 40 RGM reference strains were susceptible to amikacin, and 39 strains were susceptible to capreomycin, including M. chelonae. Tobramycin was the least active among the aminoglycosides and 9 RGM species were resistant to this drug. In previous studies [1,15,16], quinolones were found to be more active against M. fortuitum than M. abscessus, whereas in the present study, M. chelonae was more resistant to quinolones than both M. fortuitum and M. abscessus. Most strains were more susceptible to Moxifloxacin than they were to the other quinolones, and ofloxacin exhibited the lowest antimycobacterial activity.
Minocycline and doxycycline belong to the tetracycline class of antibiotics, and tigecycline is a derivative of minocycline. In a previous study, minocycline exhibited 50% activity against RGMs, and doxycycline activity was less than 3% [18]. In the present study, M. chelonae, M. abscessus, M. bolletii, M. massiliense, M. fortuitum and M. boenickei were resistant to minocycline, while M. chelonae, M. abscessus, M. bolletii and M. boenickei were resistant to doxycycline. In contrast, tigecycline displayed 100% activity and successfully inhibited all 40 RGM strains, consistent with previous reports [19,20].
Clarithromycin belongs to the macrolide class of antibiotics and this agent displayed good activity against M. chelonae and M. abscessus in recent studies [15-17]. However in the present study, M. boenickei, M. goodie, M. wolinskyi and M. flavescens were resistant to clarithromycin, suggesting these strains may carry the inducible erythromycin methylase gene erm that confers macrolide resistance [21], and this should be investigated further in the future.
Cefoxitin and imipenem are important parenteral antibiotics used in the treatment of M. abscessus infections [15,16]. In our study, we used the closely related meropenem instead of imipenem. Cefoxitin and meropenem share the same antibacterial mechanism and operate by inhibiting bacterial cell wall biosynthesis [22]. The M. abscessus complex has been divided into three subspecies, namely M. abscessus, M. massiliense and M. bolletii [23,24]. Our results showed that meropenem displayed a lower MIC value than cefoxitin, and M. abscessus and M. bolletii were more resistant to these antimicrobial agents than M. massiliense. There is increasing evidence that M. massiliense is the causative agent of soft tissue infection outbreaks and postsurgery infections [25]. The M. fortuitum group [1] has also been divided into three subspecies (M. fortuitum, M. senegalense and M. boenickei). The M. fortuitum group has been shown to be susceptible to the majority of antimicrobial agents in previous studies [16,26]. However, in the present study, M. fortuitum was resistant to tobramycin, M. senegalense was resistant to the third-line fluoroquinolones ofloxacin, ciprofloxacin and levofloxacin, and M. boenickei was more resistant to minocycline and doxycycline than the other two subspecies. The M. smegmatis group includes M. goodii and M. wolinskyi [1], and both these strains were resistant to tobramycin and clarithromycin in our study. The M. abscessus complex, M. chelonae, the M. fortuitum group, M. mucogenicum and M. neoaurum are the most frequent cause of human infections among the RGMs characterized to date [6]. The results of the present study showed that the M. abscessus group, M. chelonae, M. fortuitum, M. boenickei and M. conceptionense were the most drug-resistant of the RGM strains tested. M. chelonae and M. abscessus belong to the same complex and share some similar characteristics [27]. In our study, M. chelonae proved to be resistant to more drugs than did M. abscessus.
In summary, we measured the in vitro antimycobacterial activity of 20 selected antibacterial agents against 40 RGM reference strains. The results pave the way for future in vivo studies and provide important information for optimizing specific therapies against different RGM species.
Acknowledgements
We thank the staffs of National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention. This work was financially supported by the projects 2013ZX10003002-001 of the National Key Programme of Mega Infectious Diseases and the key project 2014SKLID104 of State Key Laboratory for Infectious Disease Prevention and Control and the science and technology innovation team support project CX201412 of Changzhi medical college.
Disclosure of conflict of interest
None.
