Abstract
Kaposi’s sarcoma (KS) is a vascular neoplasm caused by infection of endothelial or endothelial precursor cells with the Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV8). Research efforts have focused on defining the molecular events explaining how KSHV promotes pathological angiogenesis and KS tumor formation. mTOR/HIF-1 is a fundamental pathway driving these processes through the upregulation of angiogenic and inflammatory proteins, including VEGF, ANGPTL4, and ANGPT2. Interestingly, HIF-1 has also been implicated in the upregulation of metabolic genes associated with aerobic glycolysis and the growth of solid tumors. However, whether HIF-1 plays a role in regulating cell metabolism in KS remains unexplored. Here, we show that the HIF-1 metabolic effector, pyruvate kinase 2 (PKM2), is upregulated upon KSHV infection of endothelial cells and is necessary to maintain aerobic glycolysis in infected cells. We further demonstrate that PKM2 regulates KS angiogenic phenotype by acting as a coactivator of HIF-1 and increasing the levels of HIF-1 angiogenic factors, including VEGF. Indeed, inhibition of PKM2 expression blocked endothelial cell migration and differentiation and the angiogenic potential of KSHV-infected cells. We also investigated whether PKM2 regulates the angiogenic dysregulation induced by the KSHV-encoded G protein-coupled receptor (vGPCR), a viral oncogene that promotes Kaposi’s sarcomagenesis through the upregulation of HIF angiogenic factors. Interestingly, we found that PKM2 controls vGPCR-induced VEGF paracrine secretion and vGPCR oncogenesis. Our findings provide a molecular mechanism for how HIF-1 dysregulation fuels both angiogenesis and tumor metabolism in KS and support further investigations on therapeutic approaches targeting HIF-1 and PKM2 for KS treatment.
Keywords: Kaposi’s sarcoma, Kaposi’s sarcoma-associated herpesvirus, KSHV, Human herpesvirus-8, HHV-8, Hypoxia inducible factor-1, Pyruvate kinase 2, VEGF, Cell metabolism, Angiogenesis
Introduction
Although the precise molecular events that lead to the development of Kaposi’s sarcoma (KS) remain elusive, all four epidemiologic forms—classic, HIV-associated, endemic, and iatrogenic—share as a common etiology the infection with a single virus, the KS-associated human herpesvirus (KSHV or HHV-8) [1]. The classic histological features of KS lesions (e.g., proliferation of slit-like vascular channels and immature, leaky blood vessels with erythrocyte extravasation and profuse edema) are a consequence of the elaboration of angiogenic and inflammatory cytokines from KSHV-infected, spindle-shaped tumor cells [2]. These secreted factors are essential for the development and maintenance of the KS lesions through a mechanism known as paracrine neoplasia [2, 3].
Compelling data have shown that the elevated levels of angiogenic paracrine secretions and abnormal neovascularization observed in KS are due in part to the stabilization of the transcriptional activator, hypoxia inducible factor (HIF) [4–8]. HIF is composed by a constitutively expressed β subunit and an oxygen-regulated α subunit. The α subunit is unstable under normal oxygen tension (normoxia), but is stabilized by low oxygen tension (i.e., hypoxia) as well as by a number of activated oncogenes and growth factors [9]. Interestingly, recent evidence demonstrates that KSHV infection of endothelial cells is sufficient to upregulate the expression of the two homologs of the α subunit, HIF-1α and HIF-2α, leading to increased HIF-1 and HIF-2 transcriptional activity, respectively, under normoxic conditions [4–6]. Indeed, both latent and lytic KSHV proteins, including LANA1, vIRF3, and vGPCR, are able to upregulate HIF in expressing cells [4, 7, 8, 10], resulting in the increase in the levels of HIF angiogenic growth factors and cytokines (e.g., VEGF, PDGF, TGFα, TGFβ, ANGPT2, and ANGPTL4) [4, 11]. These factors bind to their respective receptors on neighboring endothelial cells, an effect that amplifies HIF-1 signaling (and the KS angiogenic phenotype) by upregulating HIF expression through the activation of the mammalian target of rapamycin (mTOR) pathway [4]. Through these autocrine and paracrine mechanisms, HIF angiogenic effectors help fuel KS tumor growth.
