Abstract
Breast cancer is one of the most common malignancies in women. Current treatment of breast cancer is mainly based on clinicopathological characteristics, and is not sufficiently customized for individual cases. The concept of genotyping in breast cancer was first proposed in 2001. Five major genotypes of breast cancer have been identified and their study has given rise to a new field of research. In our study, we investigated the expression of 13 chemokines and chemokine receptors, which play important roles in inflammation and tumor progression, in five breast cancer genotypes. Using immunohistochemistry, we found that CCL2 expression was significantly different between the different breast cancer genotypes and was negatively associated with estrogen and progesterone receptor expression. Kaplan Meier analysis showed that a low expression of CCL2 was associated with better outcome in breast cancer patients. Enzyme-linked immunosorbent assay results revealed that CCL2 expression in different breast cancer genotype cell line suspensions was significantly different.
Keywords: Breast cancer, genotype, CCL2, chemokines
Introduction
Breast cancer is a heterogeneous disease, comprising a number of distinct biological entities associated with specific morphological and immunohistochemical features and clinical behavior. For many decades, invasive breast carcinomas were only classified according to histological type, grade, and expression of hormone receptors, which does not adequately reflect the disease process. In 2001 [1], microarray-based expression analysis uncovered five genotypes of breast cancer: Luminal A subtype, Luminal B subtype, human epidermal growth factor receptor-2 (HER2) subtype, normal breast-like subtype, and basal-like subtype, which provided a new direction for research [2-4] (Table 1).
Table 1.
Five genotypes of breast cancer
| Genotypes | Characteristics | Other characteristics* |
|---|---|---|
| Basal-like subtype | ER (-), PR (-), HER2 (-) | CK5/6 (+), EGFR (+), BRCA1 mutation, 82% TP53 mutation [2] |
| HER2 subtype | HER2 (+), ER (-), PR (-) | GRB7 (+), TRAP100 [3] high expression, 71% TP53 mutation |
| Luminal A subtype | ER (+) or PR (+), HER2 (-) | GATA3 (+), FOXA1 [4] high expression |
| Luminal B subtype | ER (+) or PR (+), HER2 (+) | CGH (+), LAPTMB4 (+), NSEP-1F high expression, 40% TP53 mutation |
| Normal breast-like subtype | ER (-), PR (-), HER2 (-) | CK5/6 (-), EGFR (-), 33% TP53 mutation |
In our study, we detected CK5/6, EGFR, TP53 by IHC to identify the tissue belonging to Basal-like subtype or Normal breast-like subtype.
Chemokines comprise a superfamily of at least 46 cytokines that were initially thought to be involved in inflammation, based on their ability to bind to 18 to 22 G protein-coupled receptors to induce the directed migration of leukocytes. In addition to mediating cellular migration, chemokines and chemokine receptors have been shown to be involved in processes of malignant progression, such as proliferation, survival, adhesion, invasion, and regulation of circulating chemokine levels.
CCL2 was one of the first chemokines to be discovered, and has been extensively studied. It is secreted by a variety of cells, including fibroblasts, endothelial cells, and monocytes [5]. Some tumor cell lines such as breast cancer and prostate cancer cell lines also express the CCL2 protein. CCL2 combines not only with CCR2, but also with CCR4 and CCR10. However, CCR2 is the major receptor for CCL2, and its binding activates a series of downstream biological effects. CCR2 is mainly expressed on the cell membranes of monocytes, leukocytes, and dendritic cells.
The expression of many chemokines can be detected at the protein level in primary breast cancer tissue. However, differences in the expression of chemokines and chemokine receptors between different breast cancer genotypes have not been widely reported. In this study, we selected 205 patients with breast cancer surgically treated between 2002 and 2007, and tested the surgical specimens for the expression of 13 chemokines and chemokine receptors (CCL2, CCL5, CXCL5, CCL19, CCL3, CCL21, CXCL12, CXCL1, CXCL8, CCR5, CCR25, CCR7, and CXCR4) by immunohistochemical staining. We found that the expression of CCL2 was significantly different between the five breast cancer genotypes and was associated with the expression of estrogen receptor (ER) and progesterone receptor (PR). Analysis of follow-up data showed a statistically significant difference in overall survival between patients showing high expression of CCL2 and patients with low expression of CCL2.