References
- 1.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 2.Jang MA, Koh WJ, Huh HJ, Kim SY, Jeon K, Ki CS, Lee NY. Distribution of nontuberculous mycobacteria by multigene sequence-based typing and clinical significance of isolated strains. J Clin Microbiol. 2014;52:1207–1212. doi: 10.1128/JCM.03053-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffith DE. Mycobacterium abscessus subsp abscessus lung disease: ‘trouble ahead, trouble behind’. F1000Prime Rep. 2014;6:107. doi: 10.12703/P6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Preece CL, Perry A, Gray B, Kenna DT, Jones AL, Cummings SP, Robb A, Thomas MF, Brodlie M, O’Brien CJ, Bourke SJ, Perry JD. A novel culture medium for isolation of rapidly-growing mycobacteria from the sputum of patients with cystic fibrosis. J Cyst Fibros. 2015 doi: 10.1016/j.jcf.2015.05.002. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 5.Gray TJ, Kong F, Jelfs P, Sintchenko V, Chen SC. Improved Identification of Rapidly Growing Mycobacteria by a 16S-23S Internal Transcribed Spacer Region PCR and Capillary Gel Electrophoresis. PLoS One. 2014;9:e102290. doi: 10.1371/journal.pone.0102290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Helou G, Viola GM, Hachem R, Han XY, Raad II. Rapidly growing mycobacterial bloodstream infections. Lancet Infect Dis. 2013;13:166–74. doi: 10.1016/S1473-3099(12)70316-X. [DOI] [PubMed] [Google Scholar]
- 7.Behr MA, Falinkham JO III. Molecular epidemiology of nontuberculous mycobacteria. Future Microbiol. 2009;4:1009–1020. doi: 10.2217/fmb.09.75. [DOI] [PubMed] [Google Scholar]
- 8.Hernández-Garduño E, Elwood RK. Increasing incidence of nontuberculous mycobacteria, Taiwan, 2000-2008. Emerg Infect Dis. 2010;16:1047. doi: 10.3201/eid1606.100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bicmen C, Coskun M, Gunduz AT, Senol G, Cirak AK, Tibet G. Nontuberculous mycobacteria isolated from pulmonary specimens between 2004 and 2009: causative agent or not? New Microbiol. 2010;33:399–403. [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. Susceptibility Testing of Mycobacteria, Nocardiae, and Other aerobic actinomycetes; Approved Standard Second Edition. CLSI Document. 2011:M24–A2. [PubMed] [Google Scholar]
- 11.Barrera L, Cooreman E, de Dieu Iragena J, Drobniewski F, Duda P, Havelkova M, Hoffner S, Kam KM, Kim SJ, Labelle S, Lambregts K, Leimane V, Nunn P, Ramsay A, Raviglione M, Rich M, Ridderhof J, Rodrigues F, Rüsch-Gerdes S, Salfinger M, Scholten J, Selvakumar N, Shinnick T, Shul’gina M, Šķenders G, Sloutsky A, Small P, Van Deun A, Varaine F, Yagui M, Vincent V, Weyer K, Wright A, Zignol M World Health Organization. Policy guidance on drug-susceptibility testing (DST) of second-line antituberculosis drugs. WHO Document. 2008 [PubMed] [Google Scholar]
- 12.Shahraki AH, Heidarieh P, Bostanabad SZ, Khosravi AD, Hashemzadeh M, Khandan S, Biranvand M, Schraufnagel DE, Mirsaeidi M. “Multidrug-resistant tuberculosis” may be nontuberculous mycobacteria. Eur J Intern Med. 2015;26:279–84. doi: 10.1016/j.ejim.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li G, Lian LL, Wan L, Zhang J, Zhao X, Jiang Y, Zhao LL, Liu H, Wan K. Antimicrobial susceptibility of standard strains of nontuberculous mycobacteria by microplate Alamar Blue assay. PLoS One. 2013;8:e84065. doi: 10.1371/journal.pone.0084065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Ingen J, van der Laan T, Dekhuijzen R, Boeree M, van Soolingen D. In vitro drug susceptibility of 2275 clinical non-tuberculous Mycobacterium isolates of 49 species in The Netherlands. Int J Antimicrob Agents. 