Abstract
Objectives: This study aims to investigate the influence on human hepatocytes apoptosis and autophagy by the hepatitis C virus (HCV) core protein. Methods: QSG-7701, a human-derived non-neoplastic liver cell line, was transfected with PIRES-core vector that was a eukaryotic vector to express HCV core protein. Fluorescence microscope was used to observe the changes of nuclei in apoptosis cells by Annex in V-FITC/PI double staining. Flow cytometry was applied to detect the rate of cell apoptosis. Western blotting was used to detect the expression of HCV core protein, transcription factor nuclear factor-kappa B (NF-κB), autophagic biomarker microtubule associated protein 1 light chain 3 (LC3), and Beclin-1. Results: The apoptosis rate was significantly lower (P < 0.05) in QSG7701/core group (transfected with PIRES-core vector, (1.34±0.07)%) than in QSG7701 group (no transfection, (2.35±0.11)%) and in QSG7701 QSG7701/pcDNA3.1 group (transfected with pcDNA3.1 vector, (2.58±0.1)%). NF-κB expression was up-expressed in QSG7701/core group than in QSG7701/pcDNA3.1 group and QSG7701 group (P < 0.05). LC3-II expression and Beclin-1 expression was significant higher in QSG7701/core group than in the QSG7701/pcDNA3.1 group and QSG7701 group (P < 0.05). Conclusion: HCV core protein can repress the apoptosis and improve the autophagy of QSG7701 through up-regulating NF-κB and Beclin-1 expression.
Keywords: Cell autophagy, LC3-II, Beclin-1, NF-κB, apoptosis
Introduction
Apoptosis is programmed cell death, which plays an important role in maintaining cellular environmental homeostasis [1-3]. Several studies validated that the abnormal apoptosis was closely related to development and progression of tumor [4]. Autophagy is a eukaryotic cell process that use lysosomes to degrade intracellular damaged organelles and denatured proteins, which is another kind of non-caspase dependent programmed cell death different with apoptosis [5-7]. Autophagy is essential in cell growth, differentiation, self-renewal, aging and death, which become a hot spot in molecular biology. Autophagy plays important roles in cancer, inflammatory and degenerative diseases [8-10]. In recent years, many studies reported that HCV core protein is an important driver factor to cause malignant transformation of hepatocytes [11,12]. It is still unclear whether HCV core protein can influence hepatocytes apoptosis, autophagy, and involved signaling pathways. To further investigate the influence on hepatocytes apoptosis and autophagy induced by HCV core protein, we select a human non-neoplastic liver cell line (QSG-7701), a natural HCV host cell line, to simulate the natural state of HCV infection. The results in this study will shed light on the molecular mechanisms of HCV core protein.
Materials and methods
Cell line and vectors
QSG7701, a human-derived non-neoplastic liver cell line, was selected in this study. The cells were reserved and cultured in the central lab of Qianfoshan Hospital affiliated to Shandong University. The PIRES-core vectors that can express HCV core protein was kindly endeavored by Professor Xiaoyan Feng from Academy of Military Medical Sciences in Beijing.
Cell culture
The Freezing Tube containing QSG7701 cells was removed from liquid nitrogen, then incubated the tube into 37°C water bath immediately. After shaking to rapidly melting, the cells were centrifuged in 800-1000 rpm for 5 min. The cells were added with RPMI1640 (Gibco, American) containing 10% FBS, 100 U/ml penicillin and 100 U/ml streptomycin. The suspended cells were seeded into 50 ml sterile culture flask at 37°C within 5% CO2. The cells grown to log phase were used for further experiment.
The extraction and amplification of PIRES-core vector
The competent cells DH5α 50 μl were thawed on ice. The 2 μl PIRES-core vector was added into the bacterium suspension and incubated on ice for 30 min. Total 600 μl SOB medium was drew to culture in 200 rpm/min for 1 h. The 30 μl bacterium suspension was absorbed to evenly apply on ampicillin resistance SOB medium, and cultured overnight under 37°C incubator. The plasmid was extracted based on SDS-alkaline lysis methods.
Cell transfection
18 to 24 hours before transfection, cells were seeded in 6-well plates with the cell density about 90%~95%. The QSG7701 cells were divided into 4 groups based on transfection: blank group without any transfection (QSG7701 group), control group transfected with pcDNA3.1 (QSG7701/pcDNA3.1 group) and two experimental groups were all transfected with PIRES-core (QSG7701/core group) plasmid (one group for fluorescence detection, the other group for western blot detection).