In addition to angiogenesis, HIF has also been implicated in the regulation of other biological responses, including the reprograming of cancer cell metabolism [12]. HIF is able to promote aerobic glycolysis, the metabolic route by which cancer cells obtain energy (ATP) and biosynthetic intermediates to sustain growth (also known as the Warburg effect) [13, 14], through the transcriptional upregulation of metabolic effectors that promote increased lactate production and decreased mitochondrial oxidative phosphorylation [13, 14]. In particular, a key HIF-regulated metabolic gene that plays an essential role in aerobic glycolysis is the isoform M2 of pyruvate kinase (PKM2), an enzyme that converts phosphoenolpyruvate (PEP) into pyruvate and ADP into ATP, in the final step of the glycolytic cycle [15]. PKM2 exists in two oligomeric forms, tetrameric and dimeric. The tetrameric form of PKM2 is highly active as a glycolytic enzyme, whereas the dimeric form is nearly inactive, under physiological conditions [16]. High levels of (dimeric) inactive PKM2 in cancer cells lead to the rewire of the cellular glycolytic flux. This has been shown to occur through the PEP-dependent histidine phosphorylation of phosphoglycerate mutase (PGAM1) and the establishment of an alternate glycolytic pathway that decouples adenosine triphosphate production from PEP-mediated phosphotransfer. This change has been shown to help satisfy the special metabolic demands of these highly proliferating cells [17].
Interestingly, recent data also suggest that PKM2 may support tumor development by regulating cellular mechanisms beyond its role as a glycolytic enzyme [18–20]. PKM2 has been shown to itself be a transcriptional activator, interacting with several transcription factors including β-catenin, Oct-4, and signal transducer and activator of transcription (STAT)3, thereby supporting tumorigenesis by upregulating both survival and proliferative genes. Of note, PKM2 has also been shown to be a co-activator of HIF and be part of a HIF-related feedback mechanism: PKM2 is transcriptionally regulated by HIF-1 and can bind and increase its activity, thereby potentiating the transactivation of HIF-dependent genes [21]. Since HIF has been previously shown to play such a prominent role in KS, we set out to investigate the role of PKM2 in modulating HIF function in Kaposi’s sarcomagenesis. Here, we have found that KSHV infection leads to a HIF-dependent increase in PKM2 expression and that this upregulation contributes to the promotion of the Warburg effect as well as the angiogenic dysregulation triggered by this transcription factor in KS.
