Abstract
The macrophage migration inhibitory factor (MIF) -173G/C gene polymorphism has been implicated in the susceptibility to cancer, but the results are not conclusive. So the aim of study to investigate the association between MIF -173G/C gene polymorphism and cancer risk by a comprehensive meta-analysis. We searched the PubMed, Embase, Wanfang and China National Knowledge Internet (CNKI) databases, with the last updated search being performed on May 24, 2015. The odds ratio (OR) and 95% confidence interval (95% CI) were used to assess the association. Statistical analysis was performed by STATA 11.0 software. Finally, 7,253 participants from 15 studies were included in the meta-analysis. The results of meta-analysis indicated the significant association between MIF -173G/C gene polymorphism and cancer susceptibility, especially in Asians (C vs. G, OR: 1.22, 95% CI=1.00-1.50). In addition, the significant relationship between MIF -173G/C gene polymorphism and gastrointestinal tumors (CC+CG vs. GG, OR: 1.25, 95% CI=1.05-1.50), hematological malignancy (CC+CG vs. GG, OR: 1.27, 95% CI=1.03-1.56), gynecolgical tumors (CC vs. CG+GG, OR: 1.51, 95% CI=1.04-2.19) risk was found. However, to avoid the “false positive report”, we investigated the significant associations observed in the present meta-analysis by the false positive report probabilities (FPRPs) test. Interestingly, the results of FPRP test indicated the MIF -173G/C gene polymorphism only associated with gastrointestinal cancer and hematological malignancy risk (FPRP=0.132, 0.067 respectively) at the level of a prior probability is 0.1. Therefore, the meta-analysis suggested MIF -173G/C gene polymorphism would be a risk factor for the gastrointestinal cancer and hematological malignancy.
Keywords: Cancer, MIF, polymorphism, susceptibility, meta-analysis, FPRP
Introduction
Cancer is still a major cause of death in the world. The previous studies found that many risk factors may play important role in the pathogenesis of cancer including age, gender, life-style and environmental pollution [1]. Additionally, lots of studies focused on investigating the association between gene variants and malignant tumor susceptibility. In recent years, the macrophage migration inhibitory factor (MIF) gene which is located on chromosome 22q11.2 has been widely studied. The MIF was first found in 1950s, and it was defined as a soluble factor produced by T-lymphocytes which could inhibit the directed migration of macrophages [2]. Subsequently, many other studies suggested the MIF was also expressed on anterior pituitary cells, monocytes, eosinophils and epithelial cells etc. [3-6]. Currently, the MIF was considered a pleiotropic cytokine, and it played a major role in innate immune response. In addition, the MIF also acted as an important regulator for many other inflammatory cytokines, such as interleukin (IL)-2, IL-4 and interferon (IFN)-γ [7,8]. Furthermore, the MIF has been found that it played a critical role in the regulation of antitumor T-lymphocytes [9].
One important polymorphism named -173G/C (rs755622) has recently been indentified in MIF gene which involves a G→C substitution at base pair 173 of the 50-flanking region [10]. Previous studies indicated that the MIF -173G/C polymorphism was associated with risk of peptic ulcer diseases, systemic lupus erythematosus (SLE), polycystic ovary syndrome (PCOS) and rheumatoid arthritis (RA) [11-14]. Interestingly, a growing number of evidences suggested that MIF -173G/C polymorphism played an important role in the pathogenesis of cancer. Ramireddy et al. found the MIF -173G/C polymorphism was associated with colorectal cancer and acute myelocytic leukemia (AML) susceptibility [15,16], Yuan and colleagues reported the MIF -173G/C polymorphism could increase the risk of bladder cancer [17]. The MIF -173G/C polymorphisms may be associated with a higher risk of prostate cancer in Chinese [18]. However, there is no relationship between the MIF -173G/C polymorphism and risk of cervical cancer in Yuan’s study [19].
Due to these inconclusive reports, we performed a meta-analysis to investigate the association of the MIF -173G/C polymorphism with risk of cancer. Because the meta-analysis uses a quantitative method to combine the results from different studies with the same topic, so it is a useful technique for investigating the risk factors of genetic diseases, and can provide more reliable conclusions. To our knowledge, this is the most recent meta-analysis was conducted to assess the association between the MIF -173G/C polymorphism and cancer susceptibility.
Materials and methods
Study selection
A systematic literature search in PubMed, Embase, Wanfang Database and China National Knowledge Internet (CNKI) were carried out to identify studies involving the association between the MIF -173G/C polymorphism and cancer risk on May 24, 2015. The key words were as follows: (‘macrophage migration inhibitory factor’ or ‘MIF’) and (‘cancer’ or ‘malignancy’ or ‘tumor’, ‘neoplasm’ or ‘cancinoma’ or ‘leukemia’ or ‘myeloma’ or ‘sarcoma’ or ‘lymphoma’) and ‘polymorphism’ or ‘variant’ or ‘mutation’). There is no language restriction.