The 2 μg plasmid was diluted into 100 μl DMEM medium with high glucose and mixed well. The 6 μl Liposome transfection reagent (Beyotime, Shanghai, China) was diluted into 100 μl DMEM medium with high glucose, and mixed adequately. The solution with Liposome transfection reagent was added into plasmid solution at room temperature for 15-20 min. After adding the mixed solution to each group, RPMI1640 medium containing with 10% FBS was added.
Apoptosis detection
After transfection 15-20 minutes, fluorescent microscope was used to observe the expression of luciferase on luciferase group. The cell suspension was added into 5 µL Annexin V-FITC staining solution. After mixed adequately, the cells were incubated in dark 2-8°C for 15 min. The changes of nuclear morphometry were observed by fluorescent microscope. And the apoptosis rate was detected by FCM.
Western blot analysis
After transfection 15-20 minutes, the cells in different group were harvest to extract proteins. The expression of NF-κB protein, LC3 protein, Beclin-1 protein induced by HCV core protein on QSG7701 cells were detected by Western Blotting methods.
The AlphaEaseFC890 software was applied to analyze the image band and the expression of proteins in each group was calculated by average absorbency.
Statistical analysis
All the data were described as the mean ± SD, and difference among different group was determined by ANOVA. The Dunett-t test was used to compare the difference in subgroups. P < 0.05 was considered as statistically significant.
Results
The expression of HCV core protein and NF-κB in QSG7701 by Western Blotting
To determine the transfection efficiency of liposome method, GFP fluorescence was detected by fluorescent microscopy. As shown in Figure 1A, GFP was detected in almost of all QSG7701 cells with transfecting PIRES-core plasmid. This result indicated that we can get higher transfection efficiency cell lines by this liposome method.
Figure 1.
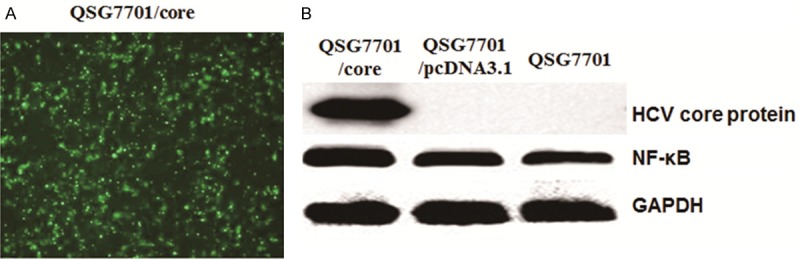
Expression of HCV core protein andNF-κBin QSG7701 cell line. A. Fluorescence detection of HCV core protein expressed in QSG7701 cell line; B. Western blot to detect the expression of HCV core protein (upper line), NF-κB (middle line) and GAPDH protein (lower line) in QSG7701 cell line by transfecting with pIRES-core, pcDNA3.1 plasmid and without any plasmid.
In order to check the expression of HCV core protein in QSG7701 cells with different transfection, Western Blotting was applied in different groups of cells. As shown in Figure 1B, HCV core protein can be detected in the QSG7701/core group, while it can be detected in QSG7701 group and QSG7701/pcDNA3.1 group. The results indicate that HCV core protein can be expressed stably in QSG7701 cells after transfection with PIRES-core plasmid.
As HCV core protein was reported to be associated with apoptosis induced by TNF-α signaling pathway, we also detected the expression of transcriptional factor NF-κB in different groups. As shown in Figure 1B, NF-κB was up-regulated in QSG7701/core group than in QSG7701/pcDNA3.1 and QSG7701 groups. We also calculated the relative intensities of each band. As shown in Table 1, NF-κB expression was significantly higher in QSG7701/core group than QSG7701/pcDNA3.1 group and QSG7701 group (P < 0.01). The results indicated that HCV core protein up-regulated NF-κB (apoptosis repressor gene) expression.
Table 1.
Expression of NF-κB in QSG7701 cell lines with transfecting PIRES-core, pcDNA3.1 and without any plasmid
Compared with QSG7701 group;
P < 0.01.