Results
Endothelial cell infection with KSHV induces increased lactate production and decreased mitochondrial respiration
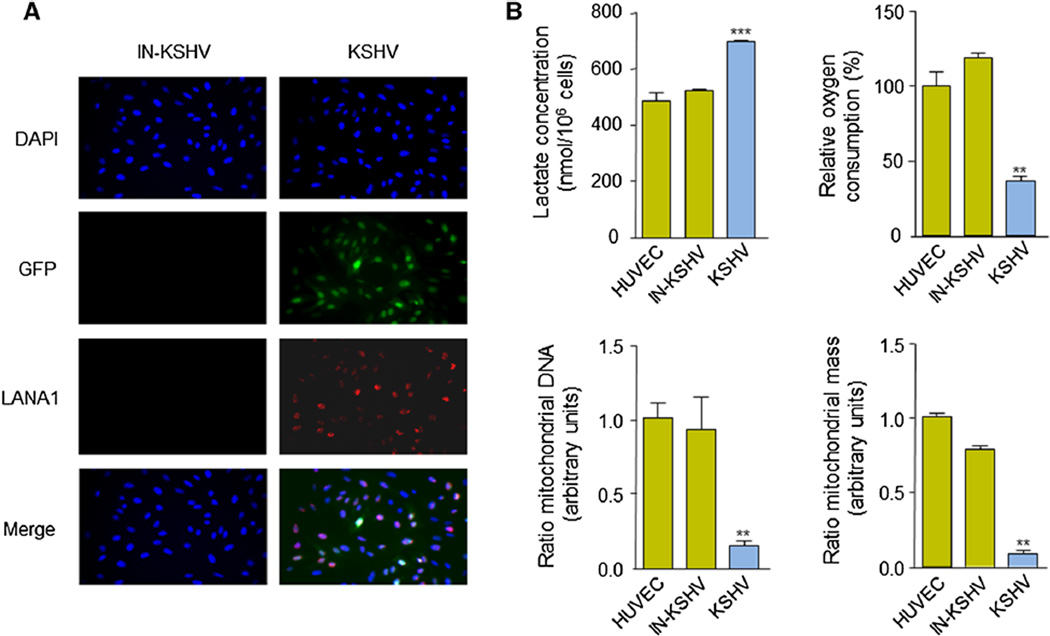
Immortalized HUVECs were infected with live or UV-inactivated recombinant KSHV (rKSHV.219), which expresses green fluorescent protein (GFP) under the EF-1α promoter and red fluorescent protein (RFP) under the promoter for the viral lytic gene PAN [22]. After selection of the infected cells with puromycin, 90 % of the cells exposed to live rKSHV.219 (KSHV) were latently infected, as judged by the expression of GFP and the latent viral protein LANA1/ORF73 (Fig. 1a). No GFP or LANA1 expression was observed in cells infected with UV-inactivated rKSHV.219 (IN-KSHV). We next examined the metabolic parameters related to glucose metabolism and oxidative phosphorylation for these cells compared to parental (HUVEC) cells. In line with prior observations [23], we found an increase in lactate production and a decrease in oxygen consumption of HUVECs infected with KSHV (Fig. 1b). We next examined the ratio of mitochondrial (mt)DNA to nuclear DNA by quantitative real-time PCR (pPCR) analysis of the DNA of the mitochondrial gene, cytochrome b, and the nuclear gene, RPL13A [24], and observed that the levels of mtDNA in KSHV-infected HUVECs were significantly lower compared to control cells (Fig. 1b). To determine whether these changes in mtDNA and nuclear DNA levels reflected a change in mitochondrial mass, we stained cells with NAO, a metachromatic dye that binds to cardiolipin in mitochondria [24]. Flow cytometry analysis of stained cells clearly demonstrated that the mitochondrial content of KSHV-infected cells is decreased compared to the one of the control cells (Fig. 1b). Collectively, these observations suggest that aerobic glycolysis is the preferred intracellular route of KSHV-infected endothelial cells to metabolize glucose.
Fig. 1.
