Abstract
Background: Metastasis suppressor-1 (MTSS1) is a novel potential metastasis suppressor gene in several types of human cancers. However, the exact function and regulatory mechanism of MTSS1 in Tongue squamous cellular carcinoma (TSCC) have not been elucidated. Material/Methods: We first confirmed the MTSS1 gene expression by using quantitative real time-PCR (qRT-PCR) and immunohistochemical staining. Then we detected the effect of MTSS1 gene on Tca8113 cells proliferation and invasion ability by using MTT, wound healing and invasion assay. Finally by using bioinformatics analysis, luciferase reporter assay and a serial method, we analyzed the targeting of miR-96 on MTSS1 and the ability of miR-96 on MTSS1 gene mediated biological alterations in Tca8113 cells. Results: Our findings showed that the expression of MTSS1 was down-regulated in both TSCC tissues and Tca8113 cells. Forced expression of MTSS1 led to inhibited cell proliferation ability, retarded wound closing and reduced trans-membrane cell numbers. MiR-96 is confirmed to be a direct target of MTSS1 gene and could regulate MTSS1 mediated Tca8113 cells proliferation and metastasis. But miR-96 could not completely restore the invasion ability of Tca8113 cells. Conclusions: MiR-96 targeting and promoting MTSS1 repression may precipitate in the TSCC tumorigenesis through bypassing cell proliferation and metastasis control.
Keywords: Cell proliferation, microRNAs, neoplasm metastasis, tongue cancer
Introduction
Tongue cancer is one of the most common malignancies in oral and maxillofacial region, with more than 98% being tongue squamous cellular carcinoma (TSCC). TSCC occurs more common in men than in women and was characterized with local infiltrating growth and cervical lymph node metastasis. Now, the main treatment strategy for TSCC is still the comprehensive treatment based on surgery. Although great progress has been made in the cancer treatment, the 5 years survival rate of TSCC was still hovering around 50% [1]. Tumor metastasis is the most significant contributor to the mortality of patients with cancer. It is thus necessary to elucidate the molecular mechanisms underlying TSCC metastasis and identify novel therapeutic targets.
Metastasis suppressor-1 (MTSS1), also known as missing in metastasis (MIM), mapped to human chromosome 8q24.1, is a novel potential metastasis suppressor gene which was initially identified as a tumor suppressor gene in non-metastatic bladder cancer cell lines [2]. Usually the expression of MTSS1 is lost in metastatic cells, but confusing in primary tumors. For example, in ovarian cancer, colorectal cancer, oesophageal cancer, prostate cancer and breast cancer, the expression of MTSS1 has been shown to be reduced [3-5], whereas up-regulation of MTSS1 expression has also been observed in hepato-cellular carcinoma and breast cancer [6,7]. Recent research showed that weak or absent staining of MTSS1 protein was observed both in kidney cancer and bladder cancer [8,9]. Our previous gene array results also indicated that MTSS1 gene was down-regulated in TSCC tissue (Fold changes = 3.527), which needs further confirmation. Functional and mechanism analysis suggested that MTSS1 protein may be related to cancer progression or tumor metastasis in a variety of organ sites, most likely through an interaction with the actin cytoskeleton or regulated by some factors including microRNAs [4,10-12]. Till now, there was no report about the role and mechanism of MTSS1 gene in TSCC.
In this study, we first confirmed the expression of MTSS1 gene in both TSCC tissues and TSCC Tca8113 cells. Then we analyzed the growth, migration and invasion ability of MTSS1 gene in Tca8113 cells. Finally, we explored the mechanism of MTSS1 induced biological alteration in cell models. Our results revealed the role and mechanism of MTSS1 gene in the TSCC progression, which further provided target for the development of novel therapeutic strategies against TSCC.