Compared with QSG7701/pcDNA3.1 group;
P < 0.01.
Influence on apoptosis rate of QSG7701 by HCV core protein
To detect the apoptosis influenced by HCV core protein, FCM with Annexin V-FITC/PI double stain was used for different groups of cells. The affected cells can be effectively distinguished into early or late stage of apoptosis and dead cells. Comparing with the lower fluorescence effects, apoptotic cells exhibit strong red fluorescence, and necrotic cells appear blue or/and red fluorescence. The apoptotic cells in early stage are located in fourth quadrant. As shown in Figure 2, the apoptosis rate was significantly different among different groups of QSG7701 cells. The rates of early apoptosis were (1.34±0.07)%, (2.58±0.13)% and (2.35±0.11)% in QSG7701/core group, QSG7701/pcDNA3.1 group and QSG7701 group respectively. The rate was significantly lower in QSG7701/core group than QSG7701/pcDNA3.1 and QSG7701 group (P < 0.05).
Figure 2.
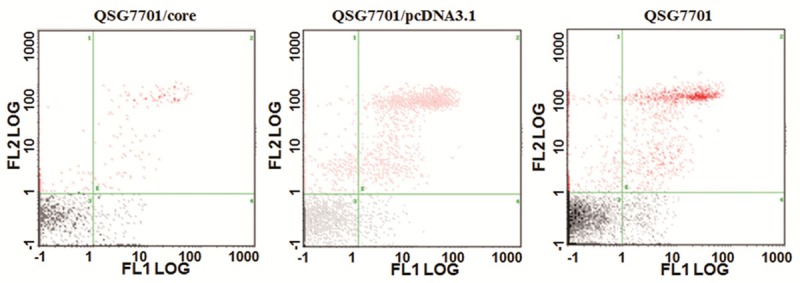
HCV core protein repressed apoptosis rate of QSG7701. Flow cytometry method to detect the apoptosis rate of QSG7701 cell line with transfecting pIRES-core, pcDNA3.1 plasmid and without any plasmid.
LC3-II and Beclin-1 expression by Western Blotting
To explore the influence on the expression of autophagy associated genes by HCV core protein, Western Blotting was applied to detect LC3-II and Beclin-1 expression. As shown in Figure 3A, 3B and Table 2, LC3-II and Beclin-1 were both up-regulated in QSG7701/core group than in QSG7701/pcDNA3.1 group and QSG7701 group (P < 0.05). The results indicated that HCV core protein might improve the cell autophagy of QSG7701 cells through regulating LC3-II and Beclin-1 expression.
Figure 3.
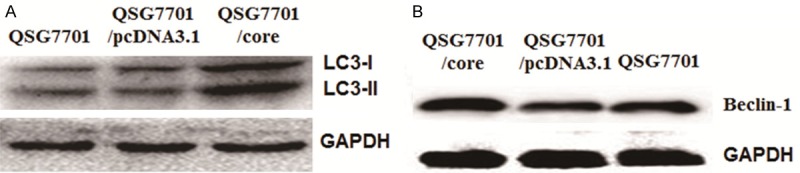
Expression of LC3-II and Beclin-1 protein in QSG7701 cell line transfecting with pIRES-core, pcDNA3.1 plasmid and without any plasmid. A. Western blot to detect the expression of LC3-II protein in QSG7701 cell line with different plasmids. GAPDH protein as the internal control. B. Western blot to detect the expression of Beclin-1protein in QSG7701 cell line with different plasmids. GAPDH protein as the internal control.
Table 2.
Expression of LC3-II and Beclin-1 protein in QSG7701 cell lines with transfecting pIRES-core, pcDNA3.1 and without any plasmid
| Group | LC3-I | LC3-II | Beclin1 |
|---|---|---|---|
| QSG7701 | 1745.23±432.65 | 6013.43±436.32 | 0.17±0.04 |
| QSG7701/pcDNA3.1 | 2134.46±384.32 | 6146.65±520.58 | 0.21±0.03 |
| QSG7701/core | 6023.54±629.43**,## | 13423.54±732.78**,## | 0.92±0.11**,## |
Compared with QSG7701 group;
P < 0.01.
Compared with QSG7701/pcDNA3.1 group;
P < 0.01.
