Abstract
Objective: This study is to characterize and identify the human Brucella strains in Xinjiang, China with multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA) scheme. Methods: Brucella strains were isolated and cultured from 62 brucellosis patients. The bacteria strains were subjected to the oxidase, catalase, rapid urease, and nitrate reduction tests, and the species identification was performed using the VITEK-2 Compact system. These Brucella strains were further identified and characterized using the 16 VNTR loci in a MLVA-16 methodology. Results: Twelve Brucella strains had been identified out of 62 patients, which were all recognized as Brucella melitensis (B. melitensis) according to the results from the VITEK-2 Compact system. Based on panel 1 (MLVA-8), these 12 Brucella isolates were clustered into three known genotypes and two new genotypes, in which 7 strains were clustered into genotype 45 (1-5-3-12-2-2-3-2), 1 strain was classified as genotype 42 (1-5-3-13-2-2-3-2), 1 stain was with genotype 62 (1-3-3-13-2-2-3-2), and the other 3 trains revealed two new genotypes, i.e., (1-5-3-12-2-3-3-2) and (1-5-3-11-2-3-3-2). Using panel 2A+2B (MLVA-16), we found that no genotypes of these strains were identical to the known genotypes, generally with differences in 2-4 loci. However, three strains shared the same genotype. Conclusion: Brucella strains in 62 brucellosis patients from Xinjiang are all identified as B. melitensis. Based on MLVA-8, two new genotypes have been discovered. These findings might contribute to the understanding of the pathogenesis and epidemiology of brucellosis in Xinjiang, China.
Keywords: Brucella, brucellosis, multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA), genotyping, Xinjiang
Introduction
Brucellosis is recognized as one of the most common zoonotic diseases worldwide [1], which is caused by bacteria of the genus Brucella. At present, approximately 0.5 million new cases are reported each year, primarily in the Mediterranean region, the Middle East, Africa, South and Central America, and Asia [2,3]. Particularly, in Central Asia, the incidence of brucellosis has been rapidly increasing [4,5], with continuous disease reports over the past decade [6-8].
In China, brucellosis has been a persistent public health problem. Since 1999, an increasing trend of the incidence of brucellosis has been observed in China, with specific geographical features [9]. Brucellosis is prevalent in northwestern China, including Xinjiang area, where living is mainly dependent on ruminant livestock [9,10]. Therefore, it is important to develop rapid and accurate genotyping methods for epidemiological investigation and disease control.
Traditional detection and identification of Brucella are mainly based on bacteriological and serological techniques. Bacteriological procedures are always tedious and time-consuming, with unsatisfactory positive results. Even though serological techniques are often fast, the frequencies of false-positive and false-negative results are rather high. Moreover, genotyping cannot be achieved. As an alternative, multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA) has been introduced and developed in recent years, which greatly facilitates genotyping and epidemiological studies [11-14]. MLVA-16 contain eight minisatellite (panel 1: Bruce 06, Bruce 08, Bruce 11, Bruce 12, Bruce 42, Bruce 43, Bruce 45, Bruce 55) and eight microsatellite markers (panel 2A: Bruce 18, Bruce 19, Bruce 21; panel 2B: Bruce 04, Bruce 07, Bruce 09, Bruce 16, Bruce 30) [15]. In this study, MLVA-16 scheme was used to genotype a collection of 12 human Brucella strains isolated from patients in Xinjiang, a geographical area with high incidence of brucellosis in China. Results from this study might contribute to the understanding of the pathogenesis and epidemiology of brucellosis in this area.
Materials and methods
Bacterial strains and culture
This study included 62 brucellosis patients from the Xinjiang Uygur Autonomous Region People’s Hospital and the First Affiliated Hospital of Xinjiang Medical University (Table 1). The Brucella strains were isolated from their blood samples. Signed informed consent was obtained from every patient and the study was approved by the ethics review board of the First Affiliated Hospital of Xinjiang Medical University Blood samples were cultured in the Bact/Alert 3D automatic blood culture system (BioMerieux, Marcy-L’Etoile, France) for 7-15 d, and then streaked onto blood plates. After 48-72-h incubation, tiny gray colonies were observed, which were Gram-negative cocci. These isolates were subjected to chemical tests, and the species identification was performed with the VITEK-2 Compact system (BioMerieux).
Table 1.