The inclusion criteria were defined as follows: (1) the design had to be a case-control study; (2) studies evaluated the association between MIF gene polymorphism and malignant tumor risk; (3) the studies should be provided available data to count the odds ratio (OR) and 95% confidence interval (CI); (4) the object of study must be human. The following exclusive items were: (1) not designed as a case-control study; (2) reviews, abstracts or overlapping studies; (3) not reported the genotype frequencies or number in the studies.
Quality score assessment
The qualities of included studies were evaluated by the Newcastle-Ottawa Scale (Case control study), The Scale to assess quality based on three aspects including selection, comparability and exposure in the study. The total scores were ranged from 0 to 9. We have assessed the quality of the studies in a consensus meeting with all authors.
Date extraction
The independent reviewers (Xiang Tong and Bing Zheng) collected the each study’s data according to the inclusive criteria. If there is a disagreement, the third author (Qiaoyi Tong) would assess those articles. First author, year of publication, ethnicity, country of origin, age, sample size, genotype distribution in cases and controls, types of cancer and genotyping method were extracted from each study.
Statistical methods
The current meta-analysis was performed with the STATA 11.0 software. We used the OR and 95% CI to investigate the strength of association between MIF -173G/C gene polymorphism and risk of cancer. The χ2 based Q-test and I-squared (I2) statistics test were used to calculate heterogeneity. The pooled OR should be counted by the random-effect model when the heterogeneity was considered statistically significant (I2 > 50% and P < 0.10), otherwise the fixed-effect model was applied. The pooled OR was estimated on the association between MIF -173G/C gene polymorphism and cancer risk in gene and allele models (CC+CG vs. GG, CC vs. CG+GG, CC vs. GG, CG vs. GG and C vs. G). To evaluate the ethnicity and types of cancer-specific effect, subgroup analysis by ethnicity groups and types of cancer was carried out.
In addition, to investigate whether an association between MIF -173G/C gene polymorphism and cancer risk is “noteworthy”, we also calculated the false positive report probabilities (FPRPs) for all significant associations were found in the current meta-analysis by prior probabilities of 0.1. In the FPRP test, we set a FPRP cut-off value of 0.2 which suggested by the previous study [20], and only the results with FPRP < 0.2 were considered “noteworthy”.
Publication bias was tested by several methods. Visual inspection of asymmetry in funnel plots was carried out. Besides, the Egger’s test was also applied to assess the publication bias. Furthermore, the Hardy-Weinberg equilibrium (HWE) was assessed by the Chi-square test in control group of each study.
Results
Study characteristics
In total, 137 articles were identified after an initial search (Figure 1). After initial reading titles and abstracts, 112 articles were excluded. The remained 25 articles were further screened for full-text view. Four of them were excluded because they were assessed the other polymorphisms of MIF gene (such as +254C/T, +656C/G etc.) rather than -173G/C polymorphism, three articles were removed for they are reviews, two articles were not included since they were not designed as case-control study, and two articles were repeated. Finally, 15 case-control studies [15-19,21-29] from 14 articles were identified in the meta-analysis. Among them, 10 papers were in English [15-18,21-25,27] and 4 articles [19,26,28,29] were in Chinese. The characteristics of included studies are listed in Tables 1, 2.
Figure 1.
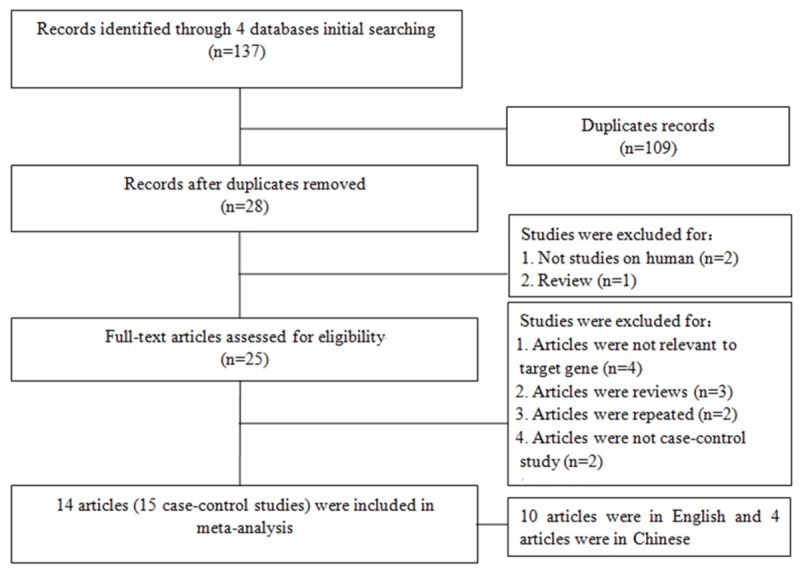
The flow diagram of included and excluded studies.
Table 1.