Materials and methods
Regents and cell lines
Dulbecco’s modified Eagle’s medium (DMEM), Ham’s F-12 medium (F12), and fetal bovine serum (FBS) were obtained from Life Technologies, Inc (Carlsbad, CA, USA). Iscove’s modified Eagle’s medium (IMEM) (phenol red-free) was purchased from Biofluids (Rockville, MD, USA). Human breast cancer cell lines, BT474 (Luminal B subtype), MCF-7 (Luminal A subtype), SKBR3 (HER2 subtype), and MDA-MB-231 (basal-like subtype) were purchased from ATCC. Normal breast-like subtype cells were routinely cultured in DMEM/F12 medium supplemented with 10% FBS. The cultures were incubated at 37°C in humidified 5% CO2.
Evaluation of immunohistochemistry (IHC)
Two hundred and five breast cancer patients (age range, 27-82 years) surgically treated at the Department of Breast Surgery, The International Peace Maternity & Child Health Hospital, Shanghai, China, were enrolled in this study. Tumors from all 205 patients were classified into five genotypes according to ER and PR status, HER2 expression, and Ki67, CK5/6, and EGFR immunochemistry (IHC) results, evaluated by pathologists following surgery. Patient follow-up ranged from 0 to 155 months. The study protocol was approved by the Ethics Committee of The International Peace Maternity & Child Health Hospital, Shanghai, China. Written informed consent was obtained from all patients. Patient details are shown in Table 2.
Table 2.
Patient’s clinical characteristics
| Case | 205 |
| Median age | 52.68±9.64 |
| Status of menses | |
| Premenopause | 97 |
| Menopause | 108 |
| Tumor size (cm) | 2.25±0.96 (0.50-7.00) |
| Pathological type | |
| Infiltrating ductal | 166 |
| Carcinoma | 39 |
| Others | |
| Nuclear grade | |
| I | 19 |
| II | 147 |
| III | 39 |
IHC, carried out for 13 chemokines and chemokine receptors, was evaluated microscopically (at 20× and 40× magnification) by two independent investigators (pathologists) who were blinded to patient outcome. IHC expression of the 13 chemokines and chemokine receptors was scored as 0, 1, 2, or 3 based on staining intensity and percentage of positive cells. Positive and negative controls were also evaluated for each IHC marker.
IHC for CCL2 shows a membranous or cytoplasmic staining pattern. To evaluate if CCL2 expression is associated with patient outcome, patients were divided into two groups: those with high expression of CCL2 (at least 25% of the cytoplasmic cells with moderate to high staining intensity) and those with low expression of CCL2 (weak staining or staining in less than 25% of the cells).
ELISA
The four cell lines (BT474, MCF-7, SKBR3, and MDA-MB-231) express many chemokines and chemokine receptors. However, based on our IHC results on 205 tumors, we only used an ELISA to investigate if there were significant differences in concentrations of CCL2 in suspension between the different breast cancer genotype cell lines.
Cells were grown for 24 h, 36 h, and 48 h before the supernatant was collected. The human CCL2 ELISA kit (eBioscience, San Diego, CA, USA) was used to measure CCL2 concentration, following the manufacturer’s instructions.
Statistical analysis
Results were reported as the mean ± standard deviation (SD) or the mean ± standard error (SE). One-way ANOVA followed by post hoc Tukey’s test, correlation analysis, and Kaplan Meier analysis were used to determine the statistical significance of differences between experimental groups.
Results
In order to study the differences in expression of chemokines and their receptors between five breast cancer genotypes in human breast cancer tissue, we examined the expression of 13 chemokines and chemokine receptors by IHC in 205 cases of breast cancer. We found that CCL2 expression (Figure 1) was significantly different between the different breast cancer genotypes (P=0.012, Table 3), while there was no statistical difference in CCL5 expression between genotypes (P=0.056, Table 3).
Figure 1.

The expression of CCL2 in 5 genotype breast cancer: A. HER2 subtype; B. Basal-like subtype; C. Luminal A subtype; D. Luminal B subtype; E. Normal breast-like subtype.