KSHV infection causes the Warburg effect in endothelial cells. a HUVECs were infected with rKSHV.219 and selected with puromycin. Expression of GFP and LANA1 (indirect immunofluorescence) of cells infected with live rKSHV.219 (KSHV) or UV-inactivated rKSHV.219 (IN-KSHV) is shown. Nuclei are visualized with DAPI staining. b Extracellular lactate concentration, relative oxygen consumption, ratio of mitochondrial DNA and mitochondrial biomass of parental HUVEC, KSHV, and IN-KSHV cell lines (calculated as described in “Materials and methods” section)
HIF promotes aerobic glycolysis in KSHV-infected endothelial cells
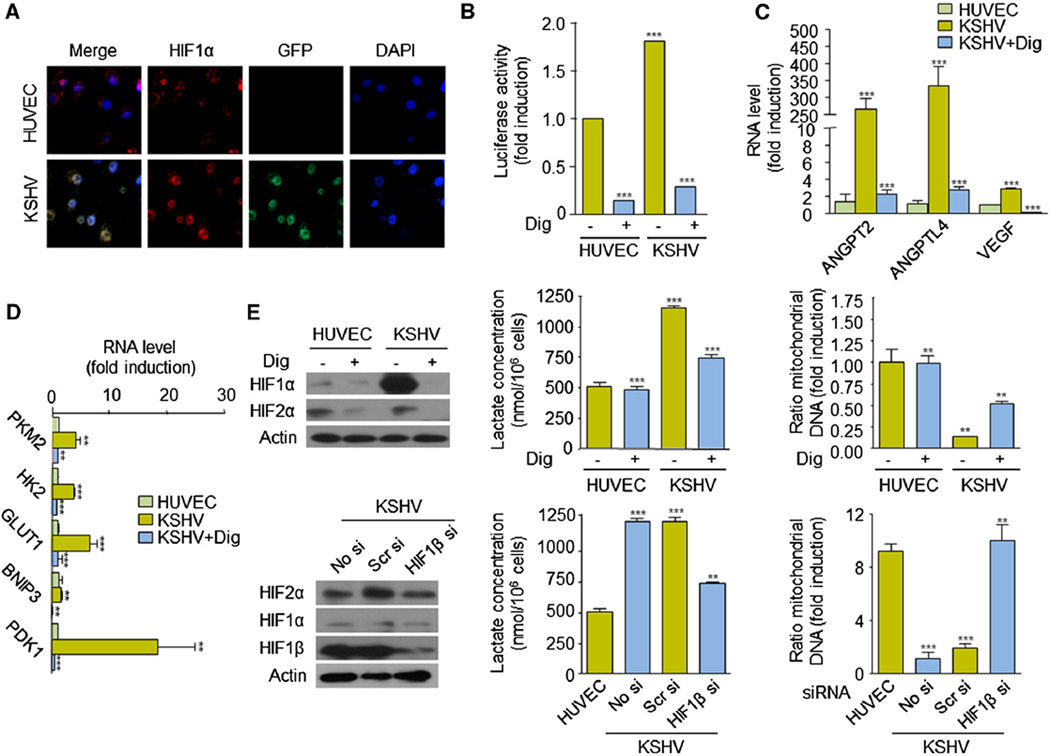
Since HIF-1α is up-regulated in endothelial cells upon KSHV infection and has previously been shown to be a master regulator of the Warburg effect in cancer cells, we next set out to investigate whether this transcriptional activator could regulate cell metabolism in KS. We first confirmed that HIF-1α was expressed and transcriptionally active in endothelial cells infected with KSHV (rKSHV.219). We observed that HIF-1α translocates to the nucleus and is able to induce transactivation from a (HIF-dependent) VEGF promoter driving expression of a luciferase reporter, in KSHV cells (Fig. 2a, b). Transcriptional up-regulation of HIF-1 angiogenic genes (e.g., VEGF, ANGPT2, and ANGPTL4) was observed in these cells (Fig. 2c). Both transcriptional effects were inhibited by pharmacologically inhibiting HIF-1α translation using digoxin (Dig) (Fig. 2b, c). Activation of HIF-1 in these cells further leads to the digoxin-sensitive upregulation of HIF-1-regulated metabolic genes, including the glucose transporter 1 (GLUT1), the glycolytic enzymes, hexokinase 2 (HK2), pyruvate kinase M2 (PKM2), and pyruvate dehydrogenase kinase-1 (PDK1), as well as the BCL2/adenovirus E1B 19 kDa interacting protein 3 (BNIP3), a protein that induces mitochondrial autophagy (Fig. 2d) [12]. In turn, inhibition of HIF-1 with digoxin resulted in a decrease in the levels of lactate production and an increase in mitochondrial DNA content (Fig. 2e). Similar effects were observed when expression of the (constitutive) HIF-1α binding partner, HIF-1β, was knocked down using a specific siRNA sequence (Fig. 2e). Collectively, these findings demonstrate that HIF-1 plays a role in defining the metabolic fate of KSHV-infected cells by increasing expression of metabolic genes that promote the Warburg effect.
Fig. 2.
