Abstract
The purpose of this study was to determine the expression of long non-coding RNA (lncRNA) FTX and analyze its prognostic and biological significance in colorectal cancer (CRC). A quantitative reverse transcription PCR was performed to detect the expression of long non-coding RNA FTX in 35 pairs of colorectal cancer and corresponding noncancerous tissues. The expression of long non-coding RNA FTX was detected in 187 colorectal cancer tissues and its correlations with clinicopathological factors of patients were examined. Univariate and multivariate analyses were performed to analyze the prognostic significance of Long Non-coding RNA FTX expression. The effects of long non-coding RNA FTX expression on malignant phenotypes of colorectal cancer cells and its possible biological significances were further determined. Long non-coding RNA FTX was significantly upregulated in colorectal cancer tissues, and low long non-coding RNA FTX expression was significantly correlated with differentiation grade, lymph vascular invasion, and clinical stage. Patients with high long non-coding RNA FTX showed poorer overall survival than those with low long non-coding RNA FTX. Multivariate analyses indicated that status of long non-coding RNA FTX was an independent prognostic factor for patients. Functional analyses showed that upregulation of long non-coding RNA FTX significantly promoted growth, migration, invasion, and increased colony formation in colorectal cancer cells. Therefore, long non-coding RNA FTX may be a potential biomarker for predicting the survival of colorectal cancer patients and might be a molecular target for treatment of human colorectal cancer.
Keywords: Colorectal cancer, long non-coding RNA, FTX, biological
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide. According to the result of IARC (International Agency for Research on Cancer), approximately 1.36 million CRC patients were diagnosed yearly worldwide and 0.6 million CRC patients died in 2012 [1]. Colorectal carcinogenesis is a multistep process characterized by genetic and epigenetic alterations that influence key cellular pathways involved in growth and development [2]. Thus, elucidation of the molecular mechanisms involved in CRC development will be helpful to exploit potential, molecular, diagnostic, and prognostic markers. Many studies have been conducted to search for disease features that can help to predict disease outcome and treatment response, but no robust tumor markers have yet been identified.
Proteins are considered to be the major molecules carrying out essential biologic actions [3], yet only 2% of the human genome contains the codes for proteins. Most genomic sequences are now understood to be transcribed, though without translation capability [4]. These non-protein coding transcripts are involved in many biologic processes and cellular activities. Recently, longnon-coding RNAs (lncRNAs) were recognized as a new class of non-coding RNAs with important biologic functions [5-10]. LncRNAs exert their actions through interactions with chromatin in the regulation of gene expression [11-13], modulation of epigenetic regulation pre- and post-transcriptionally [14-16], and influences on activities and locations of other functional molecules such as proteins and other RNA species [7,10,17-21]. Studies have shown that disruption of lncRNA action occurs in certain diseases including cancer [22-27].
We searched the literature and found no information on long non-coding RNA FTX with regard to its biological functions and associations with CRC or other diseases. To determine the clinical relevance of long non-coding RNA FTX in CRC and to assess its biologic effects on breast cancer cells, we measured long non-coding RNA FTX expression in more than 100 CRC samples to analyze its association with clinical and pathological features of CRC, independent clinical studies to confirm the findings of our clinical study, and transfected a FTX expression vector into CRC cells to assess the FTX’s effects on cell growth and migration. In this report, we describe the findings of long non-coding RNA FTX in our clinical study and in vitro experiments.
Materials and methods
Tissue collection
Paired CRC and adjacent normal colorectal tissue were obtained from 187 patients who had undergone surgical CRC resection between 2008 and 2010 at the department of gastrointestinal surgery, provincial hospital affiliated to shandong university, China. Local or systemic treatment had not been performed in these patients prior to the operation, and the clinicopathological characteristics of the patients with CRC are recorded. Samples were immediately macrodissected at the time of surgery and placed directly in RNALater stabilization solution (Qiagen, Hilden, Germany). All of the tissues were stored at -80°C until total RNA was extracted. The differentiation grade, pathological stage, grade and nodal status were appraised by an experienced pathologist. Clinicopathological characteristics including tumor-node-metastasis (TNM) staging were also scored. The non-tumorous tissues were 5 cm from the edge of the tumor, contained no obvious tumor cells and were also evaluated by the pathologist. All of the experiments were approved by the Research Ethics Committee of the Provincial Hospital Affiliated to Shandong University and written informed consent was obtained from all patients.