Characteristics of case-control studies included in meta-analysis
| Author | Year | Coutry | Ethnicity | Cases/Controls | Age | Tumor | Type |
|---|---|---|---|---|---|---|---|
| Arisawa T | 2008 | Japan | Asian | 229/428 | 63.0±10.7/54.7±18.8 | Gastric cancer | Gastrointestinal tumors |
| Cai KK | 2013 | China | Asian | 98/80 | 67.8±5.76/60.2±4.9 | Prostate cancer | Urologic tumors |
| Ding GX | 2009 | China | Asian | 259/301 | 52.0±1.5/51.6±0.8 | Prostate cancer | Urologic tumors |
| Li HX | 2012 | China | Asian | 296/319 | 44.0±16.6/44.3±15.9 | Gastric cancer | Gastrointestinal tumors |
| Meyer-Siegler KL | 2007 | America | Caucasian | 131/128 | 70.2±0.9/64.4±1.1 | Prostate cancer | Urologic tumors |
| Ramireddy L (A) | 2014 | China | Asian | 256/256 | 53.44/55.8 | AMLa | Hematological malignancies |
| Ramireddy L (C) | 2014 | China | Asian | 192/256 | 62.1/55.8 | Colorectal cancer | Gastrointestinal tumors |
| Wu S | 2011 | China | Asian | 250/147 | 49.1±9.4/48.0±10.8 | Cervical cancer | Gynecolgical tumors |
| Xue Y | 2010 | China | Asian | 346/516 | NAb | ALLc | Hematological malignancies |
| Yuan L (C) | 2012 | China | Asian | 455/447 | 46.4±8.9/45.5±9.8 | Cervical cancer | Gynecolgical tumors |
| Yuan L (O) | 2012 | China | Asian | 130/145 | 50.1±13.3/50.9±12.4 | Ovarian cancer | Gynecolgical tumors |
| Yuan QB | 2012 | China | Asian | 325/345 | NA | Bladder cancer | Urologic tumors |
| Zhou SZ (GD) | 2005 | China | Asian | 104/104 | 58.5±11.2/59.3±10.6 | Gastric cancer | Gastrointestinal tumors |
| Zhou SZ (SX) | 2005 | China | Asian | 102/102 | 59.6±10.1/61.3±9.6 | Gastric cancer | Gastrointestinal tumors |
| Ziino O | 2005 | Italy | Caucasian | 151/355 | NA | ALL | Hematological malignancies |
Acute myelocytic leukemia;
Not available;
Acute lymphocytic leukemia.
Table 2.
Distributions of MIF -173G/C genotypes in case and control group
| Author | Year | Case | Control | Method | Score | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||
| CC | CG | GG | C | G | CC | CG | GG | C | G | ||||
| Arisawa T | 2008 | 12 | 94 | 123 | 118 | 340 | 23 | 144 | 261 | 190 | 666 | PCR-SSCPd | 9 |
| Cai KK | 2013 | 18 | 43 | 37 | 79 | 117 | 6 | 32 | 42 | 44 | 116 | PCR-RFLPe | 8 |
| Ding GX | 2009 | 18 | 75 | 166 | 111 | 407 | 0 | 45 | 256 | 45 | 557 | PCR-RFLP | 8 |
| Li HX | 2012 | 27 | 101 | 168 | 155 | 437 | 12 | 114 | 193 | 138 | 500 | PCR-RFLP | 8 |
| Meyer-Siegler KL | 2007 | / | / | / | 152 | 110 | / | / | / | 57 | 199 | PCR-sequencing | 7 |
| Ramireddy L (A) | 2014 | 8 | 80 | 168 | 96 | 416 | 14 | 56 | 186 | 84 | 428 | RT-PCRf | 8 |
| Ramireddy L (C) | 2014 | 4 | 63 | 125 | 71 | 313 | 14 | 56 | 186 | 84 | 428 | RT-PCR | 8 |
| Wu S | 2011 | 91 | 117 | 42 | 299 | 201 | 39 | 68 | 40 | 146 | 148 | PCR-RFLP | 8 |
| Xue Y | 2010 | 10 | 108 | 228 | 128 | 564 | 13 | 134 | 369 | 160 | 872 | PCR-RFLP | 8 |
| Yuan L (C) | 2012 | 19 | 135 | 301 | 173 | 737 | 11 | 155 | 281 | 177 | 717 | PCR-RFLP | 8 |
| Yuan L (O) | 2012 | 1 | 40 | 89 | 42 | 218 | 4 | 61 | 80 | 69 | 221 | PCR-RFLP | 8 |
| Yuan QB | 2012 | 20 | 99 | 206 | 139 | 511 | 21 | 149 | 175 | 191 | 499 | PCR-RFLP | 7 |
| Zhou SZ (GD) | 2005 | 30 | 52 | 22 | 112 | 96 | 16 | 60 | 28 | 92 | 116 | PCR-RFLP | 8 |
| Zhou SZ (SX) | 2005 | 32 | 39 | 31 | 103 | 101 | 28 | 46 | 28 | 102 | 102 | PCR-RFLP | 8 |
| Ziino O | 2005 | 0 | 34 | 117 | 34 | 268 | 2 | 76 | 277 | 80 | 630 | DHLPCg Wave | 7 |
Polymerase chain reaction-single strand conformation polymorphism;
Polymerase chain reaction-restricted fragment length polymorphisms;
Real time-polymerase chain reaction;
Denaturing high performance liquid chromatography.