Discussion
It is still unclear about HCV pathogenesis. It is validated that HCV proteins may be important factors to cause cell cancer as HCV nucleic acid sequence is not integrated into the host DNA, while HCV is detected to infect and replicate in liver cancer tissues [13,14]. Levine et al. believed that the pathogenesis of HCV core protein may be associated with liver cell apoptosis and autophagy [15]. And in the regulation of apoptosis, NF-κB signaling pathway is a classic one involved in the processes. It was demonstrated that HCV core protein activated NF-κB to inhibit hepatocytes apoptosis through binding with the cytoplasmic portion of tumor necrosis factor receptor l (TNFRl) to activate IΚB Kinase (IKK) and accelerate IΚB (including IκBα and IΚBΒ) degradation [16]. Marusaua et al found the apoptosis effect induced by Fas and tumor necrosis factor (TNF) significantly decreased after transfection with HCV core protein, which was dependent on the increase of NF-κB transcription [17]. NF-κB expression was down-regulated in HCV infected hepatocytes when exposure to plasmacytoid dendritic cells (PDCS), and the decreased expression of NF-κB can enhance the TNF-α-induced cell death [18]. Above studies indicated that HCV core protein can regulate NF-κB signaling pathway that is highly related to the apoptosis.
Autophagy is the process that cell degrades damaged organelles and macromolecules through utilizing its own lysosome. It was found that autophagy played important roles when body was infected by pathogens [19,20]. As an important way to against pathogens in body, autophagy can effectively activate innate and acquired immune response and maintain the normal function of the immune cells, which effectively degrades pathogenic microorganisms [21]. Actually, pathogenic microorganisms can utilize autophagy to increase self-replication in some times. Improvement of autophagy may promote the proliferation of pathogenic microorganisms. It is still unclear about the relationship between infection and autophagy. LC3 located in the surface of autophagosome, which participated in the formation of autophagosome [22,23]. LC3-II binds and locates in the autophagic membranes, and amount of LC3-II represents is proportional to the number of autophagosome [24-26]. So the reaction level of autophagic cells can be inferred by detecting LC3-II expression in host cells. Through unknown ways of accelerating assembling and aggregation of autophagic vacuoles, Beclin-1 can promote the formation of autophagosome [27]. Granato suggested that over-expression of Beclin-1 can inhibit the replication of cancer cells in breast cancer [24]. Ying found that Beclin-1 was down-regulated in epithelial cells of ovarian cancer, while Beclin-1 was highly expressed in normal mammary epithelial cells [28]. These indicated that Beclin-l played an inhibitory roles in tumor replication. Li shown that HCV replication will be significantly reduced when Beclin-1 expressed was repressed [29]. There is no clear report about the relationship between autophagy and HCV infection.
In this study, we established stable cell lines to express HCV core protein based on human derived non-tumor liver cell line (QSG7701), which can truly simulate the natural state of HCV infection. Through FCM with Annexin V-FITC/PI double stain, the apoptosis rate in QSG7701/core group was significantly lower than in QSG7701/pcDNA3.1 group and QSG7701 group (P < 0.05), which indicated that HCV core protein can inhibit the apoptosis of QSG7701 cells. For NF-κB, its expression was significantly up-regulated in QSG7701/core group than in QSG7701/pcDNA3.1 group and QSG7701 group, and the apoptosis was repressed at the same time. It can be inferred that NF-κB expression was negatively correlated to apoptosis and HCV core protein may repress hepatocytes apoptosis by up-regulating NF-κB. As to the autophagy, LC3-II and Beclin-1 expression was up-regulated in QSG7701/core group than in QSG7701/pcDNA3.1 and QSG7701 groups, which indicated that HCV core protein can enhance hepatocytes autophagy through up-regulating Beclin-1.
Based on the results about hepatocytes apoptosis and autophagy, we believe that HCV core protein can promote the proliferation of host cells through interacting with multiple signaling pathways within host cells. The body will quickly respond to induce apoptosis associated signaling pathways, which inhibits cell proliferation by HCV infection to maintain cellular homeostasis. We found that HCV core protein up-regulated NF-κB expression to repress the response of host cells, which prolonged survival of host cells and benefitted to sustained replication, spread and persistent infection of HCV, cell cycle abnormalities, Dysregulation of growth, and cell differentiation and malignant transformation. At the same time HCV core protein can also increase Beclin-1 expression to enhance autophagy. Improvement of autophagy can help the body to eliminate pathogens. But in other hands, HCV can accelerate self-replication through autophagy, which led to chronic inflammation, cirrhosis and malignant transformation of hepatocytes.