Cell lines
The human colorectal cancer cell lines HT-29, SW1116, SW480, and COLO205 were obtained from American Type Culture Collection (Manassas, VA). All of the cell lines were grown and maintained in RPMI Medium 1640 (Invitrogen) Supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 mg/ml streptomycin (Invitrogen, Shanghai, China) at 37°C in a 5% CO2 atmosphere.
qRT-PCR analysis
Purified total RNA was obtained from the microdissected cells, total RNA was extracted using Trizol solution. Reverse transcription (RT) was performed in a 20-μL reaction according to the manufacturer’s recommendations (Qiagen). Real-time qRT-PCR analyses were performed using primers as follows: 5’-CAAAGCTGGTCCTGTGCCTG-3’; 5’-ATTGAGTGTGGCATCACCTCC-3’. Transcript expression levels were determined by quantifying the intensity of the PCR product normalized to U6 expression. Quantitative measurement of mRNA levels was performed using the ABI Prism 7000 (Applied Biosystems, Foster City, USA). These data were analyzed by using the comparative Ct method.
Construction and transfection of expression vector for FTX
The FTX sequences were synthesized and subcloned into the pcDNA3.1 (Invitrogen, Shanghai, China) vector. The pcDNA constructs or the empty vector were transfected into CRC cells cultured on six-well plates according to the manufacturer’s instructions. The empty vector was used as the control. The expression level of FTX was detected by qRT-PCR. SW480 CRC cell lines were used for the overexpression studies. We obtained stably transfected clones by G418 selection (Promega). A stable transfectant of the pcDNA3.1 empty vector was used as a control. For transfection, complexes of Lipofectamine 2000 (Invitrogen Corp, Carlsbad, USA) and one of the plasmids mentioned above was prepared according to the manufacturer’s instructions, and these complexes were directly mixed with cells in 24-well cell culture plates at a density of 4 × 104 cells per well. The level of FTX expression after transfection was assayed by real-time PCR.
Cell growth and soft agar colony formation assay
CRC cells (2 × 103 cells) were incubated with 100 μL of culture medium in 96-multiwell plates for one day at 37°C in 5% CO2. The cells were transfected with the plasmid for 24, 48, 72, 96, and 120 hours. Cell number was assessed using the cell counting kit-8 (CCK-8) (Dojindo, Japan). Briefly, CCK-8 (10 μL) was added to each well. After 1 h of incubation at 37°C, absorbance at 450 nm was measured using the ARVO MX plate reader (PerkinElmer, Massachusetts, USA). The number of cells was determined by the relative absorbance at 450 nm.
CRC cells were trypsinized to single-cell suspensions of 3 × 103 cells and then were plated in six-well plates in complete culture medium containing 0.3% agar layered on top of 0.6% agar. The plates were incubated at 37°C in the presence of 5% CO2 for 16 days. Colonies containing at least 50 cells were scored. The data are presented as the mean ± standard deviation of five randomly scored fields.
Invasion and migration assays
Cell invasion and migration assays were performed using SW480 cells. Cell culture was performed in transwell chambers (Corning, NY, USA). For the invasion assay, the insert membranes were coated with diluted Matrigel (San Jose, CA, USA). Cells (1 × 105) were added to the upper chamber and were cultured for 48 h. For the migration assay, the insert membranes were not coated with Matrigel but were cultured under the same conditions. Finally, the insert membranes were cut and stained with crystal violet (0.04% in water; 100 ml), and the migrated cells were counted under an inverted microscope and were photographed.