Table 3.
Thirteen chemokines and receptors expression in 5 genotype breast cancer tissue by IHC staining, CCL2 expression in different genotype is statistically significant
| Sum of squares | Mean square | F | Significance | ||
|---|---|---|---|---|---|
| CCL2 | Among groups | 8.238 | 2.746 | 3.336 | 0.023 |
| *Genotypes | 5.476 | 5.476 | 6.653 | 0.012* | |
| Within groups | 71.609 | 0.823 | |||
| CCL5 | Among groups | 2.836 | 0.709 | 1.862 | 0.125 |
| *Genotypes | 1.433 | 1.433 | 3.764 | 0.056 | |
| Within groups | 31.607 | 0.381 | |||
| CXCL5 | Among groups | 9.579 | 2.395 | 4.402 | 0.003 |
| *Genotypes | 0.404 | 0.404 | 0.742 | 0.392 | |
| Within groups | 46.244 | 0.544 | |||
| CCL19 | Among groups | 0.772 | 0.193 | 0.329 | 0.857 |
| *Genotypes | 0.356 | 0.356 | 0.607 | 0.438 | |
| Within groups | 50.414 | 0.586 | |||
| CCL3 | Among groups | 3.192 | 0.798 | 1.620 | 0.177 |
| *Genotypes | 1.105 | 1.105 | 2.244 | 0.138 | |
| Within groups | 40.888 | 0.493 | |||
| CXCL8 | Among groups | 5.477 | 1.369 | 3.116 | 0.019 |
| *Genotypes | 0.002 | 0.002 | 0.004 | 0.951 | |
| Within groups | 36.905 | 0.439 | |||
| CCR7 | Among groups | 1.683 | 0.421 | 1.176 | 0.327 |
| *Genotypes | 0.310 | 0.310 | 0.868 | 0.354 | |
| Within groups | 31.478 | 0.358 | |||
| CCL21 | Among groups | 3.303 | 0.826 | 2.066 | 0.093 |
| *Genotypes | 0.257 | 0.257 | 0.643 | 0.425 | |
| Within groups | 35.574 | 0.400 | |||
| CCR25 | Among groups | 3.171 | 1.057 | 1.676 | 0.179 |
| *Genotypes | 1.619 | 1.619 | 2.564 | 0.113 | |
| Within groups | 52.347 | 0.631 | |||
| CXCL1 | Among groups | 2.399 | 0.600 | 0.971 | 0.428 |
| *Genotypes | 1.297 | 1.297 | 2.100 | 0.151 | |
| Within groups | 52.501 | 0.618 | |||
| CXCR4 | Among groups | 2.265 | 0.566 | 0.897 | 0.469 |
| *Genotypes | 0.877 | 0.877 | 1.389 | 0.242 | |
| Within groups | 54.898 | 0.631 | |||
| CXCL12 | Among groups | 3.287 | 0.822 | 0.862 | 0.490 |
| *Genotypes | 0.420 | 0.420 | 0.440 | 0.509 | |
| Within groups | 82.919 | 0.953 | |||
| CCR5 | Among groups | 2.167 | 0.542 | 1.076 | 0.374 |
| *Genotypes | 0.269 | 0.269 | 0.535 | 0.467 | |
| Within groups | 42.304 | 0.504 |
P<0.05.
CCL2 expression in tumors was given a staining score of 0, 1, 2, or 3, and the mean staining scores of the five genotypes were as follows: HER2 subtype, 1.44; basal-like subtype, 1.2; Luminal B subtype, 0.76; Luminal A subtype, 0.64; and normal breast-like subtype, 0.60. One-way ANOVA followed by post hoc Tukey’s test revealed that mean expressions of CCL2 in 5 genotype breast cancer tissue were significantly different (Tables 3 and 4).
Table 4.
