Abstract
Purpose: In the present study, we examined both FOXM1 mRNA and protein expression by Real-time quantitative PCR (qRT-PCR) and Western blot and investigate the expression of the human FOXM1 by Immunohistochemistry (IHC), and identify their potential roles in prognosis for patients with osteosarcoma. Methods: FOXM1 mRNA and protein expression levels were detected by RT-PCR and Western blot assays, respectively. Then, IHC was performed to analyze the association of FOXM1 expression in 83 osteosarcoma tissues with clinicopathological factors and survival of patients. Results: The expression levels of FOXM1 mRNA were found to be significantly increased in osteosarcoma tissues compared to noncancerous bone tissues (P = 0.0313). Simultaneously, western blot analysis showed that the protein level of FOXM1 in osteosarcoma tissues was significantly higher than that in noncancerous bone tissues. Kaplan-Meier analysis with the log-rank test indicated that high FOXM1 expression had a significant impact on overall survival (P = 0.0001). Conclusions: Our data showed that FoxM1 was upregulated in osteosarcoma tissues, and high expression of FoxM1 was correlated with a poor prognosis of patients with osteosarcoma. FoxM1 may function as a valuable prognostic biomarker for osteosarcoma.
Keywords: Osteosarcoma, foxm1, prognosis, survival
Introduction
Osteosarcoma is the most frequently occurring primary malignant tumor of bone arising from primitive bone-forming mesenchymal cells and a leading cause of cancer-related death in young adults and adolescents, accounting for 20-35% of all malignant bone tumors, and 60% of pediatric malignant bone tumors [1,2]. Osteosarcoma predominately arises in bone during periods of rapid growth and has a predilection to affect adolescents and young adults, and if untreated it is fatal [3]. Although considerable advances in therapeutic strategies, including wide tumour excision, adjuvant chemotherapy and radiotherapy have significantly increased the survival rate of the patients with osteosarcoma to 65-75%, the survival of the osteosarcoma patients with lung metastasis and advanced clinical stage is quite poor [4]. Although clinical stage, tumor size, and metastasis have been utilized as useful prognostic factors for the patients with osteosarcoma, the sensitivity and specificity is low [5]. To improve the prognosis of the patients with osteosarcoma, it is particularly necessary to develop and indentify the novel key biomarkers for treatments of the patients with osteosarcoma.
Forkhead box protein M1 (FoxM1) belongs to the forkhead box (Fox) family of transcription factors. Fox family shares a highly conserved 100-aa DNA binding domain (the forkhead box) and comprises more than 100 members in humans, classified as FOXA to FOXR on the basis of sequence similarity [6]. Previous studies have shown that FOXM1, activated by the Ras-MAPK and hedgehog signaling pathway, played an important role in cell cycle by promoting both the transition from G1 to S phase and progression to mitosis through genes of Cdc25B, CDK1 and p27KIP [7-10]. FOXM1 has been reported to be over-expressed and play an essential role in development and progression in various malignancies, including lung, liver and breast cancer [11-15]. Until now, there have been no reports about FOXM1 in osteosarcoma and the role of FOXM1 in osteosarcoma is still unknown. In the present study, we examined both FOXM1 mRNA and protein expression by Real-time quantitative PCR (qRT-PCR) and Western blot and investigate the expression of the human FOXM1 by Immunohistochemistry (IHC), and identify their potential roles in prognosis for patients with osteosarcoma.
Materials and methods
Patients and tissue samples
A total of 83 patients who underwent surgery at Yantai Yuhuangding Hospital during June 2008 to August 2013 were selected in this research. The study was approved by the ethics committee of Yantai Yuhuangding Hospital. Written informed consent was obtained from all participants. All specimens were handled and made anonymous according to the ethical and legal standards. A total of 83 osteosarcoma tissue samples and corresponding noncancerous bone tissue samples from the same specimens were collected from the Department of Pathology, Yantai Yuhuangding Hospital. Clinicopathological characteristics of all patients were detailed in Table 1.
Table 1.