Statistical analysis
Statistical analysis was performed using SPSS15.0 software (SPSS Inc, USA). Data are expressed as the mean ± standard deviation from at least three separate experiments. Associations of long non-coding RNA FTX with clinical, and pathologic were determined using the Chi-square test or Cox proportional hazards regression model, as appropriate. Kaplan-Meier survival curves were constructed to show survival differences according to long non-coding RNA FTX expression. The survival time for either overall or time to recurrence was calculated as the time from surgery until the occurrence of death and relapse, respectively. The Mann-Whitney U test was used for comparing differences in cell counts and migration. Spearman correlation coefficients were calculated for correlation analysis. A value of P < 0.05 was considered statistically significant.
Results
Long non-coding RNA FTX was significantly upregulated in CRC tissues
To determine the status of long non-coding RNA FTX expression in CRC, a qRT-PCR assay was performed to determine the expression of long non-coding RNA FTX in 35 pairs of CRC and corresponding noncancerous tissue samples. The expression levels of long non-coding RNA FTX was 18.85±11.62 and 11.62±10.21, respectively in CRC and corresponding noncancerous tissue samples. Results showed that the relative level of long non-coding RNA FTX expression in CRC tissues was significantly upregulated, compared with that in corresponding noncancerous tissues (Figure 1, P < 0.05).
Figure 1.
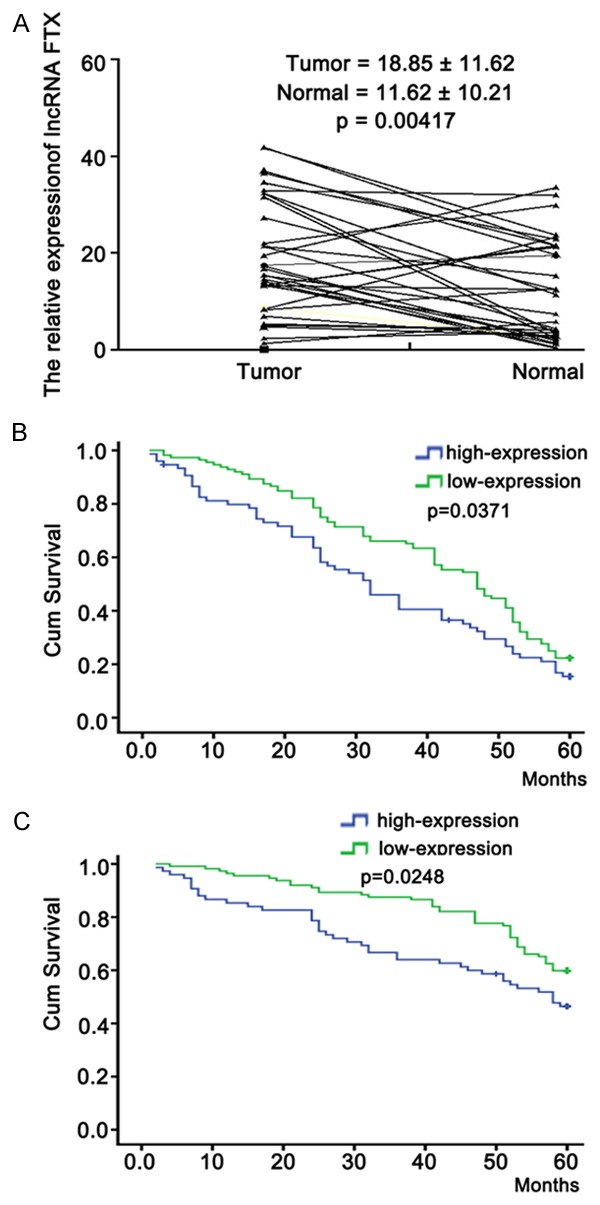
qRT-PCR detection of long non-coding RNA FTX expression in CRC tissues. A. qRT-PCR detection of long non-coding RNA FTX expression in 35 pairs of CRC (T) and corresponding noncancerous colorectal tissues (N). The mean level of long non-coding RNA FTX in 35 pairs of colorectal cancer and corresponding noncancerous tissues was 18.85±11.62 and 11.62±10.21, respectively. The relative level of long non-coding RNA FTX expression in BC tissues was significantly upregulated, compared with that in corresponding noncancerous tissues. B. Kaplan-Meier overall survival curves of patients with CRC tissues according to the level of long non-coding RNA FTX. The overall survival rate of the long non-coding RNA FTX low-expression group was significantly higher than that of the high expression group after 60 months. C. Time to recurrence of the long non-coding RNA FTX high-expression group was significantly shorter than that of the low expression group. Each experiment was performed at least in triplicate.