Material and methods
Cell culture and clinical specimens
The human TSCC cell line Tca8113 and human normal oral keratinocyte cell line (hNOK) were purchased from Shanghai Institute of Cell Biology, Chinese Academy of Sciences and were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum under an atmosphere of 95% air and 5% CO2 at 37°C. Total fifty paired TSCC tissues and adjacent normal tissues were obtained from the Affiliated Stomatology Hospital of China Medical University. All the patients signed informed consent and the studies were approved by the Ethics Committee. Tissue samples were trimmed and stored at -80°C until use.
qRT-PCR
Total RNA was extracted using Trizol reagent and cDNA was synthesized by the High Capacity cDNA Reverse Transcription Kit (# 4368814, Applied Biosystems) according to the manufacturer’s protocol. Then the MTSS1 expression level was detected with SYBR (# 4367659, Applied Biosystems) through Applied Biosystems 7500 Real-Time PCR System. The Ct value of detected gene between 18 and 30 was selected and quantified with the 2-ΔΔCt method.
Immunohistochemical staining
Fifty paired paraffin sections of TSCC and adjacent normal tissues were cut at a thickness of 6 μm. The experiment was performed by StreptAvidin peroxidase conjugated method for MTSS1 according to the operation manual. In brief, the deparaffinized and rehydrated sections were incubated with the primary polyclonal against MTSS1 (# 4385, Cell Signaling Technology) in a dilution of 1:100 at 4°C overnight and secondary antibody in 1:500 at 37°C for 30 min. Then, after thorough washing with PBS for 10 min, streptavidin for 30 min, the sections were stained with liquid Diaminobenzydine (DAB) for 5 min. Finally, the sections were photographed and the staining was assessed independently by pathologists.
Construction of MTSS1 over-expression vectors and transfection
The MTSS1 over-expression vector pEGFP-N1-MTSS1 was constructed by the company (Genescript, Nanjing, China). To brief, the whole cDNA of MTSS1 gene labeled with EcoRI-BamHI enzyme cutting sites was chemically synthesized following sub-cloned to pEGFP-N1 plasmid. Then the plasmid was purified and transfected into Tca8113 cells using LipofectamineTM 3000 Reagent (Invitrogen, USA) according to the manufacturer’s instructions. For the transient transfection, the cells were cultured with pEGFP-N1-MTSS1 plasmid 48 h and then the biological effect of MTSS1 gene on the Tca8113 cells was assayed. For the stable transfection, the cells transfected with pEGFP-N1-MTSS1 plasmid were continuous cultured and selected with G418 up to 3 w. Then the transfectants were verified for their expression of MTSS1 and successful clones were used in the MTSS1 mechanism analysis.
Biological behavior assay of MTSS1 gene in Tca8113 cells
Cell proliferation ability detected by MTT and in vitro invasion ability detected by using Transwell Chamber were finished as previously described [13]. For the Wound healing assay, Tca8113 cells were seeded into 6 well plates and when the cell confluence reached approximately 80% post-transfection, scratches were made by using a 100 μl pipette tip. After incubation for 48 h, detached cells were washed and 3 fields of observations were randomly picked for photograph.
MicroRNA targets prediction and dual-luciferase reporter assay
The miRNAs prediction of MTSS1 gene was finished by TargetScan (http://www.targetscan.org) and miRanda http://www.microrna.org). The dual-luciferase reporter plasmids, pmiR-MTSS1-wt (containing the wild-type MTSS1 3’UTR binding site in pmiR-RB-REPORTTM luciferase reporter plasmid (RiboBio Co. Ltd., China)) and pmiR-MTSS1-mut (containing the mutant MTSS1 3’UTR) were constructed by the company (Genescript, Nanjing, China) as mentioned above. For the luciferase assay, the constructed plasmid (500 ng) and miR-96 mimics (100 nM) were co-transfected into Tca8113 cells by using LipofectamineTM 3000 Reagent (Invitrogen, USA). Then the dual-luciferase reporter assay system (Promega, USA) was used to assay the luciferase activity after co-transfection Tca8113 cells 48 h according to the manufacturer’s instructions.
Western blot analysis
Total 20 mg of protein was used for western blot. Samples were resolved in SDS-PAGE gels and transferred to PVDF membranes. After blocked, the membranes were probed with primary polyclonal against MTSS1 (# 4385, Cell Signaling Technology) and beta-tubulin (# 2128, Cell signaling Technology) overnight at 4°C. Then incubated with horseradish peroxidase (HRP)-conjugated secondary anti-mouse or anti-rabbit antibody (Cell Signaling Technology, Beverly, MA). Proteins were enhanced by chemiluminescence detection kit. The protein quantification was performed using ImageJ software.