HIF regulates the Warburg effect in KSHV-infected cells. a HIF1α and GFP expression and DAPI staining in HUVEC and KSHV cells. b HUVEC and KSHV cells were transfected with pGL-VEGF/K and pCEFL Renilla luciferase and exposed to vehicle (−) or 100 nM digoxin (+) for 24 h. Luciferase activity and Renilla activity (for normalization) were determined as described in “Materials and methods” section. c, d Real-time PCR of HIF angiogenic genes (ANGPT2, ANGPTL4, VEGF) (c) and HIF metabolic genes (PKM2, HK2, GLUT1, BNIP3, PDK1) (d) in HUVEC and KSHV cells. Cells were treated with vehicle (−) or 100 nM digoxin (+) for 24 h. e Extracellular lactate concentration and ratio of mitochondrial DNA of cells exposed to vehicle (−) or 100 nM digoxin (+), or transfected with no siRNA (No si) scrambled siRNA (Scr si) or HIF1β siRNA (HIF1β si). Western blots show levels of HIF1α, HIF2α, or HIF1β in the corresponding treated/untreated or transfected/untransfected cells
PKM2 is required to maintain anerobic glycolysis in KSHV-infected cells
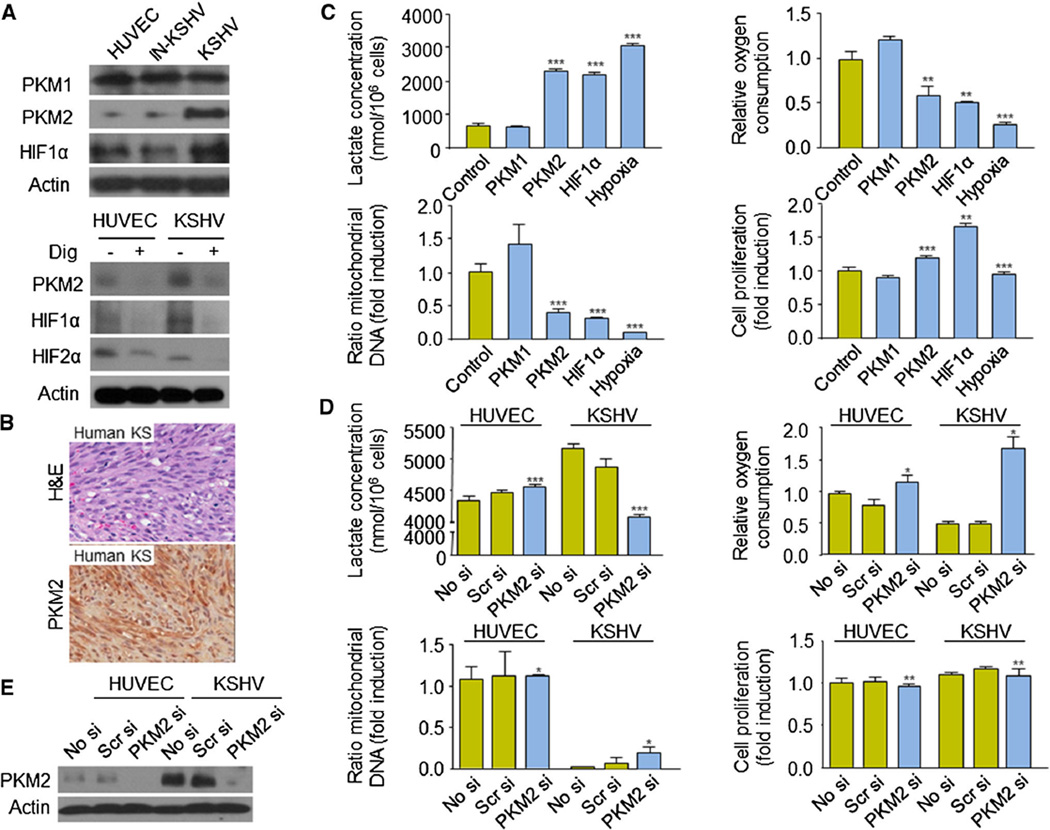
An important HIF-1 downstream effector involved in the control of cell metabolism is the M2 isoform of the pyruvate kinase enzyme (PKM2) [15, 19, 25]. Upregulation of PKM2 in cancer cells is associated with critical metabolic and transcriptional changes that allow rapid growth and proliferation. Interestingly, we observed an increase in the protein levels of PKM2, but not the related M1 isoform, PKM1, in cells infected with KSHV but not in cells infected with inactivated KSHV (Fig. 3a). Upregulation of PKM2 in KSHV cells was sensitive to the treatment of the cells with digoxin (Fig. 3a), demonstrating that HIF-1 is necessary for its expression. Indeed, expression of PKM2 was observed in tumor cells in human KS biopsies (Fig. 3b).
