Abstract
Results of published studies on the association between the miR-146a rs2910164 polymorphism and the risk of hepatocellular carcinoma (HCC) were inclusive. We performed a meta-analysis. A literature research was conducted using PubMed, Cochrane Library, Ovid, Embase, Wanfang and China National Knowledge Infrastructure (CNKI) databases, to identify studies. Statistical analyses were conducted in STATA version 11.0 (Stata Corporation, College station, TX, USA). A total of 12 publications were included in this meta-analysis. The results of this meta-analysis suggested that miR-146a rs2910164 was associated with an increased risk of HCC (OR = 1.09, 95% CI = 1.00-1.19). In sensitivity analysis, the result was still positive when excluding the studies without HWE (OR = 1.12, 95% CI = 1.01-1.23). In conclusion, this meta-analysis suggested a significant association between miR-146a rs2910164 polymorphism and HCC risk.
Keywords: Hepatocellular carcinoma, miR-146a, meta-analysis
Introduction
Hepatocellular carcinoma (HCC) accounts for 90% of cases of primary liver cancer and is the fifth most common cancer worldwide. It ranks third in mortality after gastric cancer and esophageal cancer, and half of the deaths from HCC occur in China [1]. Due to the high rate of recurrence or intrahepatic metastasis after curative resection, the overall prognosis of HCC patients remains poor despite obvious improvements in surgical techniques and perioperative management [2]. Accordingly, the development of effective therapeutic targets for HCC is urgently required.
MicroRNAs (miRNAs) are non-coding RNAs formed of 18-25 nucleotides that can cause the inhibition of gene expression at a post-transcriptional level by directly binding to the 3’-untranslational region (UTR) of mRNAs [3]. Deregulation of miRNAs, such as miR-204, miR-331, miR-125b, and miR-146a, has been demonstrated to play an important role in hepatocellular carcinoma [4,5]. Huang et al. found that miR-146a plays a vital role in the cell growth and apoptosis of HCC cells and inducing miR-146a level might be a critical targeted molecular therapy strategy for HCC [6]. Rong et al. suggested that miR-146a expression in HCC tissues was lower compared with that in adjacent non-cancerous hepatic tissues. MiR-146a expression was also related to clinical TNM stage, metastasis, portal vein tumor embolus, and number of tumor nodes [7].
The location of rs2910164 is the stem region opposite to the mature miR-146a sequence, which results in a change from G:U pair to C:U mismatch in the stem structure of the miR-146a precursor. It has previously been shown that SNPs rs2910164 in miR-146a were associated with an increased susceptibility to HCC in an Asian population. However, the results were inconsistent [8-19]. Thus, we did a meta-analysis to assess the association between rs2910164 and the risk of HCC.
Methods
Publication search
A literature research was conducted using PubMed, Cochrane Library, Ovid, Embase, Wanfang and China National Knowledge Infrastructure (CNKI) databases, to identify studies published prior to Jan 2015. Relevant studies were identified using the terms: “miR-146a or MicroRNA-146a” and “polymorphisms or variant” and “hepatocellular carcinoma or HCC”. The search was confined to humans. A manual search of references of the original articles related with this topic was used to identify additional studies. If the data or data subsets were published in more than one paper, only the paper with the largest sample size was enrolled.
Inclusion and exclusion criteria
Studies were selected for meta-analysis if they met the inclusion criteria as follows: (1) case-control study design; (2) studies that investigated the association between the miR-146a rs2910164 and the risk of HCC; (3) study subjects were HCC patients confirmed by histopathology in case group. The exclusion criteria were: (1) reviews and summaries; (2) repetitive publications; (3) no raw data of the miR-146a rs2910164 genotype.
Data extraction
Data from published studies were extracted by two authors. For each study, we collected the following information: first author, year of publication, country, ethnicity, numbers of cases and controls, and evidence of HWE.
Statistical analysis
The overall effect was measured by ORs with its 95% CI. The significance of the pooled ORs was determined by the Z test with a P value less than 0.05 considering statistically significant. The dominant model was examined to assess this association. Between-studies heterogeneity was assessed by the I2 test and the Q-statistic test. A random effects model was applied if there was heterogeneity (P < 0.05 or I2 > 50%), otherwise, a fixed effects model was employed. The publication bias was estimated by visual funnel plot inspection. To assess whether our results were substantially influenced by the presence of any individual study, we proceed a sensitivity analysis by removing the studies without HWE. Statistical analyses were conducted in STATA version 11.0 (Stata Corporation, College station, TX, USA). All the tests were two-sided.
Results
Study characteristics
According to the inclusion and exclusion criteria, a total of 12 publications were included in this meta-analysis. The 12 selected studies contained a total of 4172 HCC patients and 4901 healthy controls. Of the 12 studies, 11 studies were performed in Asians; only 1 study was performed in Caucasians. The sample sizes of the studies varied between 200-1995. Characteristics in this meta-analysis are summarized in Table 1.