Statistical analysis
All statistical analysis was performed using SPSS 16.0 software. The data were presented as mean ± SD of three independent experiments and compared using Student’s t-test and one-way ANOVA. A P-value of less than 0.05 was considered to be statistically significant and indicated by (*).
Results
MTSS1 is down-regulated in TSCC tissues and cells
In order to verify the results of TSCC gene-array, we first estimated the expression levels of MTSS1 transcript in fifty TSCC patient specimens using qRT-PCR. The results showed that lower mRNA expression level of MTSS1 was observed in TSCC tissues when compared to the paired adjacent normal tissues (P < 0.001) (Figure 1A). Further, we examined the expression of MTSS1 in Tca8113 cells and the result showed that MTSS1 was expressed at a very low level (Figure 1B). Finally, we assayed the expression pattern of MTSS1 in the human TSCC tissue sections (n = 50) using immunohistochemical analysis. Staining results showed that MTSS1 protein was expressed in the cytoplasm and nuclei of adjacent normal tissues and well differentiated TSCC tissues, while MTSS1 was not detected in the poorly differentiated TSCC tissues (Figure 1C-E). Statistically analysis revealed that the expression of MTSS1 protein was significantly reduced or absent than those in the paired adjacent normal tissues (P < 0.001). Together, all the results were consistent with our gene-array data, which indicated that MTSS1 may play some important roles in human TSCC.
Figure 1.
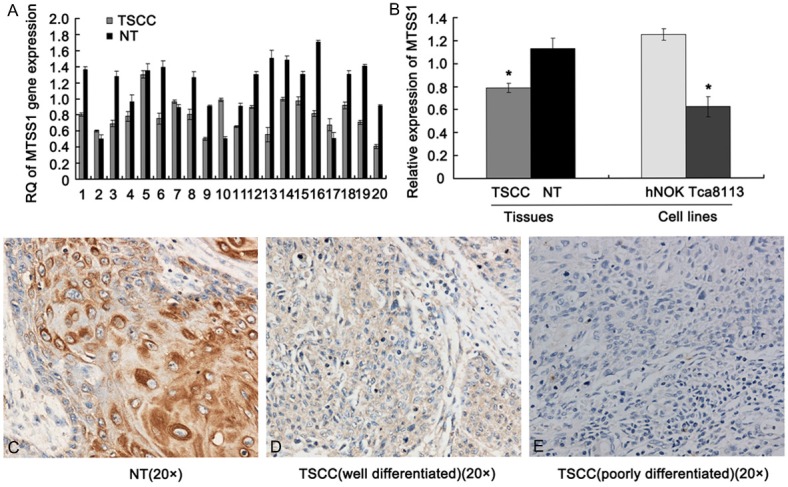
MTSS1 is down-regulated in human TSCC. A. representative qRT-PCR results of MTSS1 gene expression in fifty TSCC and adjacent normal tissues. B. Statistic analysis results of MTSS1 gene expression in TSCC tissues and Tca8113 cell line. The results were shown as mean ± SD. *P < 0.05 versus adjacent normal tissues and hNOK cell line. C-E. representative immunohistochemical staining results of MTSS1 protein in fifty TSCC and adjacent normal tissues. C is adjacent normal tissues, D is well differentiated TSCC and E is poorly differentiated TSCC (SP, original magnification 200 ×).
MTSS1 inhibits Tca8113 cells proliferation and metastasis
To investigate the impact of MTSS1 on the functions of TSCC cells, the constructed MTSS1 expression vectors were utilized to over-express MTSS1 in TSCC Tca8113 cells. Fluorescence microscope observation confirmed the successful of transfection (Figure 2A). MTT results showed significantly difference between PBS, pEGFP-N1 vector and pEGFP-N1-MTSS1 transfection groups, which revealed that MTSS1 functions as inhibitor in Tca8113 cells proliferation (Figure 2B). As for MTSS1 is a tumor metastasis suppressor gene, we future explore whether the expression of MTSS1 could decrease the invasion capacity of Tca8113 cells. As shown in Figure 2C and 2D, forced expression of MTSS1 led to retarded wound closing compared to other groups. Transwell assay was used to verify the metastasis function of MTSS1 in Tca8113 cells. After 24 h, the trans-membrane cells were fixed, stained and observed microscopically. The results demonstrated that MTSS1 over-expression led to significantly inhibition of cell invasion (Figure 2E and 2F).