Meta-analysis results
All 3,324 cases and 3,929 controls from 14 articles were included in the meta-analysis. Except for two studies reported by Ramireddy et al. [15,16] not according with the HWE, the other studies met the HWE in the control groups. The χ2 and I2 test suggested a moderate heterogeneity (I2=80.2%, P < 0.1) in the dominant model (CC+CG vs. GG), so we used a random-effect model to investigate the pooled OR. In totally analysis, no significant association between the MIF -173G/C gene polymorphism and malignant tumor susceptibility in gene models (CC+CG vs. GG, OR: 1.21, 95% CI=0.95-1.53; CC vs. CG+GG, OR: 1.32, 95% CI=0.94-1.84; CC vs. GG, OR: 1.36, 95% CI=0.93-2.00; CG vs. GG, OR: 1.15, 95% CI=0.91-1.45). However, there is a significant association between MIF -173G/C gene polymorphism and risk of cancer in allele model (C vs. G, OR: 1.32, 95% CI=1.04-1.68, P=0.02). No publication bias was checked in either the funnel plot or the Egger’s test (t=1.33, P=0.21).
Interestingly, as the results are summarized in Table 3, the statistically significant association between the MIF -173G/C gene polymorphism and cancer risk was found in Asians (C vs. G, OR: 1.22, 95% CI=1.00-1.50), but not among Caucasians. Additionally, we also conducted the subgroup analysis by types of cancer. The results suggested the MIF -173G/C gene polymorphism has a significant associated with gastrointestinal cancer (CC+CG vs. GG, OR: 1.25, 95% CI=1.05-1.50; CG vs. GG, OR: 1.21, 95% CI=1.01-1.47; C vs. G, OR: 1.23, 95% CI=1.07-1.41) (Figure 2), hematological malignancy (CC+CG vs. GG, OR: 1.27, 95% CI=1.03-1.56; CG vs. GG, OR: 1.32, 95% CI=1.07-1.65) (Figure 2), and gynecological cancer (CC vs. CG+GG, OR: 1.51, 95% CI=1.04-2.19) risk. Unfortunately, there is no association between the MIF -173G/C gene polymorphism and urologic cancer risk.
Table 3.
Summary the results of subgroup analysis from different comparative genetic models
| Gene models | Ethnicity | Type | |||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Asians | Caucasians | Gastrointestinal | Urologic | Hematological | Gynecolgical | ||
| CC+CG vs. GG | |||||||
| ORh | 1.22 | 1.03 | 1.25 | 1.50 | 1.27 | 0.96 | |
| 95% CIi | 0.95-1.57 | 0.65-1.63 | 1.05-1.50 | 0.48-4.69 | 1.03-1.56 | 0.54-1.71 | |
| CC vs. CG+GG | |||||||
| OR | 1.33 | 0.47 | 1.35 | 2.98 | 0.71 | 1.51 | |
| 95% CI | 0.95-1.87 | 0.02-9.78 | 0.82-2.23 | 0.65-13.73 | 0.38-1.35 | 1.04-2.19 | |
| CC vs. GG | |||||||
| OR | 1.39 | 0.47 | 1.37 | 3.48 | 0.79 | 1.56 | |
| 95% CI | 0.94-2.04 | 0.02-9.91 | 0.83-2.28 | 0.55-22.19 | 0.42-1.50 | 0.73-3.30 | |
| CG vs. GG | |||||||
| OR | 1.16 | 1.06 | 1.21 | 1.29 | 1.32 | 0.91 | |
| 95% CI | 0.91-1.49 | 0.67-1.68 | 1.01-1.47 | 0.46-3.63 | 1.07-1.65 | 0.55-1.50 | |
| C vs. G | |||||||
| OR | 1.22 | 2.20 | 1.23 | 2.12 | 1.16 | 0.98 | |
| 95% CI | 1.00-1.50 | 0.47-10.30 | 1.07-1.41 | 0.82-5.50 | 0.97-1.40 | 0.63-1.53 | |
Odd ratio;
Confidence interval.
Figure 2.
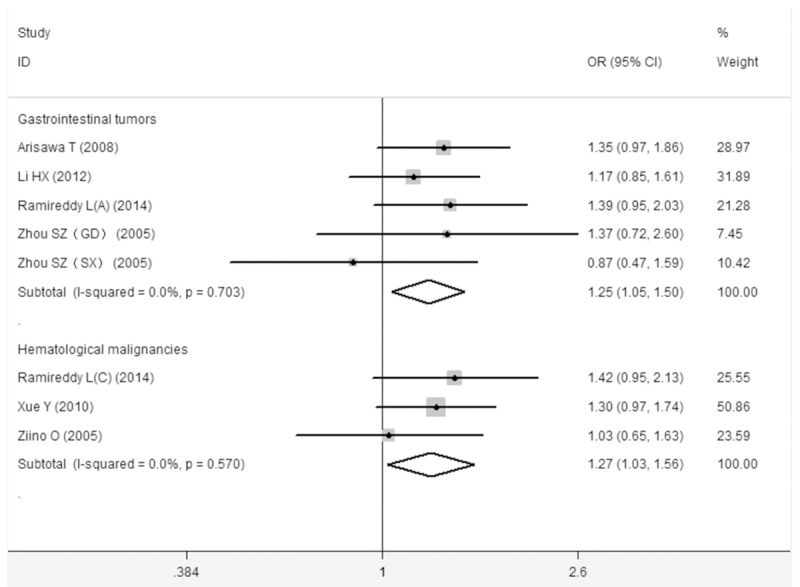
The association between the MIF -173G/C polymorphism and gastrointestinal cancer and hematological malignancy risk (CC+CG vs. GG).