Basic information of the Brucella isolates
| Strains | Isolation time | Specimen source | Patient age | Gender | Ethnicity | Symptoms | Residence |
|---|---|---|---|---|---|---|---|
| A | 2010 | Blood | 36 | Male | Kazak | Fever, aortic insufficiency | Urumqi |
| B | 2010 | Blood | 45 | Male | Han | Fever, oscheocele | Aletai |
| C | 2010 | Blood | 3 | Female | Kazak | Fever, anaemia, arthralgia | Aletai |
| D | 2010 | Blood | 14 | Male | Uighur | Fever, arthralgia | Kuqa |
| E | 2010 | Blood | 9 | Female | Hui | Fever, arthralgia | Dabancheng |
| F | 2010 | Blood | 50 | Female | Han | Fever, lumbago | Urumqi |
| G | 2010 | Blood | 46 | Male | Uighur | Fever | Keping County |
| H | 2010 | Blood | 47 | Male | Uighur | Fever, anaemia | Kuqa |
| I | 2010 | Blood | 3 | Male | Uighur | Fever, anaemia, arthralgia | Kuqa |
| J | 2011 | Blood | 56 | Male | Uighur | Fever | Turfan |
| K | 2011 | Blood | 47 | Female | Uighur | Fever | Urumqi |
| L | 2011 | Blood | 17 | Female | Kazak | Fever, anaemia | Aletai |
Oxidase test
For the oxidase test, bacterial smear was added to filter paper containing 1% N,N,N’,N’-tetramethyl-p-phenylenediamine (TMPD; Bellancom, Beijing, China). Color development was observed within 10 s. Deep purple was considered as positive.
Catalase test
For the catalase test, the bacterial cultures were scraped onto a clean slide, and a drop of 3% H2O2 was added. Plenty of bubbles indicated a positive reaction.
Rapid urease test
For the rapid urease test, bacteria were inoculated into medium containing urease. Color change from yellow to red was considered as a positive result.
Nitrate reduction test
For the nitrate reduction test, bacterial cultures were inoculated into nitrate broth prepared in-house. After incubation at 35°C for 24 h, the nitrate reagents were added. Appearance of red color in the culture medium was considered as positive results. Strains of Escherichia coli (ATCC 25922) and Staphylococcus aureus (ATCC 25923) were used as the quality control.
DNA preparation and MLVA genotyping scheme
Total genomic DNA was extracted using a MiniBEST Bacterial Genomic DNA Extraction kit (Takara, Otsu, Japan), and stored at -20°C until further use.
MLVA genotyping was performed as previously described [16]. The sixteen primer sets were shown in Table 2, including panel 1 (Bruce 06, Bruce 08, Bruce 11, Bruce 12, Bruce 42, Bruce 43, Bruce 45, and Bruce 55), panel 2A (Bruce 18, bruce19, and Bruce 21), and panel 2B (Bruce 04, Bruce 07, Bruce 09, Bruce 16, and Bruce 30). The 50 μl PCR system contained 1 μl DNA template, 1 μl primer each, 25 µl 2× EASY TAQ PCR SM, and 22 µl ddH2O. The reaction conditions were as follows: for Bruce 08, Bruce 11, Bruce 12, Bruce 42, Bruce 45, and Bruce 55 in panel 1, initial denaturation at 96°C for 5 min, and then 30 cycles of 96°C for 30 s, 60°C for 30 s, and 70°C for 1 min; for Bruce 06 and Bruce 43 in panel 1, initial denaturation at 96°C for 5 min, and then 34 cycles of 96°C for 30 s, 57°C for 30 s, and 70°C for 1 min. For panel 2A and panel 2B, initial denaturation at 95°C for 3 min, and then 34 cycles of 95°C for 30 s, 58°C for 30 s, and 70°C for 1 min. PCR products for panel 1 and panel 2A+2B were analyzed by 2% and 3% agarose gel electrophoresis, respectively. Moreover, the products were sequenced by Sangon Biotech (Shanghai, China).
Table 2.
Primer sets for MLVA