Figure 2.
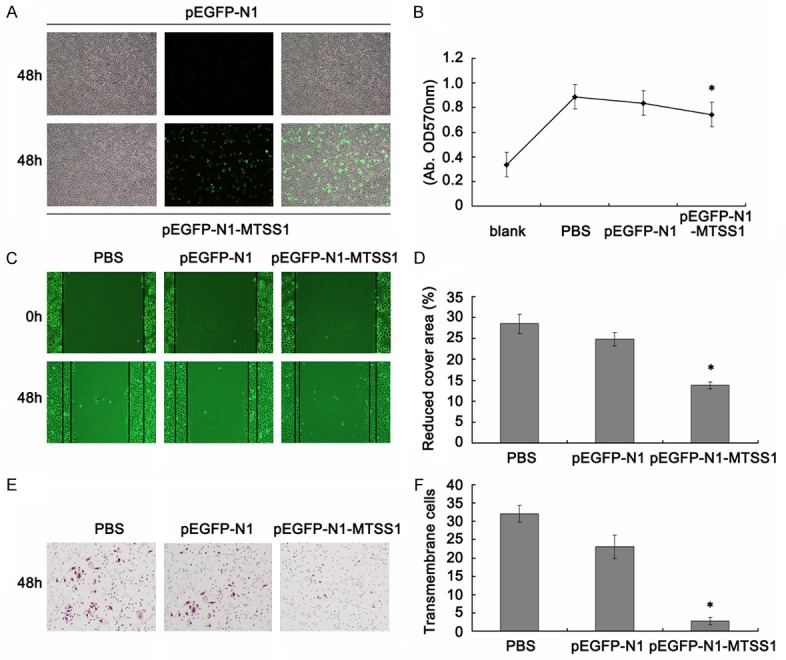
Biological effect of MTSS1 gene on Tca8113 cells. A. Fluorescence microscope observation the efficiency after pEGFP-N1-MTSS1 transfection 48 h. B. Cell proliferation ability measured by the MTT assay. Results are means of three independent experiments ± SD. C. Wound healing assay (48 h). D. Statistic analysis of reduced cover area. The results were shown as mean ± SD. *P < 0.05. E. Transwell (magnification, 200 ×). F. statistical analysis of average invasive cell number of three independent experiments ± SD. *P < 0.05.
MiR-96 functionally targets MTSS1 in Tca8113
As for the important function of MTSS1 gene in TSCC, further mechanism analysis is becoming necessary. Using the publicly available databases TargetScan and miRanda, we found two conserved miR-96 binding sites in the 3’UTRs of MTSS1 gene (261-268 bp and 1929-1935 bp). However, only site 1/miR-96 with PCT = 0.89 was predicted in both databases and highly conserved (Figure 3A).
Figure 3.
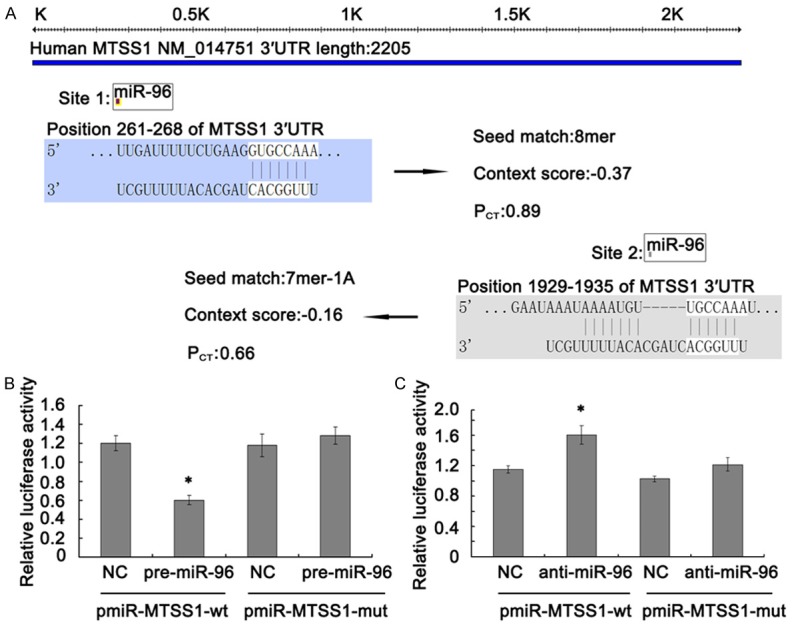
The 3’UTR of MTSS1 was a functional target of miR-96 in Tca8113 cells. A. 3’UTR region of MTSS1 mRNA is partially complementary to miR-96. B and C. Effect of miR-96 on the luciferase activity of pmiR-MTSS1-3’UTR and pmiR-MTSS1-3’UTR mutation. Firefly luciferase activity was normalized to Renilla luciferase. Each experiment was performed in triplicate. Data are shown as mean ± SD. *P < 0.05.