FPRP test results
Furthermore, we investigated the significant associations observed in the present meta-analysis by the FPRP test. As listed in Table 4, according to the results of FPRP test, we found the MIF -173G/C gene was only associated with gastrointestinal cancer and hematological malignancy risk (FPRP=0.132, 0.067 respectively). And the significant associations of overall-group, Asians-group and gynecological cancer in the present meta-analysis were proved to be false positive at the level of a prior probability is 0.1.
Table 4.
The results of FPRP test about all significant associations observed in the meta-analysis
| Gene models | OR | 95% CI | Power | P value | Prior probability=0.1 |
|---|---|---|---|---|---|
|
| |||||
| FPRP value | |||||
| CC+CG vs. GG | |||||
| Gastrointestinal | 1.25 | 1.05-1.50 | 0.975 | 0.016 | 0.132 |
| Hematological | 1.27 | 1.03-1.56 | 0.944 | 0.023 | 0.067 |
| CC vs. CG+GG | |||||
| Gynecolgical | 1.51 | 1.04-2.19 | 0.486 | 0.030 | 0.356 |
| CG vs. GG | |||||
| Gastrointestinal | 1.21 | 1.01-1.47 | 0.985 | 0.055 | 0.334 |
| Hematological | 1.32 | 1.07-1.65 | 0.869 | 0.015 | 0.132 |
| C vs. G | |||||
| Overall | 1.32 | 1.04-1.68 | 0.851 | 0.024 | 0.203 |
| Asians | 1.22 | 1.00-1.50 | 0.975 | 0.059 | 0.354 |
| Gastrointestinal | 1.23 | 1.07-1.41 | 0.998 | 0.003 | 0.026 |
Discussion
Although a number of anti-cancer drugs are developing in recent decades, the malignancies was still the top leading cause of death worldwide. Previous studies have estimated the total size of new cancer cases is expected to increase by 29% in developed countries while an increase of 73% in developing countries, and with up to 15 million new cases in 2020 [30,31]. In addition, Rastogi et al. showed the mortality rate caused by cancer will increase about 5-fold greater in the developing countries, and the global cancer mortality is expected to increase by 104% in 2020 [32]. What and how can we do?
Lots of studies focused on the aspects of pathogenesis, influence factors and prognosis of cancer. Previous studies have suggested the risk factors including unhealthy life style, environmental pollution, radiation, infection and immunity dysfunction etc. [33-37]. Furthermore, plenty of studies paid more attention to the role of host genetic variants in mechanism of cancer [38-41]. Lots of studies have reported the association between the MIF -173G/C gene polymorphism and cancer risk [18,21,25]. However, there is no well comprehensive meta-analysis to assess the association between MIF -173G/C gene polymorphism and risk of malignant tumor until now, and we conducted a meta-analysis to investigate the precise relationship. To avoid the false positive about results of the meta-analysis, we also investigated the FPRP for all significant associations shown in the current meta-analysis by set as the prior probabilities is 0.1.
By the meta-analysis, we found the MIF -173G/C gene polymorphism could increase the risk of cancer among Asians but not in Caucasians. And the mutational heterozygote could increase the risk of gastrointestinal cancer and hematological malignancy, while the homozygote could increase the gynecological cancer susceptibility. Interestingly, we just found that the MIF -173G/C gene polymorphism actually could increase the risk of gastrointestinal cancer and hematological malignancy by the FPRP test. The results of FPRP test means the significant associations of overall-group, Asians-group and gynecological cancer observed in the present meta-analysis may be a false positive association.
The results of current study are different with the previous results [42]. The following reasons may be explained the contradictory results: (1) only 5 studies were identified in previous study, the results of previous study have insufficient power to reveal a reliable association. However, there are 14 articles were included in the present meta-analysis, our results would more accurately shown the real relationship between MIF -173G/C gene polymorphism and cancer risk; (2) except for performed the totally analysis, we also conducted the subgroup meta-analyses to reduce the specific effects from the ethnicity and types of cancer. (3) Furthermore, a large number of previous significant candidate gene association studies have turned out to be “false-positive reports” [43,44]. Therefore, to assess whether the significant associations between MIF -173G/C gene polymorphism and cancer risk is “noteworthy” in the current study, we also investigated the significant associations observed in the meta-analysis by the FPRP test. Overall, the results of present study are more close to real value.