Table 1.
Characteristics of included studies
| Author | Year | Country | Ethnicity | Cases | Controls | HWE |
|---|---|---|---|---|---|---|
| Xu | 2008 | China | Asian | 479 | 504 | Yes |
| Akkiz | 2011 | Turkey | Caucasian | 222 | 222 | Yes |
| Zhang | 2011 | China | Asian | 926 | 840 | Yes |
| Zhou | 2012 | China | Asian | 186 | 483 | Yes |
| Xiang | 2012 | China | Asian | 100 | 100 | Yes |
| Kim | 2012 | Korea | Asian | 159 | 201 | Yes |
| Zhang | 2013 | China | Asian | 997 | 998 | Yes |
| Shan | 2013 | China | Asian | 172 | 185 | No |
| Kou | 2014 | China | Asian | 271 | 532 | No |
| Chu | 2014 | China | Asian | 188 | 337 | Yes |
| Cong | 2014 | China | Asian | 206 | 218 | Yes |
| Zhou | 2014 | China | Asian | 266 | 281 | No |
HWE, Hardy-Weinberg equilibrium.
Results of meta-analysis
Heterogeneity test revealed that no heterogeneity existed under allele and dominant models, and thus a fixed-effect model was used (P = 0.29). The results of this meta-analysis suggested that miR-146a rs2910164 was associated with an increased risk of HCC (OR = 1.09, 95% CI = 1.00-1.19; Figure 1). Subgroup analysis based on ethnicity indicated that the miR-146a rs2910164 increased the risk of HCC in Asian population (OR = 1.09, 95% CI = 1.00-1.19). In sensitivity analysis, the result was still positive when excluding the studies without HWE (OR = 1.12, 95% CI = 1.01-1.23; Figure 2). The funnel plot is symmetrical, suggesting no publication bias (Figure 3). Egger test further verified that no publication bias existed (P = 0.56).
Figure 1.
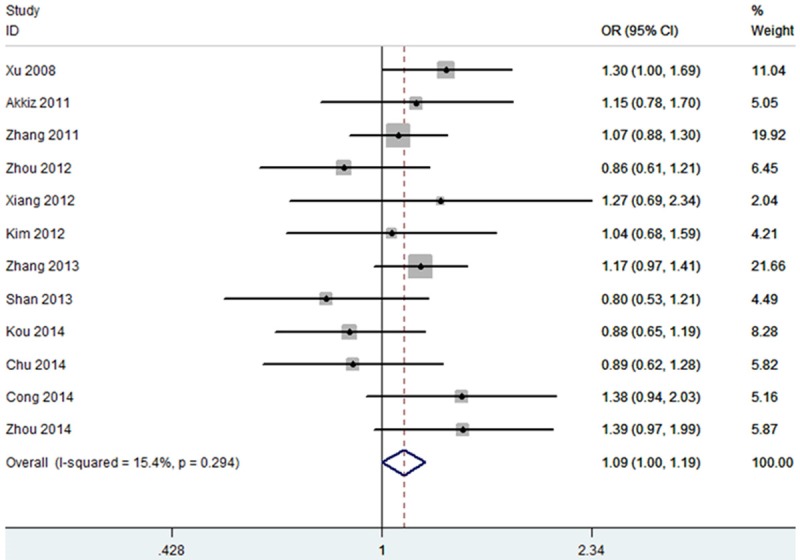
Meta-analysis for the association between miR-146a rs2910164 and HCC risk.
Figure 2.
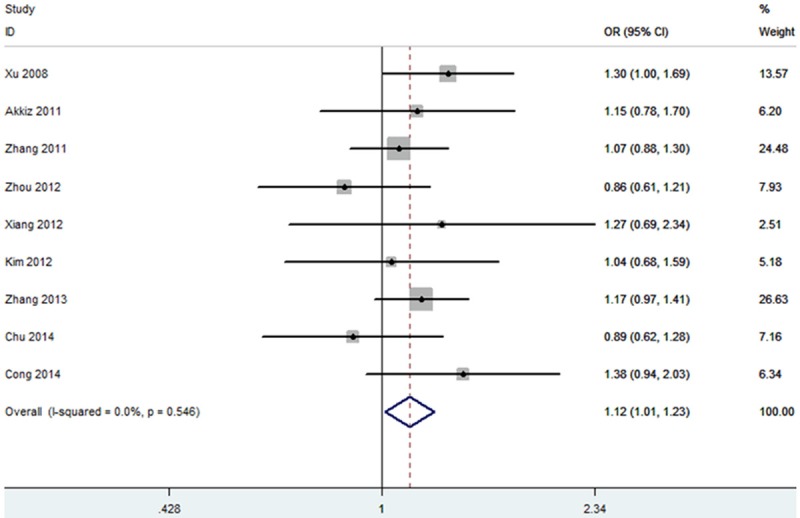
Sensitive analysis for the association between miR-146a rs2910164 and HCC risk.