| Loci | Forward primer | Reward primer |
|---|---|---|
| Bruce 06 | 5’-ATGGGATGTGGTA GGGTAATCG-3’ | 5’-ATGGGATGTGGTA GGGTAATCG-3’ |
| Bruce 08 | 5’-ATTATTCGCAGGCTCGTGATTC-3’ | 5’-ATTATTCGCAGGCTCGTGATTC-3’ |
| Bruce 11 | 5’-CTGTTGATCTGACCTTGCAACC-3’ | 5’-CTGTTGATCTGACCTTGCAACC-3’ |
| Bruce 12 | 5’-CGGTAAATCAATTGTCCCATGA-3’ | 5’-CGGTAAATCAATTGTCCCATGA-3’ |
| Bruce 42 | 5’-CATCGCCTCAACTATACCGTCA-3’ | 5’-CATCGCCTCAACTATACCGTCA-3’ |
| Bruce 43 | 5’-TCTCAA GCCCGATATGGA GAAT-3’ | 5’-TCTCAA GCCCGATATGGA GAAT-3’ |
| Bruce 45 | 5’-ATCCTTGCCTCTCCCTACCAG-3’ | 5’-ATCCTTGCCTCTCCCTACCAG-3’ |
| Bruce 55 | 5’-TCA GGCTGTTTCGTCATGTCTT-3’ | 5’-TCA GGCTGTTTCGTCATGTCTT-3’ |
| Bruce 04 | 5’-CTGACGAAGGGAAGGCAATAAG-3’ | 5’-CGATCTGGAGATTATCGGGAAG-3’ |
| Bruce 07 | 5’-GCTGACGGGGAAGAACATCTAT-3’ | 5’-ACCCTTTTTCAGTCAAGGCAAA-3’ |
| Bruce 09 | 5’-GCGGATTCGTTCTTCAGTTATC-3’ | 5’-GGGAGTATGTTTTGGTTGTACATAG-3’ |
| Bruce 16 | 5’-ACGGGAGTTTTTGTTGCTCAAT-3’ | 5’-GGCCATGTTTCCGTTGATTTAT-3’ |
| Bruce 18 | 5’-TATGTTAGGGCAATA GGGCAGT-3’ | 5’-GATGGTTGAGAGCATTGTGAAG-3’ |
| Bruce 19 | 5’-GACGACCCGGACCATGTCT-3’ | 5’-ACTTCACCGTAACGTCGTGGAT-3’ |
| Bruce 21 | 5’-CTCATGCGCAACCAAAACA-3’ | 5’-GATCTCGTGGTCGATAATCTCATT-3’ |
| Bruce 30 | 5’-TGACCGCAAAACCATATCCTTC-3’ | 5’-TATGTGCAGAGCTTCATGTTCG-3’ |
Results
Characterization and identification of the Brucella strains
All the Brucella strains isolated from 62 patients in this study were subjected to the oxidase, catalase, rapid urease, and nitrate reduction tests. The VITEK-2 Compact system was used for species identification. Totally 12 Brucella species had been identified, which were designated as isolates A to L, respectively. All the 12 strains were positive for the traditional tests. Moreover, VITEK-2 Compact system indicated that these Brucella strains were Brucella melitensis (B. melitensis). Taken together, results from these traditional detection and identification methods suggest that, the Brucella strains isolated from these brucellosis patients in Xinjiang were all identified as B. melitensis from Malta. Representative results of bacterial culture and Gram staining for isolate A were shown in Figure 1.
Figure 1.
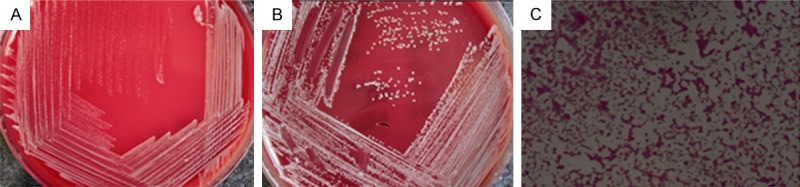
Brucella culture and Gram staining. Representative pictures of bacterial cultures at 48 h (A) and 72 h (B), as well as Gram staining (C) for isolate A were shown.
Characterization of variable-number tandem-repeat (VNTR) loci in the Brucella strains
Results from PCR amplification of all sixteen MLVA alleles were shown in Figure 2. These products were subjected to forward and reverse sequencing (Table 3), and the complete DNA sequence was obtained by overlapping the sequences with the DNAstar software. Then the Tandem Repeats Finder program was used for the repeat sequence analysis, and the VNTRs were confirmed.
Figure 2.
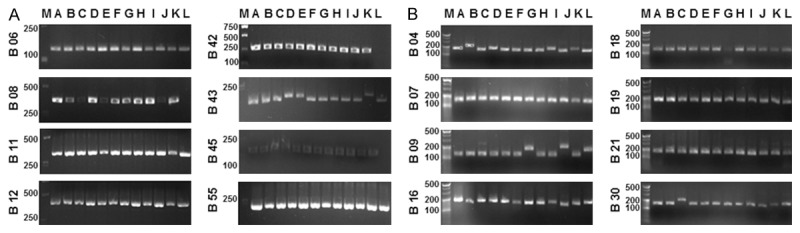
PCR analysis of the VNTRs for the Brucella strains. The 12 Brucella species were designated as isolates A to L, respectively. PCR analysis of the VNTR loci in panel 1 (A) and panel 2A+2B (B) for the Brucella strains. M, marker. Panel 1 included eight markers: Bruce 06, Bruce 08, Bruce 11, Bruce 12, Bruce 42, Bruce 43, Bruce 45, and Bruce 55. Panel 2 was composed of eight microsatellite markers: Bruce 18, bruce19, and Bruce 21 in panel 2A; and Bruce 04, Bruce 07, Bruce 09, Bruce 16, and Bruce 30 in panel 2B.
Table 3.