There were several limitations of the present meta-analysis. First, only published articles were included in a few datebases, so a publication bias may have occurred. Sencond, the complexity of cancer susceptibility in most cases probably does not depend on one single factor or on one single gene variant, but rather on many gene variants or gene-environment interaction, similar to the polygenic mode of inheritance in complex disorders. However, due to lacking of sufficient data for each included study, we failed to perform further analysis the confounding factors, such as gender, gene-environment/gene-gene interactionand age which might have influence on our pooled results. Third, the included studies of meta-analysis maily from Asians, so the results possible only applicable to the Asians. There is a need to perform larger sample size studies in other ethnic groups. What’s more, the small number of participants included in the subgroup analysis, so we must be cautious when referring to the pooled results. Despite of these limitations, we minimized the likelihood of bias through the whole process by creating a detailed protocol, by performing study identification, statistical analysis and data selection, as well as in the control of publication bias. Anyway, the reliability of the results is guaranteed.
In conclusion, the present study suggested the MIF -173G/C gene polymorphism may be an independent risk to contribute the gastrointestinal cancer and hematological malignancy susceptibility. Need to more well designed studies with larger sample size focusing on ethnicities or cancer types be conducted to confirm the results in the future.
Disclosure of conflict of interest
None.
References
- 1.Bredberg A. Cancer: more of polygenic disease and less of multiple mutations? A quantitative viewpoint. Cancer. 2011;117:440–445. doi: 10.1002/cncr.25440. [DOI] [PubMed] [Google Scholar]
- 2.Bloom BR, Bennett B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science. 1966;153:80–82. doi: 10.1126/science.153.3731.80. [DOI] [PubMed] [Google Scholar]
- 3.Makita H, Nishimura M, Miyamoto K, Nakano T, Tanino Y, Hirokawa J, Nishihira J, Kawakami Y. Effect of anti-macrophage migration inhibitory factor antibody on lipopolysaccharide-induced pulmonary neutrophil accumulation. Am J Respir Crit Care Med. 1998;158:573–579. doi: 10.1164/ajrccm.158.2.9707086. [DOI] [PubMed] [Google Scholar]
- 4.Imamura K, Nishihira J, Suzuki M, Yasuda K, Sasaki S, Kusunoki Y, Tochimaru H, Takekoshi Y. Identification and immunohistochemical localization of macrophage migration inhibitory factor in human kidney. IUBMB Life. 1996;40:1233–1242. doi: 10.1080/15216549600201883. [DOI] [PubMed] [Google Scholar]
- 5.Calandra T, Bernhagen J, Mitchell RA, Bucala R. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J Exp Med. 1994;179:1895–1902. doi: 10.1084/jem.179.6.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernhagen J, Calandra T, Mitchell RA, Martin SB, Tracey KJ, Voelter W, Manogue KR, Cerami A, Bucala R. MIF is a pituitary-derived cytokine that potentiates lethal endotoxaemia. Nature. 1993;365:756–759. doi: 10.1038/365756a0. [DOI] [PubMed] [Google Scholar]
- 7.Bacher M, Metz CN, Calandra T, Mayer K, Chesney J, Lohoff M, Gemsa D, Donnelly T, Bucala R. An essential regulatory role for macrophage migration inhibitory factor in T-cell activation. Proc Natl Acad Sci U S A. 1996;93:7849–7854. doi: 10.1073/pnas.93.15.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satoskar AR, Bozza M, Sosa MR, Lin G, David JR. Migration-inhibitory factor gene-deficient mice are susceptible to cutaneous Leishmania major infection. Infect Immun. 2001;69:906–911. doi: 10.1128/IAI.69.2.906-911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abe R, Peng T, Sailors J, Bucala R, Metz CN. Regulation of the CTL response by macrophage migration inhibitory factor. J Immunol. 2001;166:747–753. doi: 10.4049/jimmunol.166.2.747. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell R, Bacher M, Bernhagen J, Pushkarskaya T, Seldin MF, Bucala R. Cloning and characterization of the gene for mouse macrophage migration inhibitory factor (MIF) J Immunol. 1995;154:3863–3870. [PubMed] [Google Scholar]