Correlation of long non-coding RNA FTX expression with clinicopathologic factors of CRC patients
To further analyze the clinicopathological significance of long non-coding RNA FTX in CRC, the expression of long non-coding RNA FTX was determined in another 187 cases of CRC tissues. The median value of long non-coding RNA FTX in all CRC tissues was 2.00 and was used as a cutoff value, and all patients were divided into two groups high-long non-coding RNA FTX expression group (≥2.00; n = 75) and low-long non-coding RNA FTX expression group (< 2.00; n = 112). Table 1 showed the correlation between long non-coding RNA FTX expression and clinicopathological factors of CRC patients. By statistical analyses, low-long non-coding RNA FTX expression was found to be significantly correlated with differentiation grade, lymph vascular invasion, and Clinical stage (P = 0.042, 0.0361, 0.0057, and 0.031, resp.). However, there were no significant correlations between long non-coding RNA FTX expression and other clinicopathological factors of patients, such as age, sex, tumour histology, perineural invasion, tumour site, and lymph node metastasis (P = 2.85, 0.57, 1.85, 0.076, and 0.83, resp.). These data indicated that downregulation of long non-coding RNA FTX might play a critical role in CRC progression.
Table 1.
The correlations between long non-coding RNA FTX expression and clinicopathological factors of CRC patients
| Factors | High expression (n = 75) | Low expression (n = 112) | Relative FTX expression | P value | |
|---|---|---|---|---|---|
|
| |||||
| High expression | low expression | ||||
| Age (years) | 2.85 | ||||
| ≤50 | 34 | 64 | 2.54±0.21 | 0.84±1.09 | |
| >50 | 41 | 48 | 2.75±1.52 | 1.31±2.16 | |
| Sex | 0.327 | ||||
| Male | 14 | 31 | 5.62±0.35 | 1.34±2.4 | |
| Female | 28 | 57 | 8.21±2.51 | 1.84±1.04 | |
| Differentiation grade | 0.042* | ||||
| Well | 9 | 24 | 3.26±1.36 | 1.52±0.17 | |
| Moderate | 28 | 37 | 3.14±2.04 | 0.73±1.62 | |
| Poor | 38 | 51 | 15.26±0.26 | 1.68±0.04 | |
| Tumour histology | 0.361 | ||||
| Adenocarcinoma | 41 | 72 | 2.25±0.16 | 1.57±0.74 | |
| Mucinous adenocarcinoma | 34 | 40 | 3.17±3.16 | 0.73±2.58 | |
| Lymph vascular invasion | 0.0057* | ||||
| Absence | 47 | 78 | 21.16±2.05 | 1.17±0.51 | |
| Presence | 28 | 34 | 16.13±2.15 | 0.26±0.06 | |
| Perineural invasion | 0.258 | ||||
| Absence | 46 | 76 | 11.24±1.03 | 1.85±1.05 | |
| Presence | 29 | 36 | 13.16±5.12 | 1.27±0.7 | |
| Tumour site | 0.83 | ||||
| Rectum | 35 | 47 | 3.72±1.52 | 1.36±0.03 | |
| Colon | 40 | 65 | 2.15±0.05 | 1.82±1.21 | |
| Lymph node metastasis | 1.85 | ||||
| Absent | 29 | 41 | 6.26±0.15 | 0.62±0.8 | |
| Present | 46 | 71 | 2.05±4.16 | 1.32±0.27 | |
| Clinical stage | 0.031* | ||||
| I + II | 31 | 62 | 2.73±0.52 | 1.52±1.49 | |
| III | 44 | 60 | 14.83±1.26 | 0.75±0.35 | |
P < 0.05.