To further investigate whether MTSS1 was a target of miR-96, we constructed luciferase reporter plasmids containing either wild-type or mutant 3’-UTRs of MTSS1. As shown in Figure 3B, the relative luciferase activity was significantly lower in cells after 48 h co-transfection with pmiR-MTSS1-wt and pre-miR-96 as compared with the NC (P < 0.05). However, no statistically significant difference was observed between cells co-transfected with pmiR-MTSS1-mut and the pre-miR-96 or NC. On the opposite, the relative luciferase activity was significantly higher in cells after 48 h co-transfection with pmiR-MTSS1-wt and anti-miR-96 as compared with the NC (P < 0.05) (Figure 3C). These results indicated that the 3’UTR of MTSS1 was a functional target of miR-96 in Tca8113 cells.
MiR-96 regulates MTSS1 mediated Tca8113 cells proliferation and metastasis
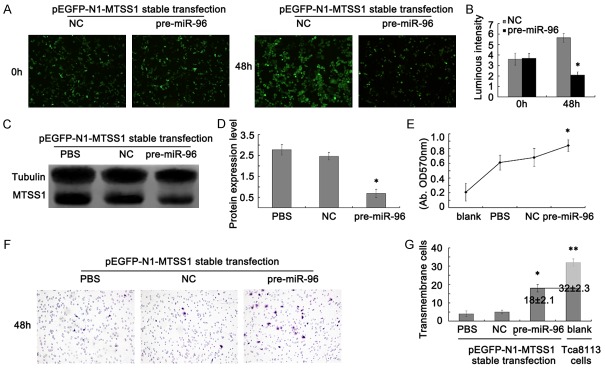
To further prove if miR-96 represses MTSS1 expression, following cell proliferation and metastasis inhibition in the Tca8113 cells, we constructed a cell model with pre-miR-96 and pEGFP-N1-MTSS1 stable transfected Tca8113 cells. After pre-miR-96 interferences pEGFP-N1-MTSS1 stable transfected Tca8113 cells 48 h, serials of assays were used to verify our hypothesis. Fluorescence microscope observation showed that the green fluorescence of pre-miR-96 transfection group was significantly weaker than that in the NC group (Figure 4A and 4B). Meanwhile, western blot assays were taken to evaluate the protein level of MTSS1. The results showed that MTSS1 was also dramatically decreased in protein level after ectopic over-expression of miR-96 (Figure 4C and 4D). Collectively, these data supported our speculation that MTSS1 is a direct target of miR-96.
Figure 4.