PCR products
| Loci | bp | A | B | C | D | E | F | G | H | I | J | K | L |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bruce 06 | 134 | 143 | 147 | 146 | 142 | 143 | 144 | 144 | 142 | 145 | 147 | 141 | 143 |
| Bruce 08 | 18 | 373 | 369 | 371 | 329 | 365 | 373 | 360 | 373 | 372 | 361 | 374 | 162 |
| Bruce 11 | 63 | 323 | 312 | 325 | 321 | 328 | 328 | 325 | 313 | 324 | 314 | 308 | 326 |
| Bruce 12 | 15 | 377 | 394 | 382 | 392 | 393 | 384 | 385 | 395 | 393 | 395 | 377 | 393 |
| Bruce 42 | 125 | 295 | 293 | 300 | 294 | 300 | 292 | 293 | 293 | 293 | 293 | 293 | 298 |
| Bruce 43 | 12 | 188 | 186 | 186 | 171 | 197 | 187 | 187 | 189 | 188 | 188 | 207 | 187 |
| Bruce 45 | 18 | 177 | 156 | 155 | 152 | 153 | 153 | 177 | 158 | 155 | 154 | 153 | 162 |
| Bruce 55 | 40 | 251 | 231 | 240 | 235 | 270 | 237 | 236 | 234 | 236 | 235 | 246 | 236 |
| Bruce 18 | 8 | 142 | 139 | 140 | 140 | 140 | 141 | 143 | 143 | 140 | 142 | 141 | 141 |
| Bruce 19 | 6 | 179 | 181 | 179 | 179 | 180 | 180 | 180 | 183 | 181 | 179 | 179 | 179 |
| Bruce 21 | 8 | 166 | 166 | 167 | 169 | 169 | 166 | 166 | 167 | 165 | 166 | 167 | 168 |
| Bruce 04 | 8 | 202 | 243 | 195 | 183 | 193 | 194 | 195 | 194 | 216 | 195 | 185 | 196 |
| Bruce 07 | 8 | 153 | 154 | 151 | 151 | 150 | 151 | 153 | 152 | 154 | 157 | 152 | 153 |
| Bruce 09 | 8 | 125 | 125 | 124 | 127 | 124 | 175 | 174 | 125 | 127 | 197 | 167 | 125 |
| Bruce 16 | 8 | 179 | 163 | 179 | 181 | 179 | 175 | 171 | 179 | 183 | 162 | 179 | 194 |
| Bruce 30 | 8 | 146 | 147 | 186 | 146 | 147 | 146 | 137 | 146 | 146 | 96 | 153 | 161 |
Genotyping and clustering of Brucella strains by MLVA
Results for the MLVA-16 genotyping assay were shown in Table 4. Resultant genotypes were compared using the Brucella 2010 MLVA database at http://minisatellites.u-psud.fr/MLVAnet/. According to panel 1, the 12 Brucella isolates were clustered into three known genotypes and two new genotypes (Figure 3). Isolates A, F, G, H, I, J, and K were clustered into genotype 45 (1-5-3-12-2-2-3-2), isolate B was classified into genotype 42 (1-5-3-13-2-2-3-2), and isolate L was with genotype 62 (1-3-3-13-2-2-3-2). Moreover, isolates C, D, and E showed new genotypes (isolate C revealing one new genotype, and isolates D and E revealing another), with differences in 1-2 loci compared with the known genotypes.
Table 4.
MLVA genotyping of the Brucella strains
| Strains | Bruce 06 | Bruce 08 | Bruce 11 | Bruce 12 | Bruce 42 | Bruce 43 | Bruce 45 | Bruce 55 | Bruce 18 | Bruce 19 | Bruce 21 | Bruce 04 | Bruce 07 | Bruce 09 | Bruce 16 | Bruce 30 | Panel 1 genotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 1 | 5 | 3 | 12 | 2 | 2 | 3 | 2 | 2 | 12 | 5 | 4 | 4 | 3 | 5 | 4 | 45 |
| B | 1 | 5 | 3 | 13 | 2 | 2 | 3 | 2 | 2 | 20 | 5 | 9 | 4 | 3 | 4 | 4 | 42 |
| C | 1 | 5 | 3 | 12 | 2 | 3 | 3 | 2 | 2 | 20 | 5 | 4 | 4 | 3 | 5 | 9 | New |
| D | 1 | 5 | 3 | 11 | 2 | 3 | 3 | 2 | 2 | 20 | 5 | 5 | 4 | 3 | 5 | 4 | New |
| E | 1 | 5 | 3 | 11 | 2 | 3 | 3 | 2 | 7 | 20 | 5 | 4 | 4 | 3 | 5 | 4 | New |
| F | 1 | 5 | 3 | 12 | 2 | 2 | 3 | 2 | 2 | 20 | 5 | 4 | 4 | 3 | 4 | 4 | 45 |
| G | 1 | 5 | 3 | 12 | 2 | 2 | 3 | 2 | 2 | 20 | 5 | 8 | 4 | 9 | 3 | 3 | 45 |
| H | 1 | 5 | 3 | 12 | 2 | 2 | 3 | 2 | 2 | 20 | 5 | 4 | 4 | 3 | 4 | 4 | 45 |
| I | 1 | 5 | 3 | 12 | 2 | 2 | 3 | 2 | 4 | 20 | 5 | 4 | 4 | 3 | 4 | 4 | 45 |
| J | 1 | 5 | 3 | 12 | 2 | 2 | 3 | 2 | 4 | 20 | 5 | 4 | 4 | 11 | 3 | 3 | 45 |
| K | 1 | 5 | 3 | 12 | 2 | 2 | 3 | 2 | 2 | 12 | 5 | 8 | 4 | 7 | 5 | 5 | 45 |
| L | 1 | 3 | 3 | 13 | 2 | 2 | 3 | 2 | 5 | 19 | 5 | 4 | 4 | 3 | 7 | 6 | 62 |
| J | 1 | 5 | 3 | 12 | 2 | 2 | 3 | 2 | 4 | 20 | 5 | 4 | 4 | 11 | 3 | 3 | 45 |
Figure 3.