Fig. 3.
PKM2 regulates KS cell metabolism. a PKM1, PKM2, and HIF1α protein levels in HUVEC, IN-KSHV, and KSHV cells and PKM2, HIF1α, and HIF2α protein levels in HUVEC and KSHV cells, treated with vehicle (−) or 100 nM digoxin (+) for 24 h. b H&E staining and PKM2 immunohistochemical staining of human AIDS-related KS biopsy. c Effect of overexpression of (pcDNA3.1-V5-His) PKM1 or PKM2 or (pCEFL myc HIF1αΔODD) HIF1α in HUVEC on lactate production, oxygen consumption, mitochondrial DNA content, and cell proliferation (calculated as described in “Materials and methods” section). Hypoxia treatment (1 % O2) was used as a control. d Effect of the expression of no siRNA (No si), scrambled siRNA (Scr si) or PKM2 siRNA (PKM2 si), in HUVEC or KSHV cells on lactate production, oxygen consumption, mitochondrial DNA content, and cell proliferation. e Corresponding levels of PKM2 upon transfection of no siRNA (No si), scrambled siRNA (Scr si) or PKM2 siRNA (PKM2 si), in HUVEC or KSHV cells
We next set out to examine whether PKM2 plays a role in the maintenance of the Warburg effect in KSHV-infected endothelial cells. For that purpose, we overexpressed the isoform PKM1 or PKM2 (pcDNA3.1-V5-His) in HUVEC and observed that PKM2 is sufficient—as it is an overexpression of a constitutively active HIF1α (pCEFL myc HIF1αΔODD)—to decrease oxidative phosphorylation in these cells, as judged by the up-regulation of the lactate production and the diminution of oxygen consumption (Fig. 3c). PKM2 also diminished the number of cell mitochondria (Fig. 3c). However, augmented PKM2 expression did not have an impact on the overall ability of the cells to proliferate (Fig. 3c). To investigate whether PKM2 is required to induce all these metabolic changes upon infection of endothelial cells with KSHV, we blocked its expression using an specific PKM2 siRNA (Fig. 3d, e). Interestingly, inhibition of PKM2 expression was able to increase oxidative phosphorylation in KSHV-infected HUVECs, but did not affect the levels of mitochondrial content neither the ability of these cells to proliferate (Fig. 3d). Collectively, these results suggest that PKM2 is an important HIF effector in the maintenance of aerobic glycolysis in cells infected with KSHV by favoring the redirection of glycolytic substrates away from mitochondrial respiration. However, other metabolic pathways must also be involved in the regulation of the mitochondrial content of KSHV-infected cells.