- 11.Sreih A, Ezzeddine R, Leng L, LaChance A, Yu G, Mizue Y, Subrahmanyan L, Pons-Estel BA, Abelson AK, Gunnarsson I. Dual effect of the macrophage migration inhibitory factor gene on the development and severity of human systemic lupus erythematosus. Arthritis Rheum. 2011;63:3942–3951. doi: 10.1002/art.30624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C, Qiao B, Zhan Y, Qi W, Chen ZJ. First evidence of genetic association between the MIF -173G/C single-nucleotide polymorphisms and polycystic ovary syndrome. Am J Reprod Immunol. 2011;66:416–422. doi: 10.1111/j.1600-0897.2011.01011.x. [DOI] [PubMed] [Google Scholar]
- 13.Shiroeda H, Tahara T, Shibata T, Nakamura M, Yamada H, Nomura T, Hayashi R, Saito T, Fukuyama T, Otsuka T. Functional promoter polymorphisms of macrophage migration inhibitory factor in peptic ulcer diseases. Int J Mol Med. 2010;26:707–711. doi: 10.3892/ijmm_00000517. [DOI] [PubMed] [Google Scholar]
- 14.Llamas-Covarrubias M, Valle Y, Bucala R, Navarro-Hernández R, Palafox-Sánchez C, Padilla-Gutiérrez J, Parra-Rojas I, Bernard-Medina A, Reyes-Castillo Z, Muñoz-Valle J. Macrophage migration inhibitory factor (MIF): genetic evidence for participation in early onset and early stage rheumatoid arthritis. Cytokine. 2013;61:759–765. doi: 10.1016/j.cyto.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramireddy L, Chen WT, Peng CT, Hu RM, Ke TW, Chiang HC, Chang SC, Tsai FJ, Lo WY. Association Between Genetic Polymorphism of the MIF Gene and Colorectal Cancer in Taiwan. J Clin Lab Anal. 2015;29:268–74. doi: 10.1002/jcla.21763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramireddy L, Lin CY, Liu SC, Lo WY, Hu RM, Peng YC, Peng CT. Association Study Between Macrophage Migration Inhibitory Factor-173 Polymorphism and Acute Myeloid Leukemia in Taiwan. Cell Biochem Biophys. 2014;70:1159–65. doi: 10.1007/s12013-014-0036-z. [DOI] [PubMed] [Google Scholar]
- 17.Yuan Q, Wang M, Wang M, Zhang Z, Zhang W. Macrophage migration inhibitory factor gene -173G>C polymorphism and risk of bladder cancer in southeast China: a case-control analysis. Mol Biol Rep. 2012;39:3109–3115. doi: 10.1007/s11033-011-1075-9. [DOI] [PubMed] [Google Scholar]
- 18.Ding G, Zhou S, Xu Z, Feng N, Song N, Wang X, Yang J, Zhang W, Wu H, Hua L. The association between MIF -173G>C polymorphism and prostate cancer in southern Chinese. J Surg Oncol. 2009;100:106–110. doi: 10.1002/jso.21304. [DOI] [PubMed] [Google Scholar]
- 19.Yuan L, Wang ML, Fu SL, Ding B, Zhang ZD, Han SP. Relationship of MIF-173 G/C polymorphsim and risk for cervical cancer. Jiangsu Medical Journal (Chinese) 2012;38:1930–1933. [Google Scholar]
- 20.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu S, Lian J, Tao H, Shang H, Zhang L. Correlation of macrophage migration inhibitory factor gene polymorphism with the risk of early stage cervical cancer and lymphatic metastasis. Oncol Lett. 2011;2:1261–1267. doi: 10.3892/ol.2011.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.la Cruz D. Functional promoter polymorphisms of the macrophage migration inhibitory factor gene in gastric carcinogenesis. Oncol Rep. 2008;19:223–228. [PubMed] [Google Scholar]
- 23.Li H, Zang J, Wang P, Dai L, Zhang J, Wang K. Gastric cancer susceptibility in gastric cancer relatives: attributable risks of Macrophage migration inhibitory factor promoter polymorphism and Helicobacter pylori. Cytokine. 2012;60:346–351. doi: 10.1016/j.cyto.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 24.Xue Y, Xu H, Rong L, Lu Q, Li J, Tong N, Wang M, Zhang Z, Fang Y. The MIF -173G/C polymorphism and risk of childhood acute lymphoblastic leukemia in a Chinese population. Leuk Res. 2010;34:1282–1286. doi: 10.1016/j.leukres.2010.03.030. [DOI] [PubMed] [Google Scholar]
- 25.Meyer-Siegler K, Vera P, Iczkowski K, Bifulco C, Lee A, Gregersen P, Leng L, Bucala R. Macrophage migration inhibitory factor (MIF) gene polymorphisms are associated with increased prostate cancer incidence. Genes Immun. 2007;8:646–652. doi: 10.1038/sj.gene.6364427. [DOI] [PubMed] [Google Scholar]
- 26.Zhou SZ, Hu PJ, Zeng ZR, Liao SY, Chen B, Chen MH. Association of macrophage migration inhibitory factor -173 locus polymorphsim and gastric cancers in China. Chinese Journal of Pathophysiology (Chinese) 2005;21:1132–1135. [Google Scholar]