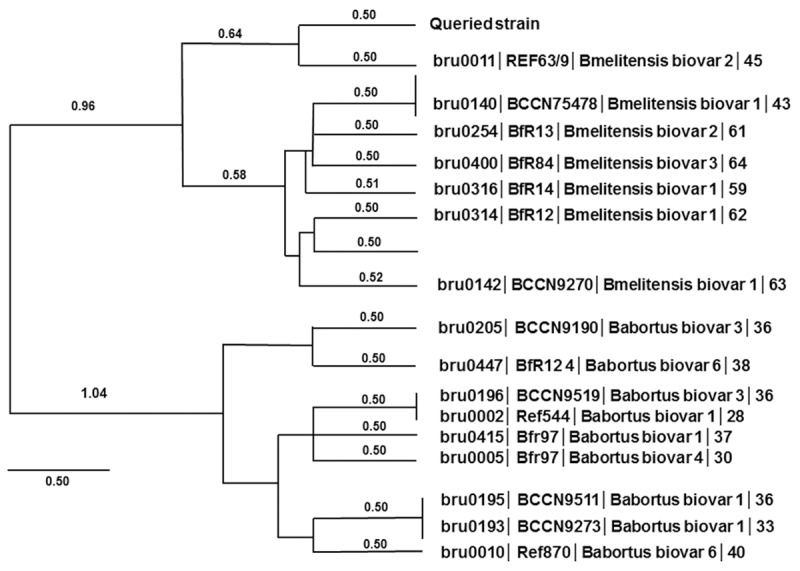
MLVA-based dendrogram and phylogenetic relationship analysis. Relationships among different Brucella genus were shown in the dendrogram. The length of the line represents the genetic distance.
Greater diversity in these Brucella isolates was observed when eight additional markers (panel 2A+2B) were included. No genotypes of these strains were found to be identical to any of the known genotypes, generally with differences in 2-4 loci. However, isolates F, H, and I shared the same genotype. Moreover, all of these 12 isolates were identified as B. melitensis, which was in line with the results from the traditional detections.
Discussion
Human brucellosis is mainly caused by direct contact with Brucella-infected animal reservoirs and/or consumption of raw animal products [17,18]. Brucella can proliferate in phagocytic cells, and cause clinical symptoms, such as undulant fever [1,19,20], orchitis [21,22], spondylitis, arthritis [23,24], endocarditis, and fatigue. Brucellosis is a systemic infection affecting human beings regardless of age and gender, and it is difficult to diagnose due to the variable clinical symptoms [25,26]. Moreover, no effective treatment methods are currently available for brucellosis, making the disease a huge economic and health burden for the society [27].
At present, Brucella can be divided into nine different species depending on the host, including B. abortus, B. canis, B. ovis, B. suis, B. neotomae, B. melitensis, B. ceti, B. pinnipedialis, and B. microti [28]. Different species are mainly distinguished by biochemical reactions, staining, and pathogen sensitivities. However, the traditional characterization and identification methods of Brucella strains are definitely cumbersome and laborious, with poor repeatability. As an alternative, emerging molecular biological techniques are more safe, rapid, specific, and sensitive. For example, the Brucella abortus-melitensis-ovis-suis (AMOS) PCR analysis has been developed. Accordingly, Brucella could be divided into four species (eight biotypes) [29]. Even more, in recent years, single nucleotide polymorphism (SNP) [30,31] analysis has been introduced into the identification of bacteria, which could cover all the currently recognized genotypes of Brucella [32].
Multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA) genotyping is mainly based on the VNTR polymorphism inside the bacterial genome, which has been applied in the investigation of tubercle bacillus (TB), leprosy bacillus, and anthrax [15]. Le Fleche et al. [16] established the original MLVA genotyping method which contains 15 VNTR loci. The technique was further improved by Al Dahouk et al. [11] to the now well-known MLVA-16 analysis, which could detect more than 500 Brucella species [33-36]. Based on the MLVA-16 scheme, a web-site Brucella database (Brucella 2010 MLVA database) has been built by collecting data from laboratories all over the world. In this study, the genotypes of 12 Brucella strains were analyzed and compared within the database. Our results showed that, according to MLVA panel 1 (MLVA-8), all the 12 isolates were B. melitensis, including 7 strains with genotype 45, 1 strain with genotype 42, 1 strain with genotype 62, and 3 strains with other two new genotypes. However, using MLVA panel 2A (MLVA-11) or 2A+2B (MLVA-16), neither of these two genotypes was found to be identical to the known genotypes.