PKM2 regulates expression of VEGF in KSHV-infected cells
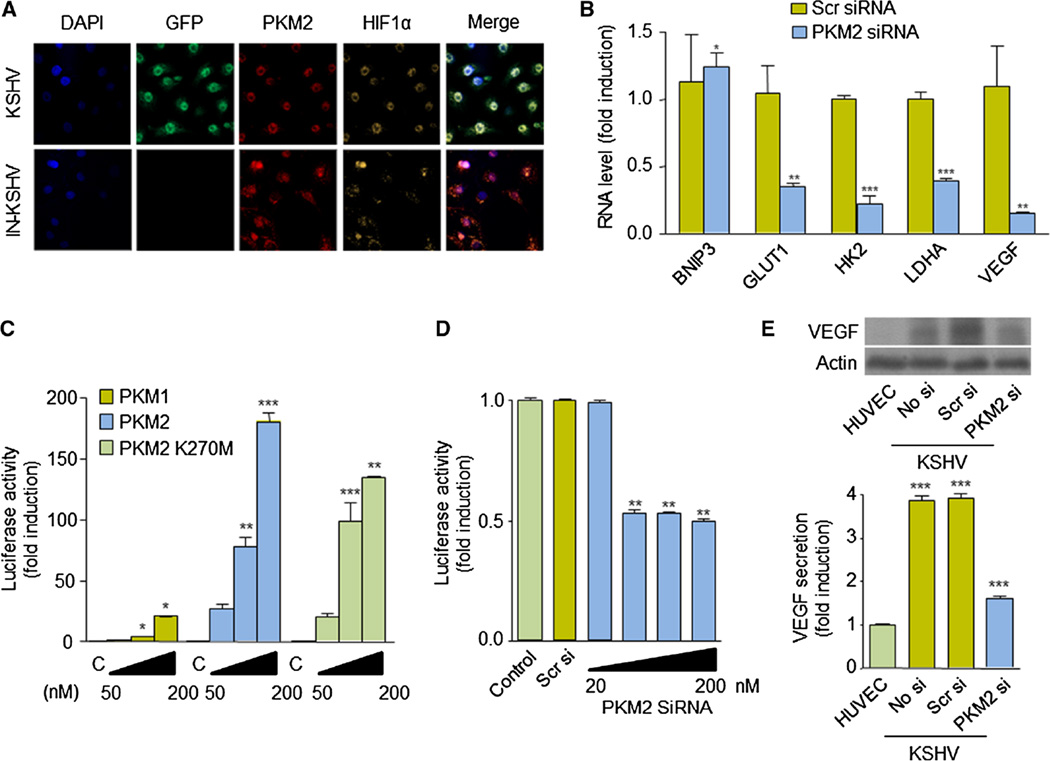
PKM2 is also able to translocate to the nucleus and regulates the transcriptional activity of transcription factors, including HIF-1α. PKM2 interacts directly with HIF-1α and is a co-activator of its function, by enhancing HIF-1 binding and p300 recruitment to hypoxia response elements (HREs) [21]. Interestingly, indirect immunofluorescence studies in KSHV-infected endothelial cells showed that PKM2 and HIF-1α localize in the nucleus of these cells but not in control IN-KSHV cells (Fig. 4a). We thus examined whether PKM2 played a role in the regulation of HIF-1-mediated gene expression in cells infected with KSHV. Interestingly, we found that inhibition of PKM2 in KSHV cells leads to a decrease in the expression of HIF-1 regulated genes, including VEGF (Fig. 4b). Forced expression of (pcDNA3.1-V5-His) PKM2—but not PKM1—in HUVECs was sufficient to promote transactivation from a VEGF promoter, an effect that was independent of PKM2 kinase activity, as it was observed also upon expression of the inactive mutant, PKM2 K270M (Fig. 4c) [21]. Conversely, inhibition of PKM2 in KSHV-infected endothelial cells using PKM2 siRNA blocked transactivation from the HIF-1-dependent VEGF promoter, as well as the expression and secretion of VEGF (Fig. 4d, e). Collectively, these results demonstrate that PKM2 is a regulator of VEGF expression in KSHV-infected endothelial cells.
Fig. 4.