MiR-96 regulates MTSS1 mediated Tca8113 cells proliferation and metastasis. A. Fluorescence microscope observation the expression of MTSS1 after pEGFP-N1-MTSS1 stable transfection in Tca8113 cells. B. Statistic Luminous intensity analysis of MTSS1 expression. The results were shown as mean ± SD. *P < 0.05. C and D. Western blot analysis results of MTSS1 protein expression in pEGFP-N1-MTSS1 stable transfected Tca8113 cells. Tubulin was used as a reference control. Quantitative analysis was done by measure the IVD of protein bands. The results were shown as mean ± SD. *P < 0.05. E. MTT assay after pre-miR-96 transfection into pEGFP-N1-MTSS1 stable expressed Tca8113 cells. The results were shown as mean ± SD. *P < 0.05. F. Transwell assay after pre-miR-96 transfection into pEGFP-N1-MTSS1 stable expressed Tca8113 cells (magnification, 200 ×). G. Statistic analysis of average invasive cell number of three independent experiments ± SD. *P < 0.05. 18 ± 2.1 and 32 ± 2.3 means trans-membrane cell numbers in pre-miR-96 transfected pEGFP-N1-MTSS1 stable expressed Tca8113 cells and Tca8113 cells transient transfected with pEGFP-N1-MTSS1 plasmid.
Next, MTT and Transwell assays were applied to detect the cell proliferation and metastasis ability changes after MTSS1 repressed with miR-96. Both the MTT and Transwell results showed significantly proliferation and transmembrane ability improved compared with the NC group. But interestingly, the transmembrane cells of pre-miR-96 interfered MTSS1 stable expression cells was 18 ± 2.1, while the transmembrane cells of Tca8113 cells was 32 ± 2.3, which revealed that transfection of miR-96 could not completely restore the invasion ability of Tca8113 cells (Figure 4E-G). Together, our results suggest that MTSS1 expression was partly regulated by miR-96, thereby inhibiting TSCC progression.
Discussion
Although MTSS1 gene was identified as a novel potential metastasis suppressor gene, its function as a tumor suppressor was in some ways controversial. Researchers reported that patients with tumors expressing reduced levels of MTSS1 had a poorer prognosis and over-expression of MTSS1 significantly suppressed the invasive, migratory, growth and adherence properties of a human breast cancer cell line, while knockdown of MTSS1 dramatically enhanced these properties [7]. Du P et al showed reduced expression level of MTSS1 in bladder cancer cells and MTSS1 over-expression significantly suppressed the aggressiveness of bladder cancer cells through inhibiting cell growth, and adhesion [9]. All these results suggest that MTSS1 gene may function as a tumor suppressor. But Mattila and his colleagues suggest that MTSS1 gene may function as a tissue-specific regulator of cytoskeletal dynamics, interact with ATP-actin monomers through its C-terminal WH2 domain, while not as a tumor suppressor gene [14]. In this study, we confirmed that MTSS1 gene was down-regulated in TSCC tissues and Tca8113 cells. Forced expression of MTSS1 in Tca8113 cells led to a reduced cell proliferation, a retarded wound closing and an inhibition of cell invasion which were tentatively indicated in other tumor cells, including breast cancer cells and oesophageal cancer cells [5,7]. Together, we conclude that MTSS1 gene playing a key role in the TSCC aggressiveness functioned as a potential tumor suppressor.
As miRNAs function as important regulators of target genes involved in normal development and disease such as cancer [15], researchers made great efforts in finding miRNAs as regulators in cancer progression and development recently years. As for MTSS1 gene, miR-182 was shown to be a directed target and contributed to the aggressive of breast cancer, ovarian cancer, prostate cancer and hepatocellular carcinoma largely by its negative regulation of MTSS1 [12,16-18]. MicroRNA-135 and miR-23a was reported to regulate MTSS1 expression and promote migration and invasion in colorectal cancer [4,19,20]. In this study, by using the TargetScan and miRanda software, we predicted miR-96 is a putative regulator of MTSS1 gene. Following detection with luciferase reporter, western blot and serials of assays, we confirmed that MTSS1 was a direct target of miR-96 in Tca8113 cells and miR-96 could promote MTSS1 repression bypassing cell proliferation and metastasis controls. Our results revealed that miR-96 is critical for the development of human TSCC, and miR-96 is one of the mechanisms of MTSS1 repression in TSCC tumorigenesis.