Association of FOXM1 expression with clinicopathological features of the patients with osteosarcoma
| Characteristics | No.of cases | FOXM1 expression | P value | |
|---|---|---|---|---|
|
| ||||
| High (n = 41) | Low (n = 42) | |||
| Age | ||||
| < 45 | 35 | 18 | 17 | 0.53 |
| ≥ 45 | 48 | 23 | 25 | |
| Gender | ||||
| Male | 59 | 31 | 28 | 0.41 |
| Female | 24 | 10 | 14 | |
| Anatomical location | ||||
| Tibia/femur | 62 | 30 | 32 | 0.79 |
| Elsewhere | 21 | 11 | 10 | |
| Tumor size (cm) | ||||
| < 8 | 41 | 14 | 27 | 0.04 |
| ≥ 8 | 42 | 27 | 15 | |
| Clinical stage | ||||
| I/II | 54 | 21 | 33 | 0.003 |
| III | 29 | 20 | 9 | |
| Pathological facture | ||||
| Present | 9 | 6 | 3 | 0.01 |
| Absent | 74 | 35 | 39 | |
| Distant metastasis | ||||
| Present | 15 | 12 | 3 | < 0.001 |
| Absent | 68 | 29 | 39 | |
Quantitative real-time RT-PCR (qRT-PCR)
Total RNA was isolated tissue using TRIZOL reagent according to the manufacturer’s protocol (Invitrogen). RNA was reverse transcribed using SuperScript First Strand cDNA System (Invitrogen) according to the manufacturer’s instructions. The primer sequences for PCR amplification were as follows: FOXM1 sense 5’-TAT TCA CAG CAT CAT CAC AGC A-3’ and antisense 5’-GAA GGC TCC TCA ACC TTA ACC T-3’; GAPDH sense 5’-ACC ACA GTC CTG CAT GCC AC-3’ and antisense 5’-TCC ACC ACC CTG TTG CTG TA-3’. The PCR amplification were performed for 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, on a Applied Biosystems 7900HT (Applied Biosystems) with 1.0 μl of cDNA and SYBR Green Real-time PCR Master Mix (Takara). Data was collected and analyzed by SDS2.3 Software (Applied Biosystems). The expression level of each candidate gene was internally normalized against that of the GAPDH. The relative quantitative value was expressed by the 2-ΔΔCt method.
Western blot assay
Total proteins from tissues were lysed in lysis buffer containing protease inhibitor. Protein concentration was determined using a Bio-Rad protein assay system (Bio-Rad). Equivalent amounts of proteins were separated by SDSPAGE, and then transferred to polyvinylidene difluoride membranes (Bio-Rad). After being blocked in Tris buffered saline (TBS) containing 5% non-fat milk, the membranes were incubated with specific primary antibodies (Abcam) at 4°C for 12 hours and then with horseradish peroxidase conjugated anti-Rabbit antibody for 2 hours at room temperature. ECL detection reagent (Amersham LifeScience, Piscataway, NJ) was used to demonstrate the results.
Immunohistochemistry
The expression of FOXM1 was detected through immunohistochemistry (IHC) analyses with 4-um-thick sections of formalin-fixed and paraffin-embedded blocks. For IHC staining, tissue sections were dewaxed in xylene and rehydrated gradually with graded ethanol. For antigen retrieval, all the sections were incubated by microwave oven in citrate buffer solution (pH 6.0) for 20 minutes. Endogenous peroxidase was inactivated by 0.3% hydrogen peroxide in methanol for 15 min. After that, tissue slides were incubated with rabbit polyclonal antibody against human FOXM1 (dilution 1:100, Epitomics, US) at 4°C overnight and then visualized antibody binding sites with the SP peroxidase detection system. Finally, sections were incubated in 3,3’-diaminobenzidine tetrahydrochloride for 3-10 minutes and re-stained with 0.1% hematoxylin. In every case, negative control reaction was set with PBS replacing FOXM1 antibody, while the known positive-stained section was used as positive control. The results of IHC were evaluated by two pathologists independently with no knowledge of clinic-pathological features. The results of immunostaining were evaluated and scored semi-quantitatively by two pathologists who were blinded to the patients’ clinical data. The evaluation of the immunostaining results was based on a double scoring system (staining intensity multiplied by staining area). Staining intensity was scored as 0 for no staining, 1 for definite but weak staining, 2 for moderate staining and 3 for strong staining. The staining area was scored as 1 for staining of < 35%, 2 for 35-75%, and 3 for > 75% of the tumor cells. High expression of FOXM1 was defined as an immunostaining score of ≥ 4, whereas low expression of FOXM1 was defined as a score of < 4.