Means of CCL2 expression in 5 genotype breast cancer tissue (One-way ANOVA followed by post-hoc Tukey’s test)
| Genotypes | Means | N | Standard deviation | Min | Max |
|---|---|---|---|---|---|
| HER2 subtype | 1.44 | 31 | 1.094 | 0 | 3 |
| Basal-like subtype | 1.20 | 34 | 1.095 | 0 | 3 |
| Luminal B subtype | 0.76 | 53 | 0.723 | 0 | 3 |
| Luminal A subtype | 0.64 | 67 | 0.908 | 0 | 3 |
| Normal breast-like subtype | 0.60 | 20 | 0.895 | 0 | 3 |
| Total | 0.83 | 205 | 0.931 | 0 | 3 |
Correlation analysis showed that CCL2 expression was significantly associated with the expression of both ER and PR in breast tumor tissues, and more strongly associated with the expression of PR (correlation coefficient =-0.299, P=0.002) (Table 5).
Table 5.
CCL2 expression is significantly associated with ER and PR expression in breast cancer tissue
| ER | PR | HER2 | CCL2 | ||
|---|---|---|---|---|---|
| ER | Pearson correlation significance (2-tailed) | 1 | 0.731** | -0.153 | -0.228* |
| 0.000 | 0.088 | 0.018 | |||
| N | 205 | 205 | 205 | 205 | |
| PR | Pearson correlation significance (2-tailed) | 0.731** | 1 | -0.061 | -0.299** |
| 0.000 | 0.497 | 0.002 | |||
| N | 205 | 205 | 205 | 205 | |
| HER2 | Pearson correlation significance (2-tailed) | -0.153 | -0.061 | 1 | 0.170 |
| 0.088 | 0.497 | 0.105 | |||
| N | 205 | 205 | 205 | 205 | |
| CCL2 | Pearson correlation significance (2-tailed) | -0.228* | -0.299** | 0.170 | 1 |
| 0.018 | 0.002 | 0.105 | |||
| N | 205 | 205 | 205 | 205 |
P<0.01;
P<0.05.
Kaplan Meier analysis showed that the overall survival between the groups with high CCL2 expression and low CCL2 expression was significantly different (P=0.002, Figure 2A). The differences between overall survival of five different genotypes patients with high CCL2 expression were statistically significant (P=0.000, Figure 2B), and the differences between overall survival of five different genotypes patients with low CCL2 expression were also statistically significant (P=0.000, Figure 2C).
Figure 2.
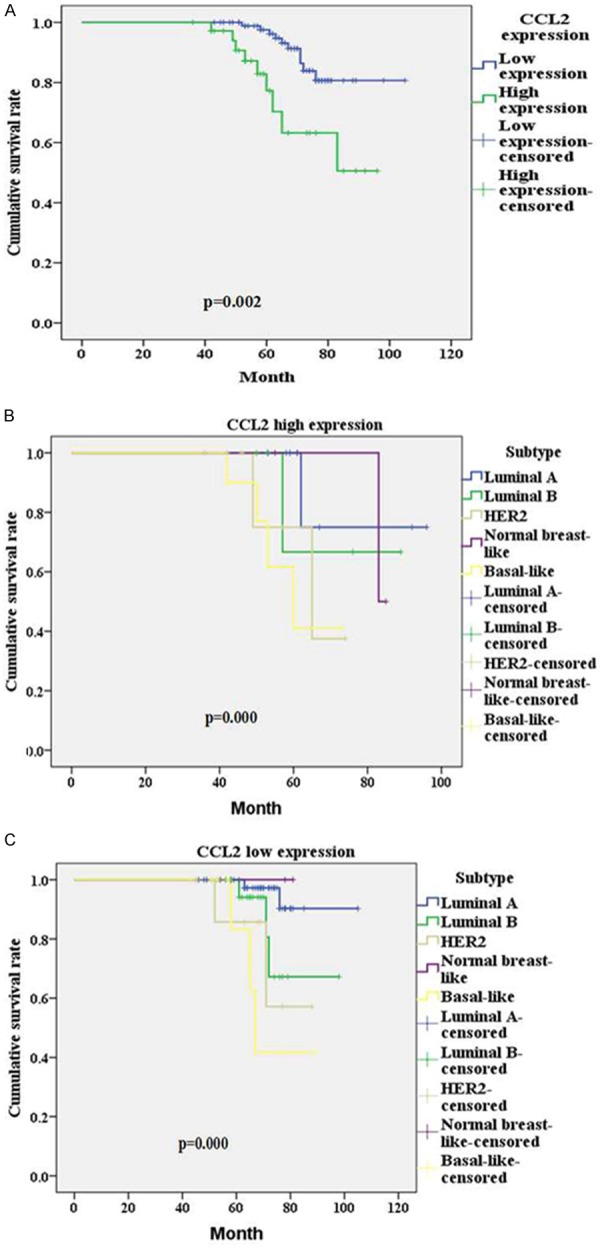
Kaplan Meier analysis: A. The overall survival of CCL2 high expressing group and CCL2 low expressing group; B. The overall survival of 5 subtype patients in CCL2 high expressing group; C. The overall survival of 5 subtype patients in CCL2 low expressing group.