Within the Brucella 2010 MLVA database, the “Eastern Mediterranean” genotype 42 (1-5-3-13-2-2-3-2) has been reported in Xinjiang, China [37], and the genotype 45 has been found in Turkey [35]. Unique genotypes could be reported, in comparison with the known genotypes from different regions. In this study, greater discriminations have been observed in MLVA panel 2 compared with panel 1, with only three strains identical in the genotype, indicating that distinct genotypes might be expected in the same region. However, the regional differences were limited within 1-2 repeats, suggesting their close phylogenetic relationship. All the 12 strains in this study were B. melitensis, without other species, which might be the dominant species in Xinjiang, in line with previous findings that B. melitensis is the predominant strain associated with human brucellosis outbreaks in China [38]. Particularly, in these brucellosis patients, twelve of them were engaged in pastoral livestock or with close contact with cattle. Moreover, out of the three child patients, two had a history of drinking raw milk. Of course, further in-depth studies are needed to address this issue.
In conclusion, in this study, 12 Brucella strains were identified out of bacterial cultures from 62 patients in Xinjiang, all were recognized as B. melitensis. Using panel 1 (MLVA-8), these 12 Brucella isolates were clustered into three known genotypes and two new genotypes. According to panel 2A+2B (MLVA-16), none of these two genotypes were identical to the known genotypes. These findings might contribute to the understanding of the pathogenesis of brucellosis in Xinjiang, China.
Acknowledgements
This study was supported by the Natural Science Foundation of XinJiang (NO 2015211C100). We express our gratitude to Buka Samten for the critical reading and helpful discussion of the manuscript.
Disclosure of conflict of interest
None.
Abbreviations
- VNTR
variable-number tandem-repeat
- MLVA
multiple-locus variable-number tandem-repeat analysis
- TMPD
N,N,N’,N’-tetramethyl-p-phenylenediamine
- AMOS
abortus-melitensis-ovis-suis
- SNP
single nucleotide polymorphism
- TB
tubercle bacillus
References
- 1.Whatmore AM. Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect Genet Evol. 2009;9:1168–1184. doi: 10.1016/j.meegid.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Buzgan T, Karahocagil MK, Irmak H, Baran AI, Karsen H, Evirgen O, Akdeniz H. Clinical manifestations and complications in 1028 cases of brucellosis: a retrospective evaluation and review of the literature. Int J Infect Dis. 2010;14:e469–e478. doi: 10.1016/j.ijid.2009.06.031. [DOI] [PubMed] [Google Scholar]
- 3.Skalsky K, Yahav D, Bishara J, Pitlik S, Leibovici L, Paul M. Treatment of human brucellosis: systematic review and meta-analysis of randomised controlled trials. BMJ. 2008;336:701–704. doi: 10.1136/bmj.39497.500903.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos EV. The new global map of human brucellosis. Lancet Infect Dis. 2006;6:91–99. doi: 10.1016/S1473-3099(06)70382-6. [DOI] [PubMed] [Google Scholar]
- 5.Gwida M, Al Dahouk S, Melzer F, Rösler U, Neubauer H, Tomaso H. Brucellosis-regionally emerging zoonotic disease? Croat Med J. 2010;51:289–295. doi: 10.3325/cmj.2010.51.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guler S, Kokoglu OF, Ucmak H, Gul M, Ozden S, Ozkan F. Human brucellosis in Turkey: different clinical presentations. J Infect Dev Ctries. 2014;8:581–588. doi: 10.3855/jidc.3510. [DOI] [PubMed] [Google Scholar]
- 7.Mantur BG, Amarnath SK. Brucellosis in India-a review. J Biosci. 2008;33:539–547. doi: 10.1007/s12038-008-0072-1. [DOI] [PubMed] [Google Scholar]
- 8.Kumar A. Brucellosis: need of public health intervention in rural India. Prilozi. 2010;31:219–231. [PubMed] [Google Scholar]