2010;35:169–73. doi: 10.1016/j.ijantimicag.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 15.Park S, Kim S, Park EM, Kim H, Kwon OJ, Chang CL, Lew WJ, Park YK, Koh WJ. In vitro antimicrobial susceptibility of Mycobacterium abscessus in Korea. J Korean Med Sci. 2008;23:49–52. doi: 10.3346/jkms.2008.23.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang SS, Lye DC, Jureen R, Sng LH, Hsu LY. Rapidly growing mycobacteria in Singapore, 2006-2011. Clin Microbiol Infect. 2015;21:236–241. doi: 10.1016/j.cmi.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Broda A, Jebbari H, Beaton K, Mitchell S, Drobniewski F. Comparative drug resistance of Mycobacterium abscessus and M. chelonae isolates from patients with and without cystic fibrosis in the United Kingdom. J Clin Microbiol. 2013;51:217–223. doi: 10.1128/JCM.02260-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang TS, Lee SS, Hsueh PR, Tsai HC, Chen YS, Wann SR, Leu HS, Ko WC, Yan JJ, Yuan SZ, Chang FY, Lu JJ, Wang JH, Wang HK, Liu YC. Antimicrobial resistance of rapidly growing mycobacteria in western Taiwan: SMART program 2002. J Formos Med Assoc. 2008;107:281–287. doi: 10.1016/s0929-6646(08)60088-1. [DOI] [PubMed] [Google Scholar]
- 19.Huang CW, Chen JH, Hu ST, Huang WC, Lee YC, Huang CC, Shen GH. Synergistic activities of tigecycline with clarithromycin or amikacin against rapidly growing mycobacteria in Taiwan. Int J Antimicrob Agents. 2013;41:218–223. doi: 10.1016/j.ijantimicag.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Fernández-Roblas R, Martín-de-Hijas NZ, Fernández-Martínez AI, García-Almeida D, Gadea I, Esteban J. In vitro activities of tigecycline and 10 other antimicrobials against nonpigmented rapidly growing mycobacteria. Antimicrob Agents Chemother. 2008;52:4184–4186. doi: 10.1128/AAC.00695-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bastian S, Veziris N, Roux AL, Brossier F, Gaillard JL, Jarlier V, Cambau E. Assessment of clarithromycin susceptibility in strains belonging to the Mycobacterium abscessus group by erm (41) and rrl sequencing. Antimicrob Agents Chemother. 2011;55:775–781. doi: 10.1128/AAC.00861-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai YK, Liou CH, Fung CP, Lin JC, Siu LK. Single or in combination antimicrobial resistance mechanisms of Klebsiella pneumoniae contribute to varied susceptibility to different carbapenems. PLoS One. 2013;8:e79640. doi: 10.1371/journal.pone.0079640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howard ST. Recent progress towards understanding genetic variation in the Mycobacterium abscessus complex. Tuberculosis (Edinb) 2013;93(Suppl):S15–20. doi: 10.1016/S1472-9792(13)70005-2. [DOI] [PubMed] [Google Scholar]
- 24.Lee SH, Yoo HK, Kim SH, Koh WJ, Kim CK, Park YK, Kim HJ. The drug resistance profile of Mycobacterium abscessus group strains from Korea. Ann Lab Med. 2014;34:31–37. doi: 10.3343/alm.2014.34.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raiol T, Ribeiro GM, Maranhão AQ, Bocca AL, Silva-Pereira I, Junqueira-Kipnis AP, Brigido Mde M, Kipnis A. Complete genome sequence of Mycobacterium massiliense. J Bacteriol. 2012;194:5455. doi: 10.1128/JB.01219-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gayathri R, Therese KL, Deepa P, Mangai S, Madhavan HN. Antibiotic susceptibility pattern of rapidly growing mycobacteria. J Postgrad Med. 2010;56:76–78. doi: 10.4103/0022-3859.65278. [DOI] [PubMed] [Google Scholar]
- 27.Sampaio JL. Prokaryotic taxonomy rules and nomenclature changes in the Mycobacterium chelonae-abscessus group. Future Microbiol. 2010;5:1457. doi: 10.2217/fmb.10.111. [DOI] [PubMed] [Google Scholar]