In order to study the expression of CCL2 in the different breast cancer genotypes in vitro, we detected the CCL2 concentration in suspension of different genotype breast cancer cell lines after 24 h incubation for 3 times by using ELISA. The column bar graph through one-way ANVOA analysis indicated that the difference between individual cell lines was statistically significant (P=0.000, Figure 3). The means of CCL2 concentration in MCF7, BT474, MDA-MB-468, and SKBR3 were 1310.56, 1389.13, 1720.80, and 1799.19 respectively, which meant that after 24 incubation, different genotype breast cancer cell lines expressed significantly different amount of CCL2.
Figure 3.
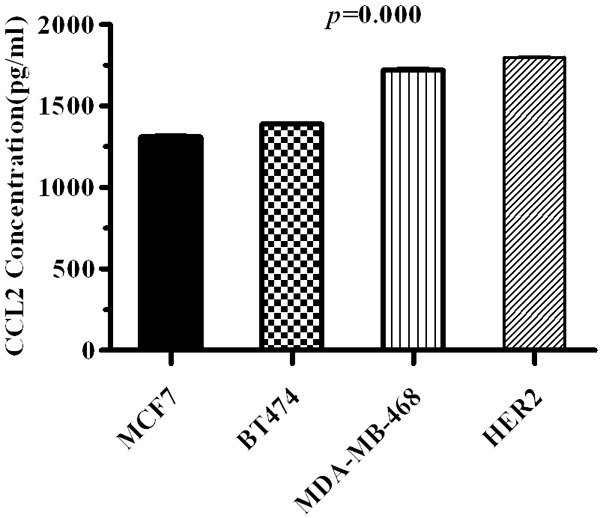
CCL2 concentrations in suspension of different genotype breast cancer cell lines after 24 h incubation.
Discussion
Many studies have detected CCL2 expression at the protein level in primary tumor cells, regional lymph nodes, and metastatic sites. It has been confirmed that CCL2 expression is associated with tumor malignancy [6]. More than 50% of breast cancer tissue shows enhanced expression of CCL2 [7], and prospective research has further revealed that high expression of CCL2 is closely related to advanced tumor stage, lymph node metastasis [8], and early recurrence [9]. In this study, our results demonstrate that CCL2 expression is significantly different between the five breast cancer genotypes. This is based on the results of IHC staining of 205 paraffin embedded breast cancer tissues and ELISA detection in cell line suspensions.
The tumor microenvironment consists of tumor cells, stromal cells, cytokines, chemokines, and other components. Carcinoma-associated-fibroblasts (CAFs) arise from fibroblasts existing in normal tissue, which have been stimulated by soluble signal molecules. They are activated fibroblasts and the most important host cells at the tumor-host interface. CAFs affect the development or reverse of malignant disease through the secretion of a variety of biological factors, which interact with constituents of the microenvironment [10]. An increase in CCL2 expression in breast tumor tissue is related with tumor-associated macrophage (TAMs) infiltration and microvascular density increase [7,9].
CCL2 is synthesized and secreted rapidly by fibroblasts and vascular endothelial cells when vascular endothelial growth factor (VEGF) is increased in the tumor microenvironment or in a hypoxic state. Through its interaction with its main receptor, CCR2, CCL2 recruits large numbers of monocytes to the tumor tissue, and initiates the following biological processes: (1) chemo-attraction of inflammation cells that infiltrate tumor tissue [11], (2) promotion of tumor cell growth and survival [12], (3) induction of tumor angiogenesis [13,14], (4) inhibition of anti-tumor immunity [15,16], and (5) promotion of tumor invasion and metastasis [11].