Statistical analysis
Statistical analysis was conducted using the Statistical Package for the Social Sciences 13.0 for Windows (SPSS Inc., Chicago, IL, USA). The chi-square test was used to assess FOXM1 expression with respect to clinicopathological parameters. The survival curves of the patients were determined using the Kaplan-Meier method and Cox regression, and the log-rank test was used for statistical evaluations. Data were expressed as the mean and standard deviation and analyzed using one-way analysis of variance. P < 0.05 was considered to indicate a significant difference.
Results
Expression of FOXM1 in osteosarcoma tissues and corresponding noncancerous bone tissues
The expression levels of FOXM1 mRNA were found to be significantly increased in osteosarcoma tissues compared to noncancerous bone tissues (P = 0.0313, Figure 1).
Figure 1.
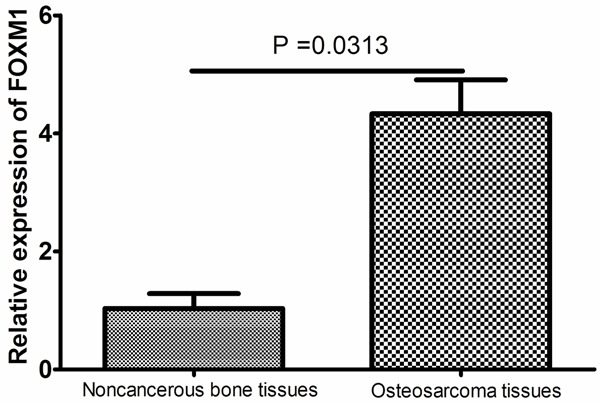
The expression of FOXM1 mRNA in the human osteosarcoma tissue and the corresponding noncancerous bone tissues. The expression levels of FOXM1 mRNA were found to be significantly increased in osteosarcoma tissues compared to noncancerous bone tissues (P = 0.0313).
Simultaneously, western blot analysis showed that the protein level of FOXM1 in osteosarcoma tissues was significantly higher than that in noncancerous bone tissues (Figure 2). IHC showed that the FOXM1 protein was mainly expressed in the cytoplasm of malignant cells (shown in Figure 3). The samples were divided into two expression groups as described previously. FOXM1 was highly expressed in 41 of the 83 (49.4%) patients with osteosarcoma.
Figure 2.
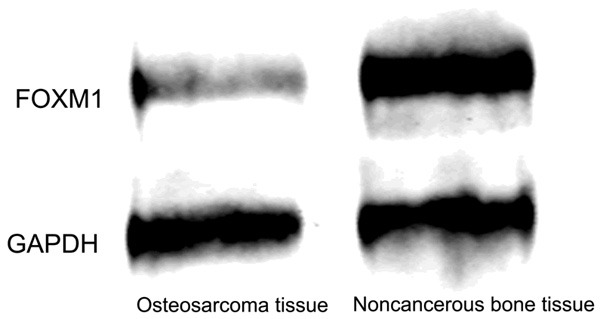
The expression of FOXM1 protein in the human osteosarcoma tissue and the corresponding noncancerous bone tissues. Western blot analysis showed that the protein level of FOXM1 in osteosarcoma tissues was significantly higher than that in noncancerous bone tissues.
Figure 3.