Correlation of long non-coding RNA FTX expression with prognosis of CRC patients
Overall survival curves and time to recurrence (TTR) curves were plotted according to the expression level of long non-coding RNA FTX by the Kaplan-Meier method. As in Figure 1B and 1C, Kaplan-Meier survival analysis indicated that high long non-coding RNA FTX group had a remarkable shorter overall and time to recurrence than low long non-coding RNA FTX group (P < 0.05). The median survival months were 33.32±2.78 and 41.60±1.63, respectively (Figure 1B, P < 0.05).
Correlation between long non-coding RNA FTX expression and prognosis of CRC patients
Univariate and multivariate analyses of factors related to prognosis of CRC patients were shown in Table 2. Univariate regression analysis indicated that status of long non-coding RNA FTX expression (relative risk (RR), 1.261; 95% confidence interval (CI), 1.593-2.478; P = 0.0007) was significantly correlated with OS of BC patients, along with differentiation grade (P = 0.018), and clinical stage (P = 0.038). Furthermore, multivariate regression analysis indicated that status of long non-coding RNA FTX expression (RR, 2.371; 95% CI, 1.42-2.739; P = 0.00041) was an independent predictor for the prediction of OS, as well as differentiation grade (RR, 1.273; 95% CI, 2.83-4.278; P = 0.029) and clinical stage (RR, 1.05; 95% CI, 1.581-2.372; P = 0.018).
Table 2.
Univariate and multivariate analysis for OS of CRC patients
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
| ||||
| RR (95% CI) | P value | RR (95% CI) | P value | |
| Age (year) | 0.625 (0.462-2.512) | 1.57 | ||
| Sex | 0.452 (0.721-1.835) | 0.76 | ||
| Tumour site | 0.937 (0.682-2.317) | 2.65 | ||
| Tumour histology | 0.720 (0.839-3.371) | 3.06 | ||
| Clinical stage | 2.326 (1.417-3.282) | 0.038* | 1.05 (1.581-2.372) | 0.018* |
| Lymph node metastasis | 0.472 (0.621-2.371) | 1.75 | ||
| long non-coding RNA FTX expression | 1.261 (1.593-2.478) | 0.0007* | 2.371 (1.42-2.739) | 0.00041* |
| Lymph vascular invasion | 0.571 (0.489-1.804) | 0.47 | ||
| Differentiation grade | 1.427 (1.904-2.074) | 0.018* | 1.273 (2.83-4.278) | 0.029* |
| Perineural invasion | 1.581 (1.342-2.581) | 0.053 | ||
P < 0.05.
HR: hazard ratio; 95% CI: 95% confidence interval.
Upregulation of long non-coding RNA FTX promotes growth and reduces colony formation
At first, we screened long non-coding RNA FTX expression in four CRC cell lines by real-time PCR. Different long non-coding RNA FTX expressing levels were found in CRC cell lines (Figure 2A). To analyze the effects of long non-coding RNA FTX expression on malignant phenotypes of CRC cells, long non-coding RNA FTX vector (SW480/FTX) or control vector (SW480/C) was stably transfected into CRC cells (SW480), which was named SW480/FTX (or SW480/C). After transfection, we examined expression of long non-coding RNA FTX on SW480 cells by real-time PCR. The expressing level of long non-coding RNA FTX was up-regulated (P < 0.05, Figure 2B) in SW480 cells. Results from CCK-8 assays indicated that upregulation of long non-coding RNA FTX could significantly promote growth of CRC cells (P < 0.01, Figure 2C). The capacity of colony formation in SW480/FTX cells was significantly increased, in comparison with that in the control cells (P < 0.05, Figure 2D). These data showed that upregulation of long non-coding RNA FTX could promote growth and reduced the capacity of colony formation in CRC cells.
Figure 2.