The interaction of CCL2 and CCR2 causes an increase in Ca2+ flow, cyclic adenosine monophosph-ate (CAMP) inhibition, and phospholipase-c and phosphatidylinositol3-kinase (PI3-k) activation. Studies have shown that CCL2/CCR2 regulates the proliferation and biological behavior of breast cancer cells through the Smad3 and p42/44MAPK pathways [17]. CCL2 also causes crosstalk between tumor cells and stromal fibroblasts and the induction of NOTCH1 expression, which promotes the progression of tumor stem cell-related diseases [18].
Chavey et al. found that CCL2 expression in breast cancer tissue correlated with a lack of ER and PR expression, indicative of a poor prognosis [19]. CCL2 expression by TAMs and/or tumor cells was strongly associated with the expression of membrane type 1-matrix metalloproteinase, tumor necrosis factor-α, thymidine phosphorylase, and other angiogenic factors [7,9]. Ghilardi et al. [20] reported that the CCL2-2518A/G promoter region polymorphism found in monocytes, influences CCL2 transcriptional activity and the level of CCL2 secretion. The presence of at least one G in the CCL2 allele is a marker of significantly increased breast cancer metastasis risk. In our study, we demonstrated that the expression of CCL2 in different breast cancer genotypes was negatively associated with ER and PR expression. When comparing overall survival, we found that the tumors with low CCL2 expression had better survival than those with high expression.
Together, these studies suggest that CCL2 is involved in promotion and progression of breast cancer, that CCL2 is associated with the expression of ER and PR, and that the expression of CCL2 is significantly different between different breast cancer genotypes. Due to differential gene expression between cavity epithelial cells and basement epithelial cells in the five genotypes, the degree of ER, PR, HER2, CK5/6, EGFR, P53, GATA3, FOXA1, GRB7, TRAP100, CGH, LAPTMB4, and NSEP-1F expression is different. These differences will in turn result in differences in downstream signal pathway conduction. Therefore, further studies to elucidate the role of CCL2 in different signaling pathways in the different breast cancer genotypes need to be carried out.
Investigating the regulation of CCL2/CCR2 signaling in CAFs and TAMs and elucidating the different CCL2 signal transduction pathways in the different breast cancer genotypes is expected to contribute to new and individually targeted therapy for the different breast cancer genotypes.
Acknowledgements
This research was supported by National Natural Science Foundation of China (No. 81001172).
Disclosure of conflict of interest
None.
References
- 1.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez-Pinilla SM, Sarrio D, Honrado E, Moreno-Bueno G, Hardisson D, Calero F, Benitez J, Palacios J. Vimentin and laminin expression is associated with basal-like phenotype in both sporadic and BRCA1-associated breast carcinomas. J Clin Pathol. 2007;60:1006–1012. doi: 10.1136/jcp.2006.042143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, Foekens JA, Martens JW. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68:3108–3114. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 4.Badve S, Turbin D, Thorat MA, Morimiya A, Nielsen TO, Perou CM, Dunn S, Huntsman DG, Nakshatri H. FOXA1 expression in breast cancer--correlation with luminal subtype A and survival. Clin Cancer Res. 2007;13:4415–4421. doi: 10.1158/1078-0432.CCR-07-0122. [DOI] [PubMed] [Google Scholar]
- 5.Van Coillie E, Van Damme J, Opdenakker G. The MCP/eotaxin subfamily of CC chemokines. Cytokine Growth Factor Rev. 1999;10:61–86. doi: 10.1016/s1359-6101(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 6.Steiner JL, Murphy EA. Importance of chemokine (CC-motif) ligand 2 in breast cancer. Int J Biol Markers. 2012;27:e179–e185. doi: 10.5301/JBM.2012.9345. [DOI] [PubMed] [Google Scholar]