Figure 3.
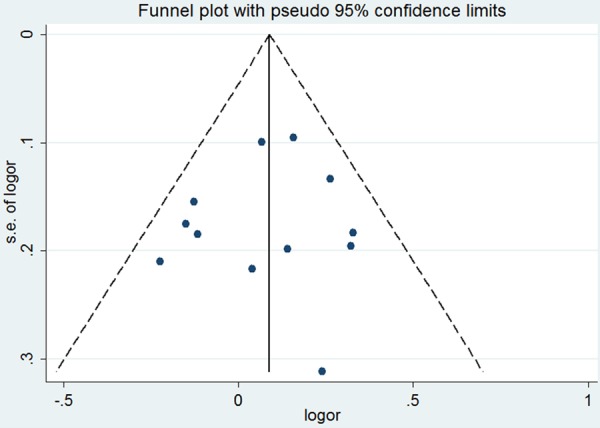
Funnel plot for the association between miR-146a rs2910164 and HCC risk.
Discussion
In the present study, we found that miR-146a rs2910164 was associated with an increased risk of HCC. In addition, we also noticed that miR-146a rs2910164 increased the risk of HCC in Asian population. However, only one study used Caucasians. Thus, more studies are needed to confirm the result of this meta-analysis.
HCC represents a major form of primary liver malignancy in adults worldwide. The tumorigenesis and development of HCC is typical of a multistage process. Major risk factors for HCC include infection with HBV or HCV, alcoholic liver disease, and most probably nonalcoholic fatty liver disease [20]. The progression is considered to deregulate genes that are critical to biological cellular procedures such as cell cycle control, apoptosis, cell migration, and metastasis [21]. However, the sensitivity and specificity of these markers remain imperfect [22]. Therefore, new biomarkers are needed to help to understand the causes of hepatocarcinogenesis and to predict response possibilities towards different therapeutic methods.
MiR-146a rs2910164 polymorphism which locates in the passenger strand of miR-146a, can disturb the secondary structure and maturation of miR-146a [8]. Xie et al. suggested that digestive tract neoplasms might associate with miR-146a variant [23]. Sun and coworkers indicated that up-regulation of miR-146a expression in tissues was related to carcinogenesis and deterioration of papillary thyroid carcinoma [24].
In conclusion, this meta-analysis suggested a significant association between miR-146a rs2910164 polymorphism and HCC risk.
Disclosure of conflict of interest
None.
References
- 1.Nordenstedt H, White DL, El-Serag HB. The changing pattern of epidemiology in hepatocellular carcinoma. Dig Liver Dis. 2010;42(Suppl 3):S206–14. doi: 10.1016/S1590-8658(10)60507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844–55. doi: 10.1136/gutjnl-2013-306627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang X, Liang M, Dittmar R, Wang L. Extracellular microRNAs in urologic malignancies: chances and challenges. Int J Mol Sci. 2013;14:14785–99. doi: 10.3390/ijms140714785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui ZH, Shen SQ, Chen ZB, Hu C. Growth inhibition of hepatocellular carcinoma tumor endothelial cells by miR-204-3p and underlying mechanism. World J Gastroenterol. 2014;20:5493–504. doi: 10.3748/wjg.v20.i18.5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsang FH, Au V, Lu WJ, Shek FH, Liu AM, Luk JM, Fan ST, Poon RT, Lee NP. Prognostic marker microRNA-125b inhibits tumorigenic properties of hepatocellular carcinoma cells via suppressing tumorigenic molecule eIF5A2. Dig Dis Sci. 2014;59:2477–87. doi: 10.1007/s10620-014-3184-5. [DOI] [PubMed] [Google Scholar]
- 6.Huang S, He R, Rong M, Dang Y, Chen G. Synergistic effect of MiR-146a mimic and cetuximab on hepatocellular carcinoma cells. Biomed Res Int. 2014;2014:384121. doi: 10.1155/2014/384121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rong M, He R, Dang Y, Chen G. Expression and clinicopathological significance of miR-146a in hepatocellular carcinoma tissues. Ups J Med Sci. 2014;119:19–24. doi: 10.3109/03009734.2013.856970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akkiz H, Bayram S, Bekar A, kgöllü E, Usküdar O, Sandıkçı M. No association of premicroRNA-146a rs2910164 polymorphism and risk of hepatocellular carcinoma development in Turkish population: a case-control study. Gene. 2011;486:104–109. doi: 10.1016/j.gene.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge YY, Yang JR, Su H, Zhuang SM. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29:2126–2131. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- 10.Zhang XW, Pan SD, Feng YL, Liu JB, Dong J, Zhang YX, Chen JG, Hu ZB, Shen HB. [Relationship between genetic polymorphism in microRNAs precursor and genetic predisposition of hepatocellular carcinoma] . Zhonghua Yu Fang Yi Xue Za Zhi. 2011;45:239–243. [PubMed] [Google Scholar]