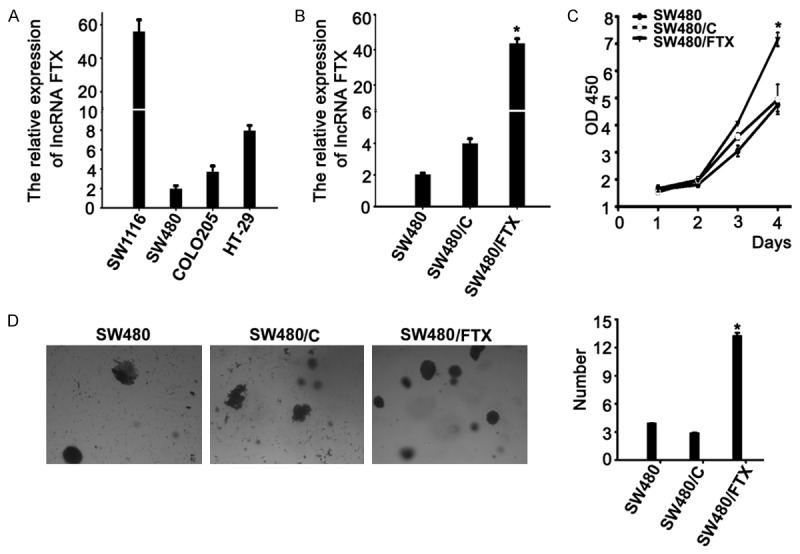
Effect of long non-coding RNA FTX transfection on SW480 cells. A. The different expression levels of long non-coding RNA FTX in four colorectal cancer cell lines. B. Expression of long non-coding RNA FTX was upregulated in SW480 cells upon FTX vector transfection relative to the control. C. Overexpression of long non-coding RNA FTX promote to cell growth as determined by the CCK-8 assay. D. Colony formation rates were significantly different between FTX-transfected cells and controls in SW480 cells. *P < 0.05. Each bar represents the mean value ± standard deviation from three independent experiments.
Upregulation of long non-coding RNA FTX significantly promotes migration and invasion of CRC cells
Next, we explored the effects of long non-coding RNA FTX expression on migration and invasion of CRC cells. Migration assay indicated that the migration of SW480/FTX cells was markedly increased in comparison with that of SW480/C cells (P < 0.05, Figure 3A). Similarly, matrigel invasion assays indicated that the invasion of SW480/FTX cells was significantly reduced in comparison with that of SW480/NC cells (P < 0.05, Figure 3B). Thus, upregulation of long non-coding RNA FTX could increase the capacity of migration and invasion in CRC cells.
Figure 3.
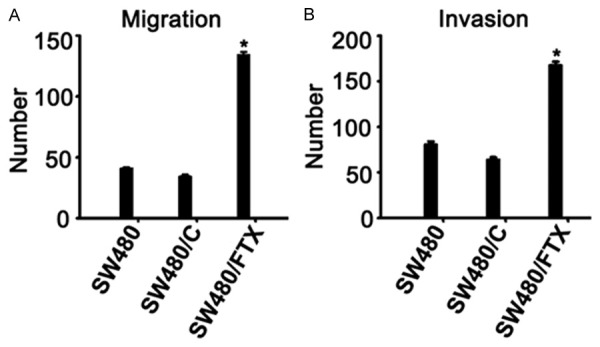
Upregulation of long non-coding RNA FTX significantly promotes migration and invasion of CRC cells. A. Cell migratory ability was promoted by migration assays in SW480 cells. B. Cell invasive ability was increased in the transwell assay in SW480 cells. *P < 0.05. Each bar represents the mean value ± standard deviation from three independent experiments.