- 9.Li YJ, Li XL, Liang S, Fang LQ, Cao WC. Epidemiological features and risk factors associated with the spatial and temporal distribution of human brucellosis in China. BMC Infect Dis. 2013;13:547. doi: 10.1186/1471-2334-13-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Yin F, Zhang T, Yang C, Zhang X, Feng Z, Li X. Spatial analysis on human brucellosis incidence in mainland China: 2004-2010. BMJ Open. 2014;4:e004470. doi: 10.1136/bmjopen-2013-004470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al Dahouk S, Flèche PL, Nöckler K, Jacques I, Grayon M, Scholz HC, Tomaso H, Vergnaud G, Neubauer H. Evaluation of Brucella MLVA typing for human brucellosis. J Microbiol Methods. 2007;69:137–145. doi: 10.1016/j.mimet.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 12.Her M, Kang SI, Cho DH, Cho YS, Hwang IY, Heo YR, Jung SC, Yoo HS. Application and evaluation of the MLVA typing assay for the Brucella abortus strains isolated in Korea. BMC Microbiol. 2009;9:230. doi: 10.1186/1471-2180-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alvarez J, Sáez JL, García N, Serrat C, Pérez-Sancho M, González S, Ortega MJ, Gou J, Carbajo L, Garrido F, Goyache J, Domínguez L. Management of an outbreak of brucellosis due to B. melitensis in dairy cattle in Spain. Res Vet Sci. 2011;90:208–211. doi: 10.1016/j.rvsc.2010.05.028. [DOI] [PubMed] [Google Scholar]
- 14.García-Yoldi D, Le Fleche P, De Miguel MJ, Muñoz PM, Blasco JM, Cvetnic Z, Marín CM, Vergnaud G, López-Goñi I. Comparison of multiple-locus variable-number tandem-repeat analysis with other PCR-based methods for typing Brucella suis isolates. J Clin Microbiol. 2007;45:4070–4072. doi: 10.1128/JCM.01096-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kattar MM, Jaafar RF, Araj GF, Le Flèche P, Matar GM, Abi Rached R, Khalife S, Vergnaud G. Evaluation of a multilocus variable-number tandem-repeat analysis scheme for typing human Brucella isolates in a region of brucellosis endemicity. J Clin Microbiol. 2008;46:3935–3940. doi: 10.1128/JCM.00464-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Flèche P, Jacques I, Grayon M, Al Dahouk S, Bouchon P, Denoeud F, Nöckler K, Neubauer H, Guilloteau LA, Vergnaud G. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 2006;6:9. doi: 10.1186/1471-2180-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franco MP, Mulder M, Gilman RH, Smits HL. Human brucellosis. Lancet Infect Dis. 2007;7:775–786. doi: 10.1016/S1473-3099(07)70286-4. [DOI] [PubMed] [Google Scholar]
- 18.Arimi SM, Koroti E, Kang’ethe EK, Omore AO, McDermott JJ. Risk of infection with Brucella abortus and Escherichia coli O157:H7 associated with marketing of unpasteurized milk in Kenya. Acta Trop. 2005;96:1–8. doi: 10.1016/j.actatropica.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Forland F, De Carvalho Gomes H, Nokleby H, Escriva A, Coulombier D, Giesecke J, Jansen A. Applicability of evidence-based practice in public health: risk assessment on Q fever under an ongoing outbreak. Euro Surveill. 2012;17:20060. [PubMed] [Google Scholar]
- 20.Dean AS, Bonfoh B, Kulo AE, Boukaya GA, Amidou M, Hattendorf J, Pilo P, Schelling E. Epidemiology of brucellosis and q Fever in linked human and animal populations in northern togo. PLoS One. 2013;8:e71501. doi: 10.1371/journal.pone.0071501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purwar S, Metgud SC, Gokale SK. Exceptionally high titres in atypical presentation of occult epididymo-orchitis due to brucellosis. J Med Microbiol. 2012;61:443–445. doi: 10.1099/jmm.0.034892-0. [DOI] [PubMed] [Google Scholar]
- 22.Erdem H, Elaldi N, Ak O, Gulsun S, Tekin R, Ulug M, Duygu F, Sunnetcioglu M, Tulek N, Guler S, Cag Y, Kaya S, Turker N, Parlak E, Demirdal T, Ataman Hatipoglu C, Avci A, Bulut C, Avci M, Pekok A, Savasci U, Kaya S, Sozen H, Tasbakan M, Guven T, Bolukcu S, Cesur S, Sahin-Horasan E, Kazak E, Denk A, Gonen I, Karagoz G, Haykir Solay A, Alici O, Kader C, Senturk G, Tosun S, Turan H, Baran AI, Ozturk-Engin D, Bozkurt F, Deveci O, Inan A, Kadanali A, Sayar MS, Cetin B, Yemisen M, Naz H, Gorenek L, Agalar C. Genitourinary brucellosis: results of a multicentric study. Clin Microbiol Infect. 2014;20:847–53. doi: 10.1111/1469-0691.12680. [DOI] [PubMed] [Google Scholar]
- 23.Hasanoglu I, Guven T, Maras Y, Guner R, Tasyaran MA, Acikgoz ZC. Brucellosis as an aetiology of septic arthritis. Trop Doct. 2014;44:48–49. doi: 10.1177/0049475513512645. [DOI] [PubMed] [Google Scholar]