- 7.Saji H, Koike M, Yamori T, Saji S, Seiki M, Matsushima K, Toi M. Significant correlation of monocyte chemoattractant protein-1 expression with neovascularization and progression of breast carcinoma. Cancer. 2001;92:1085–1091. doi: 10.1002/1097-0142(20010901)92:5<1085::aid-cncr1424>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 8.Lebrecht A, Grimm C, Lantzsch T, Ludwig E, Hefler L, Ulbrich E, Koelbl H. Monocyte chemoattractant protein-1 serum levels in patients with breast cancer. Tumour Biol. 2004;25:14–17. doi: 10.1159/000077718. [DOI] [PubMed] [Google Scholar]
- 9.Ueno T, Toi M, Saji H, Muta M, Bando H, Kuroi K, Koike M, Inadera H, Matsushima K. Significance of macrophage chemoattractant protein-1 in macrophage recruitment, angiogenesis, and survival in human breast cancer. Clin Cancer Res. 2000;6:3282–3289. [PubMed] [Google Scholar]
- 10.Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle. 2006;5:1597–1601. doi: 10.4161/cc.5.15.3112. [DOI] [PubMed] [Google Scholar]
- 11.Mantovani A, Schioppa T, Porta C, Allavena P, Sica A. Role of tumor-associated macrophages in tumor progression and invasion. Cancer Metastasis Rev. 2006;25:315–322. doi: 10.1007/s10555-006-9001-7. [DOI] [PubMed] [Google Scholar]
- 12.Song G, Ouyang G, Bao S. The activation of Akt/PKB signaling pathway and cell survival. J Cell Mol Med. 2005;9:59–71. doi: 10.1111/j.1582-4934.2005.tb00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salcedo R, Ponce ML, Young HA, Wasserman K, Ward JM, Kleinman HK, Oppenheim JJ, Murphy WJ. Human endothelial cells express CCR2 and respond to MCP-1: direct role of MCP-1 in angiogenesis and tumor progression. Blood. 2000;96:34–40. [PubMed] [Google Scholar]
- 14.Ma J, Wang Q, Fei T, Han JD, Chen YG. MCP-1 mediates TGF-beta-induced angiogenesis by stimulating vascular smooth muscle cell migration. Blood. 2007;109:987–994. doi: 10.1182/blood-2006-07-036400. [DOI] [PubMed] [Google Scholar]
- 15.Vitiello PF, Shainheit MG, Allison EM, Adler EP, Kurt RA. Impact of tumor-derived CCL2 on T cell effector function. Immunol Lett. 2004;91:239–245. doi: 10.1016/j.imlet.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Gu L, Tseng S, Horner RM, Tam C, Loda M, Rollins BJ. Control of TH2 polarization by the chemokine monocyte chemoattractant protein-1. Nature. 2000;404:407–411. doi: 10.1038/35006097. [DOI] [PubMed] [Google Scholar]
- 17.Fang WB, Jokar I, Zou A, Lambert D, Dendukuri P, Cheng N. CCL2/CCR2 chemokine signaling coordinates survival and motility of breast cancer cells through Smad3 protein- and p42/44 mitogen-activated protein kinase (MAPK)-dependent mechanisms. J Biol Chem. 2012;287:36593–36608. doi: 10.1074/jbc.M112.365999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuyada A, Chow A, Wu J, Somlo G, Chu P, Loera S, Luu T, Li AX, Wu X, Ye W, Chen S, Zhou W, Yu Y, Wang YZ, Ren X, Li H, Scherle P, Kuroki Y, Wang SE. CCL2 mediates cross-talk between cancer cells and stromal fibroblasts that regulates breast cancer stem cells. Cancer Res. 2012;72:2768–2779. doi: 10.1158/0008-5472.CAN-11-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chavey C, Bibeau F, Gourgou-Bourgade S, Burlinchon S, Boissiere F, Laune D, Roques S, Lazennec G. Oestrogen receptor negative breast cancers exhibit high cytokine content. Breast Cancer Res. 2007;9:R15. doi: 10.1186/bcr1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghilardi G, Biondi ML, La Torre A, Battaglioli L, Scorza R. Breast cancer progression and host polymorphisms in the chemokine system: role of the macrophage chemoattractant protein-1 (MCP-1)-2518 G allele. Clin Chem. 2005;51:452–455. doi: 10.1373/clinchem.2004.041657. [DOI] [PubMed] [Google Scholar]