Discussion
The conventional view of gene regulation in biology has centered on protein-coding genes until the discovery of thousands of lncRNAs. Numerous reports of dysregulated lncRNA expression across numerous cancer types suggest that abnormal lncRNA expression may be a major contributor to tumorigenesis. The aberrant expressions of specific lncRNAs in cancer could mark the spectrum of disease progression and these lnRNAs may serve as independent biomarkers for diagnosis and prognosis [28,29]. More recently, lncRNAs have been implicated in CRC pathogenesis [30,31]. However, the prognostic values of lncRNAs in CRC have not yet been investigated. A better understanding of the genetic and molecular characteristics of CRC is urgently needed for early diagnosis, choice of the appropriate treatment and an improved prognosis for patients with CRC. Recent studies have demonstrated that a class of non-protein-coding RNAs (ncRNAs), which are known as long non-coding RNAs (lncRNAs), participates in cell fate determination and human disease pathogenesis [7,9,18,32]. An increasing body of evidence has suggested that lncRNAs are key regulators in several biological processes and are increasingly recognized as diagnostic or prognostic cancer biomarkers, including in CRC. For example, Ma Y et al identified colorectal cancer-associated lncRNA (CCAL) as a key regulator of CRC progression. Patients whose tumours had high CCAL expression had a shorter overall survival and a worse response to adjuvant chemotherapy than patients whose tumours had low CCAL expression. CCAL promoted CRC progression by targeting activator protein 2α (AP-2α), which in turn activated Wnt/β-catenin pathway. CCAL induced multidrug resistance (MDR) through activating Wnt/β-catenin signalling by suppressing AP-2α and further upregulating MDR1/P-gp expression. In addition, we found that histone H3 methylation and deacetylases contributed to the upregulation of CCAL in CRC [33]. BRAF activated non-coding RNA (BANCR), a long non-coding RNA (lncRNA) expression was significantly down-regulated in colorectal cancer tissues compared with normal tissues, and overexpression of BANCR suppressed colorectal cancer cell growth in vitro and in vivo. We also determined that pCDNA-BANCR-mediated colorectal cancer cell proliferation was associated with induction of G0/G1 cell-cycle arrest and apoptosis enhancement through regulation of p21, and its effects were most likely posttranscriptional [25].
LncRNAs FTX is located upstream of XIST, within the X-inactivation center (XIC). It produces a spliced long non-coding RNA that is thought to positively regulate the expression of XIST, which is essential for the initiation and spread of X-inactivation [34]. Up to date, there is no report of lncRNAs FTX on CRC. In our study, we first found that lncRNAs FTX was significantly unregulated in CRC tissues compared with adjacent normal tissues. Our data indicated that a low expression of this lncRNAs is correlated with differentiation grade, lymph vascular invasion, and clinical stage. However, there were no significant correlations between long non-coding RNA FTX expression and other clinicopathological factors of patients, such as age, sex, tumour histology, perineural invasion, tumour site, and lymph node metastasis, indicating that lncRNA FTX may be a promising prognostic biomarker for CRC patients. Overall survival curves and time to recurrence (TTR) curves were plotted according to the expression level of long non-coding RNA FTX by the Kaplan-Meier method. Kaplan-Meier survival analysis indicated that high long non-coding RNA FTX group had a remarkable shorter overall and time to recurrence than low long non-coding RNA FTX group. Multivariate regression analysis indicated that status of long non-coding RNA FTX expression was an independent predictor for the prediction of OS, as well as differentiation grade and clinical stage. Additionally, we speculated that FTX may play a significant role in tumor biology. First, we chose representative CRC cell lines and investigated FTX expression in CRC cell lines. We found that the HT-29 was high expression, whereas there was a low expression in SW480 cell lines. To further determine the roles of long non-coding RNA FTX in CRC development, we analyzed the effects of long non-coding RNA FTX on malignant phenotypes of CRC cells. From this study, restoring long non-coding RNA FTX expression in human CRC cells, we then determined whether long non-coding RNA FTX expression influences tumor-like characteristics such as proliferation, colony formation, migration and invasion. Indeed, the upregulation of long non-coding RNA FTX promotes cell proliferation and increases colony formation in SW480 cell lines, whereas the ectopic expression of long non-coding RNA FTX significantly enhances cell migration and invasion in SW480 cell lines.