- 24.Baldi PC, Giambartolomei GH. Pathogenesis and pathobiology of zoonotic brucellosis in humans. Rev Sci Tech. 2013;32:117–125. doi: 10.20506/rst.32.1.2192. [DOI] [PubMed] [Google Scholar]
- 25.Herrick JA, Lederman RJ, Sullivan B, Powers JH, Palmore TN. Brucella arteritis: clinical manifestations, treatment, and prognosis. Lancet Infect Dis. 2014;14:520–526. doi: 10.1016/S1473-3099(13)70270-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akhvlediani T, Clark D V, Chubabria G, Zenaishvili O, Hepburn MJ. The changing pattern of human brucellosis: clinical manifestations, epidemiology, and treatment outcomes over three decades in Georgia. BMC Infect Dis. 2010;10:346. doi: 10.1186/1471-2334-10-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ducrotoy MJ, Bertu WJ, Ocholi RA, Gusi AM, Bryssinckx W, Welburn S, Moriyón I. Brucellosis as an emerging threat in developing economies: lessons from Nigeria. PLoS Negl Trop Dis. 2014;8:e3008. doi: 10.1371/journal.pntd.0003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scholz HC, Nöckler K, Göllner C, Bahn P, Vergnaud G, Tomaso H, Al Dahouk S, Kämpfer P, Cloeckaert A, Maquart M, Zygmunt MS, Whatmore AM, Pfeffer M, Huber B, Busse HJ, De BK. Brucella inopinata sp. nov., isolated from a breast implant infection. Int J Syst Evol Microbiol. 2010;60:801–808. doi: 10.1099/ijs.0.011148-0. [DOI] [PubMed] [Google Scholar]
- 29.Matope G, Bhebhe E, Muma JB, Skjerve E, Djønne B. Characterization of some Brucella species from Zimbabwe by biochemical profiling and AMOS-PCR. BMC Res Notes. 2009;2:261. doi: 10.1186/1756-0500-2-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engelsma KA, Veerkamp RF, Calus MPL, Windig JJ. Consequences for diversity when animals are prioritized for conservation of the whole genome or of one specific allele. J Anim Breed Genet. 2014;131:61–70. doi: 10.1111/jbg.12052. [DOI] [PubMed] [Google Scholar]
- 31.Gopaul KK, Koylass MS, Smith CJ, Whatmore AM. Rapid identification of Brucella isolates to the species level by real time PCR based single nucleotide polymorphism (SNP) analysis. BMC Microbiol. 2008;8:86. doi: 10.1186/1471-2180-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Surucuoglu S, El S, Ural S, Gazi H, Kurutepe S, Taskiran P, Yurtsever SG. Evaluation of real-time PCR method for rapid diagnosis of brucellosis with different clinical manifestations. Pol J Microbiol. 2009;58:15–19. [PubMed] [Google Scholar]
- 33.Maquart M, Le Flèche P, Foster G, Tryland M, Ramisse F, Djønne B, Al Dahouk S, Jacques I, Neubauer H, Walravens K, Godfroid J, Cloeckaert A, Vergnaud G. MLVA-16 typing of 295 marine mammal Brucella isolates from different animal and geographic origins identifies 7 major groups within Brucella ceti and Brucella pinnipedialis. BMC Microbiol. 2009;9:145. doi: 10.1186/1471-2180-9-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang SI, Heo EJ, Cho D, Kim JW, Kim JY, Jung SC, Her M. Genetic comparison of Brucella canis isolates by the MLVA assay in South Korea. J Vet Med Sci. 2011;73:779–786. doi: 10.1292/jvms.10-0334. [DOI] [PubMed] [Google Scholar]
- 35.Kiliç S, Ivanov IN, Durmaz R, Bayraktar MR, Ayaslioglu E, Uyanik MH, Aliskan H, Yasar E, Bayramoglu G, Arslantürk A, Vergnaud G, Kantardjiev TV. Multiple-locus variable-number tandem-repeat analysis genotyping of human Brucella isolates from Turkey. J Clin Microbiol. 2011;49:3276–3283. doi: 10.1128/JCM.02538-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorneles EMS, Santana JA, Alves TM, Pauletti RB, Mol JP da S, Heinemann MB, Lage AP. Genetic stability of Brucella abortus isolates from an outbreak by multiple-locus variable-number tandem repeat analysis (MLVA16) BMC Microbiol. 2014;14:186. doi: 10.1186/1471-2180-14-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aftab H, Dargis R, Christensen JJ, Le Flèche P, Kemp M. Imported brucellosis in Denmark: molecular identification and multiple-locus variable number tandem repeat analysis (MLVA) genotyping of the bacteria. Scand J Infect Dis. 2011;43:536–538. doi: 10.3109/00365548.2011.562531. [DOI] [PubMed] [Google Scholar]
- 38.Deqiu S, Donglou X, Jiming Y. Epidemiology and control of brucellosis in China. Vet Microbiol. 2002;90:165–182. doi: 10.1016/s0378-1135(02)00252-3. [DOI] [PubMed] [Google Scholar]