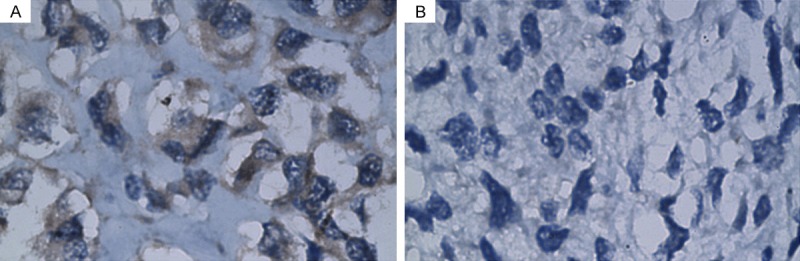
Immunostaining of FOXM1 protein in osteosarcoma tissue samples. Osteosarcoma tissues showed varied stainability for FOXM1 (A: Positive expression; B: Negative expression).
Association between FOXM1 expression and clinicopathologic parameters
To evaluate the significance of FOXM1 expression in osteosarcoma, we investigated the relationship between FOXM1 immunostaining and clinicopathologic features (shown in Table 1). Overall, there was no significant relationship between FOXM1 expression and gender (P = 0.53), age (P = 0.41), or anatomical location (P = 0.79). However, significant correlations were found between FOXM1 expression and tumor size (P = 0.04), clinical stage (P = 0.003), pathological facture (P = 0.01), and distant metastasis (P < 0.001).
Prognostic significance of FOXM1 expression in survival of patients with osteosarcoma
In the present study, 5 patients were lost to follow-up and were excluded from the survival analyses. The remaining 78 patients with adequate follow-up data were followed for 11-69 months. Multivariate analysis revealed that FOXM1 expression and clinical stage were independently associated with the overall survival (HR = 5.991, 95% CI: 4.286-6.541; P = 0.001; HR = 6.782, 95% CI: 4.009-7.994; P = 0.002; respectively, shown in Table 2). Kaplan-Meier analysis with the log-rank test indicated that high FOXM1 expression had a significant impact on overall survival (P = 0.0001; Figure 4).
Table 2.
Cox regression model for multivariate analysis of prognostic factors for osteosarcoma
| Variable | Overall survival | ||
|---|---|---|---|
|
| |||
| HR | 95% CI | P-value | |
| Age (≥ 45 vs. < 45) | 1.191 | 0.921-5.228 | 0.411 |
| Gender (male vs. female) | 1.006 | 0.273-6.159 | 0.523 |
| Anatomical location (tibia/femur vs. elsewhere) | 0.812 | 0.235-3.996 | 0.114 |
| Tumor size (≥ 8 vs. < 8) | 2.334 | 0.734-3.991 | 0.091 |
| Clinical stage (III vs. I/II) | 6.782 | 4.009-7.994 | 0.002 |
| Pathological facture (present vs. absent) | 3.127 | 0.998-7.119 | 0.062 |
| Distant metastasis (present vs. absent) | 6.119 | 0.987-8.991 | 0.053 |
| FOXM1 expression (high vs. low) | 5.991 | 4.286-6.541 | 0.001 |
HR = Hazard ratio; CI = confidence interval.
Figure 4.
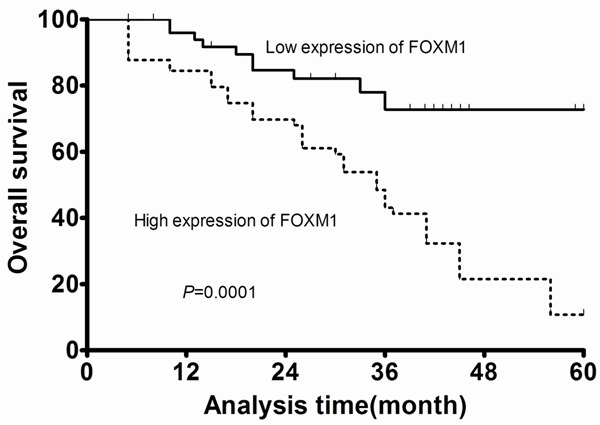
Kaplan-Meier survival analyses of patients with osteosarcoma.