Taken together, we have found that long non-coding RNA FTX is markedly up-regulated in human CRC. Low-long non-coding RNA FTX expression was significantly correlated with differentiation grade, lymph vascular invasion, and clinical stage. Multivariate regression analysis indicated that status of long non-coding RNA FTX expression was an independent predictor for the prediction of OS, as well as differentiation grade and clinical stage. Additionally, upregulation of Low-long non-coding RNA FTX could significantly promote growth, increase colony formation, and enhance migration and invasion of CRC cells. Thus, long non-coding RNA FTX could be a potential prognostic biomarker and therapeutic target for CRC. It implied that long non-coding RNA cannot be overlooked as a class of molecules on regulating biological functions and on human colorectal cancer therapy. Our data enhance understanding the functions of long non-coding RNAs on CRC. Of course, this study has several limits. First, as the number of patients in this study is small, a larger case population is needed to confirm the prognostic value of Low-long non-coding RNA FTX expression in CRC. Second, further studies are needed to identify other undefined long non-coding RNA FTX targets, which may also affect cellular phenotypes at other levels.
Acknowledgements
This work was supported in part by grants from the National Youthful Science Foundation of China (81101858), the China Postdoctoral Science Foundation (2014M560561), the Youthful Science Foundation of Shandong Province of China (BS2010YY060). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Stiegelbauer V, Perakis S, Deutsch A, Ling H, Gerger A, Pichler M. MicroRNAs as novel predictive biomarkers and therapeutic targets in colorectal cancer. World J Gastroenterol. 2014;20:11727–35. doi: 10.3748/wjg.v20.i33.11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 4.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–23. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 5.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152–61. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mourtada-Maarabouni M, Hasan AM, Farzaneh F, Williams GT. Inhibition of human T-cell proliferation by mammalian target of rapamycin (mTOR) antagonists requires noncoding RNA growth-arrest-specific transcript 5 (GAS5) Mol Pharmacol. 2010;78:19–28. doi: 10.1124/mol.110.064055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pickard MR, Williams GT. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: implications for chemotherapy. Breast Cancer Res Treat. 2014;145:359–70. doi: 10.1007/s10549-014-2974-y. [DOI] [PubMed] [Google Scholar]
- 10.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–6. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Molecular Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 12.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–70. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–30. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 15.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 16.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, Triller A, Spector DL, Bessis A. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–93. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–25. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su X, Malouf GG, Chen Y, Zhang J, Yao H, Valero V, Weinstein JN, Spano JP, Meric-Bernstam F, Khayat D, Esteva FJ. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014;5:9864–76. doi: 10.18632/oncotarget.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–74. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–42. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 22.Fitzpatrick GV, Soloway PD, Higgins MJ. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet. 2002;32:426–31. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- 23.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–7. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 24.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–63. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 25.Shi Y, Liu Y, Wang J, Jie D, Yun T, Li W, Yan L, Wang K, Feng J. Downregulated Long Noncoding RNA BANCR Promotes the Proliferation of Colorectal Cancer Cells via Downregualtion of p21 Expression. PLoS One. 2015;10:e0122679. doi: 10.1371/journal.pone.0122679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–3. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 27.Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, Spector DL. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–23. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Sun S, Pu JK, Tsang AC, Lee D, Man VO, Lui WM, Wong ST, Leung GK. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Zhang XQ, Sun S, Lam KF, Kiang KM, Pu JK, Ho AS, Lui WM, Fung CF, Wong TS, Leung GK. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol Dis. 2013;58:123–31. doi: 10.1016/j.nbd.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 31.Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3’ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169–75. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- 32.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, Yang Z, Liu W, Zhang H, Chen N, Wang H, Wang H, Qin H. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2alpha. Gut. 2015 doi: 10.1136/gutjnl-2014-308392. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 34.Zhao Q, Li T, Qi J, Liu J, Qin C. The miR-545/374a cluster encoded in the Ftx lncRNA is overexpressed in HBV-related hepatocellular carcinoma and promotes tumorigenesis and tumor progression. PLoS One. 2014;9:e109782. doi: 10.1371/journal.pone.0109782. [DOI] [PMC free article] [PubMed] [Google Scholar]