- 27.Ziino O, D‘Urbano L, De Benedetti F, Conter V, Barisone E, De Rossi G, Basso G, Aricò M. The MIF -173G/C polymorphism does not contribute to prednisone poor response in vivo in childhood acute lymphoblastic leukemia. Leukemia. 2005;19:2346–2347. doi: 10.1038/sj.leu.2403973. [DOI] [PubMed] [Google Scholar]
- 28.Yuan L, Wang ML, Ni J, Yue C, Ding B, Zhang ZD & Han SP. Relationship of MIF-173 G/C polymorphsim and susceptibility of ovarian cancer. Jiangsu Medical Journal (Chinese) 2012;37:2791–2794. [Google Scholar]
- 29.Cai KK. Study on Expression of MIF gene in Human Prostate Cancer and Relationship between its single nucleotide polymorphisms and risk of Prostate Cancer. Tianjing Medical Universtity (Chinese) 2013 [Google Scholar]
- 30.Mathers C, Bernard C, Iburg K, Inoue M, Fat D, Shibuya K, Stein C, Parkin D, Pisani P, Ferlay J. Global cancer statistics. CA Cancer J Clin. 1999;49:33–64. doi: 10.3322/canjclin.49.1.33. [DOI] [PubMed] [Google Scholar]
- 31.Salminen E, Izewska J, Andreo P. IAEA’s role in the global management of cancer-focus on upgrading radiotherapy services. Acta Oncologica. 2005;44:816–824. doi: 10.1080/02841860500341355. [DOI] [PubMed] [Google Scholar]
- 32.Rastogi T, Hildesheim A, Sinha R. Opportunities for cancer epidemiology in developing countries. Nat Rev Cancer. 2004;4:909–917. doi: 10.1038/nrc1475. [DOI] [PubMed] [Google Scholar]
- 33.Brownson RC, Alavanja MC, Caporaso N, Simoes EJ, Chang JC. Epidemiology and prevention of lung cancer in nonsmokers. Epidemiol Rev. 1998;20:218–236. doi: 10.1093/oxfordjournals.epirev.a017982. [DOI] [PubMed] [Google Scholar]
- 34.Boffetta P. Human cancer from environmental pollutants: the epidemiological evidence. Mutat Res. 2006;608:157–162. doi: 10.1016/j.mrgentox.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 35.March HC. Leukemia in radiologists in a 20 year period. Am J Med Sci. 1950;220:282–286. doi: 10.1097/00000441-195022030-00007. [DOI] [PubMed] [Google Scholar]
- 36.zur Hausen H. Papillomaviruses in human cancers. Proc Assoc Am Physicians. 1999;111:581–587. doi: 10.1046/j.1525-1381.1999.99723.x. [DOI] [PubMed] [Google Scholar]
- 37.Liyanage UK, Moore TT, Joo H-G, Tanaka Y, Herrmann V, Doherty G, Drebin JA, Strasberg SM, Eberlein TJ, Goedegebuure PS. Prevalence of regulatory T cells is increased in peripheral blood and tumor microenvironment of patients with pancreas or breast adenocarcinoma. J Immunol. 2002;169:2756–2761. doi: 10.4049/jimmunol.169.5.2756. [DOI] [PubMed] [Google Scholar]
- 38.Hedditch EL, Gao B, Russell AJ, Lu Y, Emmanuel C, Beesley J, Johnatty SE, Chen X, Harnett P, George J. ABCA Transporter Gene Expression and Poor Outcome in Epithelial Ovarian Cancer. J Natl Cancer Inst. 2014;106:dju149. doi: 10.1093/jnci/dju149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo XY, Chen YP, Song W, Zhao B, Cao B, Wei QQ, Ou RW, Yang Y, Yuan LX, Shang HF. An association analysis of the rs1572931 polymorphism of the RAB7L1 gene in Parkinson’s disease, amyotrophic lateral sclerosis and multiple system atrophy in China. Eur J Neurol. 2014;21:1337–43. doi: 10.1111/ene.12490. [DOI] [PubMed] [Google Scholar]
- 40.Li CJ, Dai Y, Fu YJ, Tian JM, Li JL, Lu HJ, Duan F, Li QW. Correlations of IFN-γ genetic polymorphisms with susceptibility to breast cancer: a meta-analysis. Tumour Biol. 2014;35:6867–77. doi: 10.1007/s13277-014-1856-6. [DOI] [PubMed] [Google Scholar]
- 41.Chang Z, Zhou H, Liu Y. Promoter methylation and polymorphism of E-cadherin gene may confer a risk to prostate cancer: a meta-analysis based on 22 studies. Tumour Biol. 2014;35:10503–13. doi: 10.1007/s13277-014-2323-0. [DOI] [PubMed] [Google Scholar]
- 42.Vera PL, Meyer-Siegler KL. Association between macrophage migration inhibitory factor promoter region polymorphism (-173G/C) and cancer: a meta-analysis. BMC Research Notes. 2011;4:395. doi: 10.1186/1756-0500-4-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dickersin K, Berlin JA. Meta-analysis: state-of-the-science. Epidemiol Rev. 1992;14:154–176. doi: 10.1093/oxfordjournals.epirev.a036084. [DOI] [PubMed] [Google Scholar]
- 44.Morgan TM, Krumholz HM, Lifton RP, Spertus JA. Nonvalidation of reported genetic risk factors for acute coronary syndrome in a large-scale replication study. JAMA. 2007;297:1551–1561. doi: 10.1001/jama.297.14.1551. [DOI] [PubMed] [Google Scholar]