As we all known, miR-96, miR-183 and miR-182 belong to the same miR-183 family and share the same transcription start site (chr7: 129207158), may be coordinately expressed and play roles together during tumorigenesis [21]. A recent report showed that the up-regulation of microRNA-182 and microRNA-96 targeting forkhead box O3 played a significant role in the pro-proliferation effect of leptin on ovarian cancer cells [22]. Knocking down of miR-182 and miR-183, both highly over-expressed in ductal carcinoma in situ (preinvasive breast cancer), increased the expression of CBX7, DOK4, NMT2 and EGR1 [23]. In addition, miR-96, miR-182 and miR-183 over-expressed in endometrial cancer and functioned as an oncogene through repressing FOXO1 expression. Then aberrant miR-183 family expression resulted in deregulated cell cycle control and impaired apoptotic responses [24]. In this study, we also predicted miR-182 and miR-183 binding sites in the 3’UTR of MTSS1 transcripts by two miRNAs prediction software (TargetScan and miRanda). Together with our present findings that transfection of pre-miR-96 was insufficient to fully restore the invasion ability mediated by MTSS1 gene in Tca8113 cells. We speculated that as miR-183 family members, miR-182 and/or miR-183 may also regulate MTSS1 expression in a miR-96-depend directly or indirectly manner, which need further exploring.
In summary, we identified MTSS1 gene was down-regulated in TSCC and the ability of miR-96 to promote MTSS1 repression may precipitate in the TSCC tumorigenesis through bypassing cell proliferation and metastasis control.
Conclusions
MTSS1 gene was down-regulated in TSCC and the ability of miR-96 to promote MTSS1 repression may precipitate in the TSCC tumorigenesis through bypassing cell proliferation and metastasis control. MiR-183 family members, miR-182 and/or miR-183 weather regulate MTSS1 expression in a miR-96-depend directly or indirectly manner need further exploring.
Acknowledgements
This work was supported by the National Nature Science Foundation of China (81301834) and Nature Science Foundation of Liaoning province (2013021094).
Disclosure of conflict of interest
None.
References
- 1.Krishna Rao SV, Mejia G, Roberts-Thomson K, Logan R. Epidemiology of oral cancer in Asia in the past decade--an update (2000-2012) Asian Pac J Cancer Prev. 2013;14:5567–5577. doi: 10.7314/apjcp.2013.14.10.5567. [DOI] [PubMed] [Google Scholar]
- 2.Lee YG, Macoska JA, Korenchuk S, Pienta KJ. MIM, a potential metastasis suppressor gene in bladder cancer. Neoplasia. 2002;4:291–294. doi: 10.1038/sj.neo.7900231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isaksson HS, Sorbe B, Nilsson TK. Whole genome expression profiling of blood cells in ovarian cancer patients-prognostic impact of the CYP1B1, MTSS1, NCALD, and NOP14. Oncotarget. 2014;5:4040–4049. doi: 10.18632/oncotarget.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu W, Wang Z, Yang P, Yang J, Liang J, Chen Y, Wang H, Wei G, Ye S, Zhou Y. MicroRNA-135b regulates metastasis suppressor 1 expression and promotes migration and invasion in colorectal cancer. Mol Cell Biochem. 2014;388:249–259. doi: 10.1007/s11010-013-1916-z. [DOI] [PubMed] [Google Scholar]
- 5.Xie F, Ye L, Chen J, Wu N, Zhang Z, Yang Y, Zhang L, Jiang WG. The impact of Metastasis Suppressor-1, MTSS1, on oesophageal squamous cell carcinoma and its clinical significance. J Transl Med. 2011;9:95. doi: 10.1186/1479-5876-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma S, Guan XY, Lee TK, Chan KW. Clinicopathological significance of missing in metastasis B expression in hepatocellular carcinoma. Hum Pathol. 2007;38:1201–1206. doi: 10.1016/j.humpath.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Parr C, Jiang WG. Metastasis suppressor 1 (MTSS1) demonstrates prognostic value and anti-metastatic properties in breast cancer. Eur J Cancer. 2009;45:1673–1683. doi: 10.1016/j.ejca.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 8.Du P, Ye L, Li H, Yang Y, Jiang WG. The tumour suppressive role of metastasis suppressor-1, MTSS1, in human kidney cancer, a possible connection with the SHH pathway. J Exp Ther Oncol. 2012;10:91–99. [PubMed] [Google Scholar]