- 11.Zhou J, Lv R, Song X, Li D, Hu X, Ying B, Wei Y, Wang L. Association between two genetic variants in miRNA and primary liver cancer risk in the Chinese population. DNA Cell Biol. 2012;31:524–530. doi: 10.1089/dna.2011.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Wang R, Ma YY, Chen LQ, Jin BH, Yu H, Wang JC, Gao CF, Liu J. Association between single nucleotide polymorphisms in miRNA196a-2 and miRNA146a and susceptibility to hepatocellular carcinoma in a Chinese population. Asian Pac J Cancer Prev. 2013;14:6427–6431. doi: 10.7314/apjcp.2013.14.11.6427. [DOI] [PubMed] [Google Scholar]
- 13.Shan YF, Huang YH, Chen ZK, Huang KT, Zhou MT, Shi HQ, Song QT, Yu ZP, Deng AM, Zhang QY. miR-499A>G rs3746444 and miR-146a G>C expression and hepatocellular carcinoma risk in the Chinese population. Genet Mol Res. 2013;12:5365–5371. doi: 10.4238/2013.November.7.11. [DOI] [PubMed] [Google Scholar]
- 14.Kou JT, Fan H, Han D, Li L, Li P, Zhu J, Ma J, Zhang ZH, He Q. Association between four common microRNA polymorphisms and the risk of hepatocellular carcinoma and HBV infection. Oncol Lett. 2014;8:1255–1260. doi: 10.3892/ol.2014.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu YH, Hsieh MJ, Chiou HL, Liou YS, Yang CC, Yang SF, Kuo WH. MicroRNA gene polymorphisms and environmental factors increase patient susceptibility to hepatocellular carcinoma. PLoS One. 2014;9:e89930. doi: 10.1371/journal.pone.0089930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong N, Chen H, Bu WZ, Li JP, Liu N, Song JL. miR-146a G>C polymorphisms and risk of hepatocellular carcinoma in a Chinese population. Tumour Biol. 2014;35:5669–5673. doi: 10.1007/s13277-014-1750-2. [DOI] [PubMed] [Google Scholar]
- 17.Zhou B, Dong LP, Jing XY, Li JS, Yang SJ, Wang JP, Zhao LF. Association between miR-146aG>C and miR-196a2C>T polymorphisms and the risk of hepatocellular carcinoma in a Chinese population. Tumour Biol. 2014;35:7775–7780. doi: 10.1007/s13277-014-2020-z. [DOI] [PubMed] [Google Scholar]
- 18.Kim WH, Min KT, Jeon YJ, Kwon CI, Ko KH, Park PW, Hong SP, Rim KS, Kwon SW, Hwang SG, Kim NK. Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene. 2012;504:92–97. doi: 10.1016/j.gene.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Xiang Y, Fan S, Cao J, Huang S, Zhang LP. Association of the microRNA-499 variants with susceptibility to hepatocellular carcinoma in a Chinese population. Mol Biol Rep. 2012;39:7019–7023. doi: 10.1007/s11033-012-1532-0. [DOI] [PubMed] [Google Scholar]
- 20.Kudo M. Hepatocellular carcinoma in 2011 and beyond: from the pathogenesis to molecular targeted therapy. Oncology. 2011;81(Suppl 1):1–10. doi: 10.1159/000333252. [DOI] [PubMed] [Google Scholar]
- 21.Hu L, Chen G, Yu H, Qiu X. Clinicopathological significance of RASSF1A reduced expression and hypermethylation in hepatocellular carcinoma. Hepatol Int. 2010;4:423–32. doi: 10.1007/s12072-010-9164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48:2047–63. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- 23.Xie M, Li Y, Wu J, Wu J. A risk of digestive tract neoplasms susceptibility in miR-146a and miR-196a2. Fam Cancer. 2015;14:229–39. doi: 10.1007/s10689-014-9776-6. [DOI] [PubMed] [Google Scholar]
- 24.Sun M, Fang S, Li W, Li C, Wang L, Wang F, Wang Y. Associations of miR-146a and miR-146b expression and clinical characteristics in papillary thyroid carcinoma. Cancer Biomark. 2015;15:33–40. doi: 10.3233/CBM-140431. [DOI] [PubMed] [Google Scholar]