Discussion
Forkhead transcription factor is a new family of transcription factors, which was officially unified in 2000 [16]. FoxM1 transcription factor is expressed in actively dividing cells and is critical for cell cycle progression. It was first identified as a proliferation-specific transcription factor, which is expressed in various tumor cell lines and embryonic tissues. Previous work has linked FoxM1 upregulation to a variety of cancers, including cancers of the liver, gastric, prostate, brain, breast, lung, esophagus, colon, pancreas and nervous system, marking it as a proto-oncogene. Genome-wide gene expression profiling of cancers has independently and consistently identified FoxM1 as one of the most commonly upregulated genes in human solid tumor [17]. Importantly, its expression is often correlated with poor prognosis and chemotherapy resistance [18]. However, little is known about its expression pattern and biological significance in osteosarcoma. In the current study, we showed that FoxM1 expression determined by real-time quantitative PCR and Western blot was significantly higher in osteosarcoma tissues compared to noncancerous bone tissues. Immunohistochemical analysis also confirmed that tumor tissues exhibited abundant FoxM1 expression, in contrast to noncancerous bone tissues which displayed absence or lower FoxM1 expression. To investigate whether FoxM1 expression might be associated with the progression of osteosarcoma, the FoxM1 expression levels and the clinicopathologic characteristics of 83 patients with osteosarcoma were compared by immunohistochemistry. We found that high FoxM1 expression is significantly correlated with tumor size, clinical stage, pathological facture, and distant metastasis, suggesting that its expression might be important for the acquirement of malignant potential in osteosarcoma. Furthermore, the results of Kaplan-Meier analyses shown that osteosarcoma tissues with high FoxM1 expression tend to have shorter overall survival. Finally, the multivariate analysis demonstrated that FoxM1 overexpression was a statistically significant risk factor affecting overall survival in osteosarcoma patients, suggesting that FoxM1 expression could be a valuable marker of tumor progression and prognosis of osteosarcoma. These findings are in agreement with results from studies in other human cancers overexpressing FoxM1.
To our knowledge, this is the first study to investigate the clinical significance of FoxM1 in patients with osteosarcoma, however, the precise mechanism of FoxM1 in osteosarcoma tumorigenesis and progression is still not understood. In Liu D et al’s study, FoxM1 expression was knocked down by small interfering RNA (siRNA) in bladder cancer cells; and proliferation, migration and invasion were assayed. They found that down-regulation of FoxM1 inhibited cell proliferation, migration and invasion [19]. Interestingly, Li X et al found that FOXM1 was revealed to alter microtubule dynamics in response to the treatment of docetaxel, and the drug resistance could be reversed with FOXM1 inhibitor thiostrepton treatment. Similar to their results, Millour J et al found that FOXM1 was a key mediator of the mitogenic functions of estrogen receptor alpha and estrogen in breast cancer cells, and that the deregulation of FOXM1 may contribute to anti-estrogen insensitivity [14]. The precise mechanism of FoxM1 in osteosarcoma tumorigenesis and progression needs to be investigated in future studies.
In conclusion, our data showed for the first time that FoxM1 was upregulated in osteosarcoma tissues, and that high expression of FoxM1 was correlated with a poor prognosis of patients with osteosarcoma. FoxM1 may function as a valuable prognostic biomarker for osteosarcoma. The precise mechanism of FoxM1 in osteosarcoma tumorigenesis and progression needs to be investigated in future studies.
Acknowledgements
This study is supported by The National Natural Science Foundation of China (81460406), and The National Natural Science Foundation of the Jiangxi Province (2013BAB205044).
Disclosure of conflict of interest
None.