- 9.Du P, Ye L, Ruge F, Yang Y, Jiang WG. Metastasis suppressor-1, MTSS1, acts as a putative tumour suppressor in human bladder cancer. Anticancer Res. 2011;31:3205–3212. [PubMed] [Google Scholar]
- 10.Woodings JA, Sharp SJ, Machesky LM. MIM-B, a putative metastasis suppressor protein, binds to actin and to protein tyrosine phosphatase delta. Biochem J. 2003;371:463–471. doi: 10.1042/BJ20021962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong J, Shaik S, Wan L, Tron AE, Wang Z, Sun L, Inuzuka H, Wei W. SCF β-TRCP targets MTSS1 for ubiquitination-mediated destruction to regulate cancer cell proliferation and migration. Oncotarget. 2013;4:2339–2353. doi: 10.18632/oncotarget.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirata H, Ueno K, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, Hinoda Y, Dahiya R. MicroRNA-182-5p promotes cell invasion and proliferation by down regulating FOXF2, RECK and MTSS1 genes in human prostate cancer. PLoS One. 2013;8:e55502. doi: 10.1371/journal.pone.0055502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y, Fu W, Chen H, Shang C, Zhong M. miR-24 functions as a tumor suppressor in Hep2 laryngeal carcinoma cells partly through down-regulation of the S100A8 protein. Oncol Rep. 2012;27:1097–1103. doi: 10.3892/or.2011.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattila PK, Salminen M, Yamashiro T, Lappalainen P. Mouse MIM, a tissue-specific regulator of cytoskeletal dynamics, interacts with ATP-actin monomers through its C-terminal WH2 domain. J Biol Chem. 2003;278:8452–8459. doi: 10.1074/jbc.M212113200. [DOI] [PubMed] [Google Scholar]
- 15.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163–6169. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- 16.Lei R, Tang J, Zhuang X, Deng R, Li G, Yu J, Liang Y, Xiao J, Wang HY, Yang Q, Hu G. Suppression of MIM by microRNA-182 activates RhoA and promotes breast cancer metastasis. Oncogene. 2014;33:1287–1296. doi: 10.1038/onc.2013.65. [DOI] [PubMed] [Google Scholar]
- 17.Xu X, Ayub B, Liu Z, Serna VA, Qiang W, Liu Y, Hernando E, Zabludoff S, Kurita T, Kong B, Wei JJ. Anti-miR182 reduces ovarian cancer burden, invasion, and metastasis: an in vivo study in orthotopic xenografts of nude mice. Mol Cancer Ther. 2014;13:1729–1739. doi: 10.1158/1535-7163.MCT-13-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Li J, Shen J, Wang C, Yang L, Zhang X. MicroRNA-182 downregulates metastasis suppressor 1 and contributes to metastasis of hepatocellular carcinoma. BMC Cancer. 2012;12:227. doi: 10.1186/1471-2407-12-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou W, Li X, Liu F, Xiao Z, He M, Shen S, Liu S. MiR-135a promotes growth and invasion of colorectal cancer via metastasis suppressor 1 in vitro. Acta Biochim Biophys Sin (Shanghai) 2012;44:838–846. doi: 10.1093/abbs/gms071. [DOI] [PubMed] [Google Scholar]
- 20.Jahid S, Sun J, Edwards RA, Dizon D, Panarelli NC, Milsom JW, Sikandar SS, Gümüs ZH, Lipkin SM. miR-23a promotes the transition from indolent to invasive colorectal cancer. Cancer Discov. 2012;2:540–553. doi: 10.1158/2159-8290.CD-11-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu X, Dong Z, Li Y, Yang Y, Yuan Z, Qu X, Kong B. The upregulation of signal transducer and activator of transcription 5-dependent microRNA-182 and microRNA-96 promotes ovarian cancer cell proliferation by targeting forkhead box O3 upon leptin stimulation. Int J Biochem Cell Biol. 2013;45:536–545. doi: 10.1016/j.biocel.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Hannafon BN, Sebastiani P, de las Morenas A, Lu J, Rosenberg CL. Expression of microRNA and their gene targets are dysregulated in preinvasive breast cancer. Breast Cancer Res. 2011;13:R24. doi: 10.1186/bcr2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myatt SS, Wang J, Monteiro LJ, Christian M, Ho KK, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S, Lam EW. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2010;70:367–377. doi: 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]