Through the investigation the influence on apoptosis and autophagy of hepatocytes induced by HCV core protein, we further revealed the mechanisms of HCV pathogenesis and the relationship with apoptosis and autophagy. The results in this study may present indication about clinical treatment for cancer, degenerative diseases and pathogen infection.
Disclosure of conflict of interest
None.
References
- 1.Li HF, Wang XA, Xiang SS, Hu YP, Jiang L, Shu YJ, Li ML, Wu XS, Zhang F, Ye YY, Weng H, Bao RF, Cao Y, Lu W, Dong Q, Liu YB. Oleanolic acid induces mitochondrial-dependent apoptosis and G0/G1 phase arrest in gallbladder cancer cells. Drug Des Devel Ther. 2015;9:3017–3030. doi: 10.2147/DDDT.S84448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X, Sun J, Hu S, Liu J. Icariin Induced B16 Melanoma Tumor Cells Apoptosis, Suppressed Tumor Growth and Metastasis. Iran J Public Health. 2014;43:847–848. [PMC free article] [PubMed] [Google Scholar]
- 3.Yang B, Lu Y, Zhang A, Zhou A, Zhang L, Zhang L, Gao L, Zang Y, Tang X, Sun L. Doxycycline Induces Apoptosis and Inhibits Proliferation and Invasion of Human Cervical Carcinoma Stem Cells. PLoS One. 2015;10:e0129138. doi: 10.1371/journal.pone.0129138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasprzak A, Adamek A. Role of hepatitis C virus proteins (C, NS3, NS5A) in hepatic oncogenesis. Hepatol Res. 2008;38:1–26. doi: 10.1111/j.1872-034X.2007.00261.x. [DOI] [PubMed] [Google Scholar]
- 5.Gu ZT, Li L, Wu F, Zhao P, Yang H, Liu YS, Geng Y, Zhao M, Su L. Heat stress induced apoptosis is triggered by transcription-independent p53, Ca(2+) dyshomeostasis and the subsequent Bax mitochondrial translocation. Sci Rep. 2015;5:11497. doi: 10.1038/srep11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sikdar S, Mukherjee A, Khuda-Bukhsh AR. Anti-lung cancer potential of pure esteric-glycoside condurangogenin A against nonsmall-cell lung cancer cells in vitro via p21/p53 mediated cell cycle modulation and DNA damage-induced apoptosis. Pharmacogn Mag. 2015;11:S73–85. doi: 10.4103/0973-1296.157698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang JY, Lin MT, Zhou MJ, Yi T, Tang YN, Tang SL, Yang ZJ, Zhao ZZ, Chen HB. Combinational Treatment of Curcumin and Quercetin against Gastric Cancer MGC-803 Cells in Vitro. Molecules. 2015;20:11524–11534. doi: 10.3390/molecules200611524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren SX, Shen J, Cheng AS, Lu L, Chan RL, Li ZJ, Wang XJ, Wong CC, Zhang L, Ng SS, Chan FL, Chan FK, Yu J, Sung JJ, Wu WK, Cho CH. Correction: FK-16 Derived from the Anticancer Peptide LL-37 Induces Caspase-Independent Apoptosis and Autophagic Cell Death in Colon Cancer Cells. PLoS One. 2015;10:e0131750. doi: 10.1371/journal.pone.0131750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao HJ, Liu PF, Li PW, Huang ZY, Yu FB, Lei T, Chen Y, Cheng Y, Mu QC, Huang HY. Ligustrazine monomer against cerebral ischemia/reperfusion injury. Neural Regen Res. 2015;10:832–840. doi: 10.4103/1673-5374.156991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramachandran A, Lebofsky M, Yan HM, Weinman SA, Jaeschke H. Hepatitis C virus structural proteins can exacerbate or ameliorate acetaminophen-induced liver injury in mice. Arch Toxicol. 2015;89:773–783. doi: 10.1007/s00204-015-1498-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruggieri V, Mazzoccoli C, Pazienza V, Andriulli A, Capitanio N, Piccoli C. Hepatitis C virus, mitochondria and auto/mitophagy: exploiting a host defense mechanism. World J Gastroenterol. 2014;20:2624–2633. doi: 10.3748/wjg.v20.i10.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka N, Moriya K, Kiyosawa K, Koike K, Aoyama T. Hepatitis C virus core protein induces spontaneous and persistent activation of peroxisome proliferator-activated receptor alpha in transgenic mice: implications for HCV-associated hepatocarcinogenesis. Int J Cancer. 2008;122:124–131. doi: 10.1002/ijc.23056. [DOI] [PubMed] [Google Scholar]