References
- 1.Broadhead ML, Clark JC, Myers DE, Dass CR, Choong PF. The molecular pathogenesis of osteosarcoma: a review. Sarcoma. 2011;2011:959248. doi: 10.1155/2011/959248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 3.Bielack SS, Kempf-Bielack B, Delling G, Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M, Winkelmann W, Zoubek A, Jürgens H, Winkler K. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 2002;20:776–790. doi: 10.1200/JCO.2002.20.3.776. [DOI] [PubMed] [Google Scholar]
- 4.Ta HT, Dass CR, Choong PF, Dunstan DE. Osteosarcoma treatment: state of the art. Cancer Metastasis Rev. 2009;28:247–263. doi: 10.1007/s10555-009-9186-7. [DOI] [PubMed] [Google Scholar]
- 5.Bispo Junior RZ, Camargo OP. Prognostic factors in the survival of patients diagnosed with primary non-metastatic osteosarcoma with a poor response to neoadjuvant chemotherapy. Clinics. 2009;64:1177–1186. doi: 10.1590/S1807-59322009001200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma RY, Tong TH, Leung WY, Yao KM. Raf/MEK/MAPK signaling stimulates the nuclear translocation and transactivating activity of FOXM1. Methods Mol Biol. 2010;647:113–123. doi: 10.1007/978-1-60761-738-9_6. [DOI] [PubMed] [Google Scholar]
- 8.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, Ackerson T, Costa RH. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katoh Y, Katoh M. Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr Mol Med. 2009;9:873–886. doi: 10.2174/156652409789105570. [DOI] [PubMed] [Google Scholar]
- 10.Costa RH. FoxM1 dances with mitosis. Nat Cell Biol. 2005;7:108–110. doi: 10.1038/ncb0205-108. [DOI] [PubMed] [Google Scholar]
- 11.Balli D, Zhang Y, Snyder J, Kalinichenko VV, Kalin TV. Endothelial cell-specific deletion of transcription factor FoxM1 increases urethane-induced lung carcinogenesis. Cancer Res. 2011;71:40–50. doi: 10.1158/0008-5472.CAN-10-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calvisi DF, Pinna F, Ladu S, Pellegrino R, Simile MM, Frau M, De Miglio MR, Tomasi ML, Sanna V, Muroni MR, Feo F, Pascale RM. Forkhead box M1B is a determinant of rat susceptibility to hepatocarcinogenesis and sustains ERK activity in human HCC. Gut. 2009;58:679–687. doi: 10.1136/gut.2008.152652. [DOI] [PubMed] [Google Scholar]
- 13.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, Michalopoulos G, Becich M, Monzon FA. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millour J, Constantinidou D, Stavropoulou AV, Wilson MS, Myatt SS, Kwok JM, Sivanandan K, Coombes RC, Medema RH, Hartman J, Lykkesfeldt AE, Lam EW. FOXM1 is a transcriptional target of ERalpha and has a critical role in breast cancer endocrine sensitivity and resistance. Oncogene. 2010;29:2983–2995. doi: 10.1038/onc.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai B, Kang SH, Gong W, Liu M, Aldape KD, Sawaya R, Huang S. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26:6212–6219. doi: 10.1038/sj.onc.1210443. [DOI] [PubMed] [Google Scholar]
- 16.Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- 17.Uddin S, Ahmed M, Hussain A, Abubaker J, Al-Sanea N, AbdulJabbar A, Ashari LH, Alhomoud S, Al-Dayel F, Jehan Z, Bavi P, Siraj AK, Al-Kuraya KS. Genome-wide expression analysis of Middle Eastern colorectal cancer reveals FOXM1 as a novel target for cancer therapy. Am J Pathol. 2011;178:537–547. doi: 10.1016/j.ajpath.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwok JM, Peck B, Monteiro LJ, Schwenen HD, Millour J, Coombes RC, Myatt SS, Lam EW. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mol Cancer Res. 2010;8:24–34. doi: 10.1158/1541-7786.MCR-09-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu D, Zhang Z, Kong CZ. High FOXM1 expression was associated with bladder carcinogenesis. Tumour Biol. 2013;34:1131–1138. doi: 10.1007/s13277-013-0654-x. [DOI] [PubMed] [Google Scholar]