- 14.Mutso M, Nikonov A, Pihlak A, Zusinaite E, Viru L, Selyutina A, Reintamm T, Kelve M, Saarma M, Karelson M, Merits A. RNA Interference-Guided Targeting of Hepatitis C Virus Replication with Antisense Locked Nucleic Acid-Based Oligonucleotides Containing 8-oxo-dG Modifications. PLoS One. 2015;10:e0128686. doi: 10.1371/journal.pone.0128686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Developmental Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 16.Park J, Kang W, Ryu SW, Kim WI, Chang DY, Lee DH, Park do Y, Choi YH, Choi K, Shin EC, Choi C. Hepatitis C virus infection enhances TNFalpha-induced cell death via suppression of NF-kappaB. Hepatology. 2012;56:831–840. doi: 10.1002/hep.25726. [DOI] [PubMed] [Google Scholar]
- 17.Marusawa H, Hijikata M, Chiba T, Shimotohno K. Hepatitis C virus core protein inhibits Fas- and tumor necrosis factor alpha-mediated apoptosis via NF-kappaB activation. J Virol. 1999;73:4713–4720. doi: 10.1128/jvi.73.6.4713-4720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dental C, Florentin J, Aouar B, Gondois-Rey F, Durantel D, Baumert TF, Nunes JA, Olive D, Hirsch I, Stranska R. Hepatitis C virus fails to activate NF-kappaB signaling in plasmacytoid dendritic cells. J Virol. 2012;86:1090–1096. doi: 10.1128/JVI.05444-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puri P, Chandra A. Autophagy modulation as a potential therapeutic target for liver diseases. J Clin Exp Hepatol. 2014;4:51–59. doi: 10.1016/j.jceh.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, James Ou JH. Hepatitis C virus and autophagy. Biol Chem. 2015;396:1215–22. doi: 10.1515/hsz-2015-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K. Autophagy and apoptosis in liver injury. Cell Cycle. 2015;14:1631–1642. doi: 10.1080/15384101.2015.1038685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Granato M, Lacconi V, Peddis M, Di Renzo L, Valia S, Rivanera D, Antonelli G, Frati L, Faggioni A, Cirone M. Hepatitis C virus present in the sera of infected patients interferes with the autophagic process of monocytes impairing their in-vitro differentiation into dendritic cells. Biochim Biophys Acta. 2014;1843:1348–1355. doi: 10.1016/j.bbamcr.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Mohl BP, Tedbury PR, Griffin S, Harris M. Hepatitis C virus-induced autophagy is independent of the unfolded protein response. J Virol. 2012;86:10724–10732. doi: 10.1128/JVI.01667-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das GC, Hollinger FB. Molecular pathways for glucose homeostasis, insulin signaling and autophagy in hepatitis C virus induced insulin resistance in a cellular model. Virology. 2012;434:5–17. doi: 10.1016/j.virol.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Sun T, Li X, Zhang P, Chen WD, Zhang HL, Li DD, Deng R, Qian XJ, Jiao L, Ji J, Li YT, Wu RY, Yu Y, Feng GK, Zhu XF. Acetylation of Beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat Commun. 2015;6:7215. doi: 10.1038/ncomms8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ying H, Qu D, Liu C, Ying T, Lv J, Jin S, Xu H. Chemoresistance is associated with Beclin-1 and PTEN expression in epithelial ovarian cancers. Oncol Lett. 2015;9:1759–1763. doi: 10.3892/ol.2015.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Chen B, Wu Y, Jin F, Xia Y, Liu X. Genetic and epigenetic silencing of the beclin 1 gene in sporadic breast tumors. BMC Cancer. 2010;10:98. doi: 10.1186/1471-2407-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]