VEGF expression is regulated by PKM2 in KSHV-infected cells. a Immunofluorescent staining of PKM2 and HIF1α in IN-KSHV and KSHV cells. b Real-time PCR analysis of HIF downstream genes (BNIP3, GLUT1, HK2, LDHA, VEGF) in KSHV cells expressing (control) scrambled siRNA or PKM2 siRNA. c, d HUVEC (c) or KSHV (d) cells were transfected with increasing doses of PKM1, PKM2, and PKM2 K270M expression vector (pcDNA3.1-V5-His) (c) or increasing doses of PKM2 siRNA (d), along with pGL-VEGF/K and pCEFL Renilla luciferase. Luciferase activity and Renilla activity (for normalization) were determined as described in “Materials and methods” section. e Levels of VEGF protein expression and VEGF secretion of HUVEC and KSHV expressing no siRNA (No si), scrambled siRNA (Scr si) or PKM2 siRNA (PKM2 si)
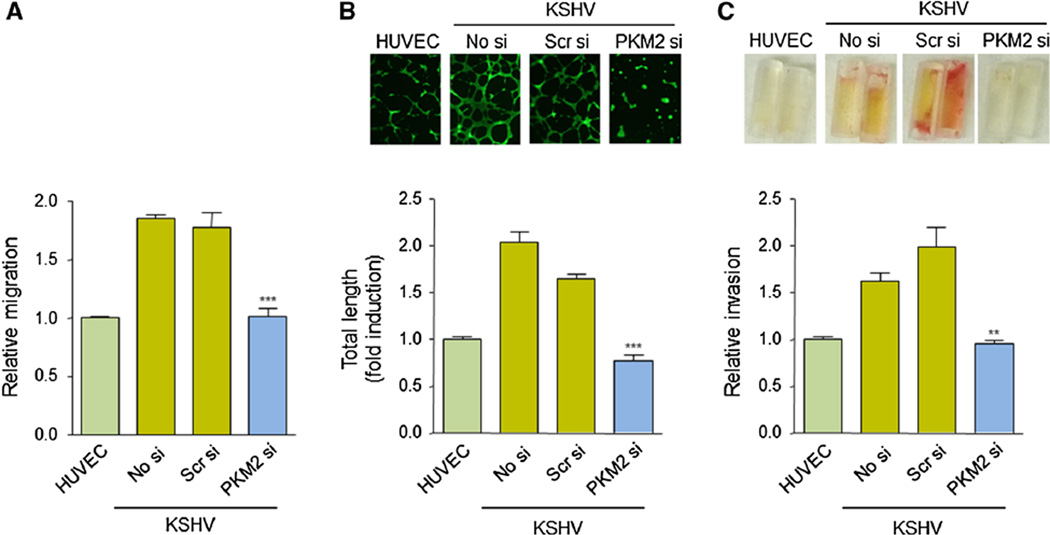
PKM2 promotes the angiogenic potential of KSHV-infected cells
We next examined whether the PKM2-dependent regulation of angiogenic factors, including VEGF, influenced the angiogenicity of KSHV-infected cells. To this end, we performed a modified, Boyden chamber assay in KSHV cells expressing or not PKM2 siRNA. We observed that downregulation of PKM2 expression in those cells impairs cell migration (Fig. 5a). When we investigated the role of PKM2 in regulating the ability of KSHV cells to form capillary structures (endothelial cell tube formation assay), we observed that PKM2 siRNA expression in KSHV cells resulted in an inhibition of endothelial cell differentiation (Fig. 5b). Furthermore, to elucidate the contribution of PKM2 to the angiogenic capability of KSHV-infected cells in vivo, we used the directed in vivo angiogenesis assay (DIVAA) [26]. For this purpose, basement membrane extract (BME)-filled angioreactors containing supernatants from KSHV cells expressing or not PKM2 were implanted in immunosuppresed mice. Figure 5c shows that the inhibition of PKM2 expression in KSHV cells blocks the development of blood vessels in vivo. Collectively, these results demonstrate that PKM2 plays a critical role in regulating the angiogenic potential of KSHV-infected endothelial cells and their paracrine secretions.
Fig. 5.
Inhibition of PKM2 expression blocks KSHV-infected cell angiogenicity. a, b Cell migration (modified Boyden chamber assay) assay (a) and tube formation assay (matrigel assay) (b) in HUVEC or KSHV cells expressing no siRNA (No si), scrambled siRNA (Scr si) or PKM2 siRNA (PKM2 si). c Conditioned media from HUVEC or KSHV cells expressing no siRNA (No si), scrambled siRNA (Scr si) or PKM2 siRNA (PKM2 si) was used to induce blood vessel development in BME-filled angioreactors implanted in nude mice (DIVAA assay)
PKM2 promotes the tumorigenic potential of the KSHV oncogene, vGPCR
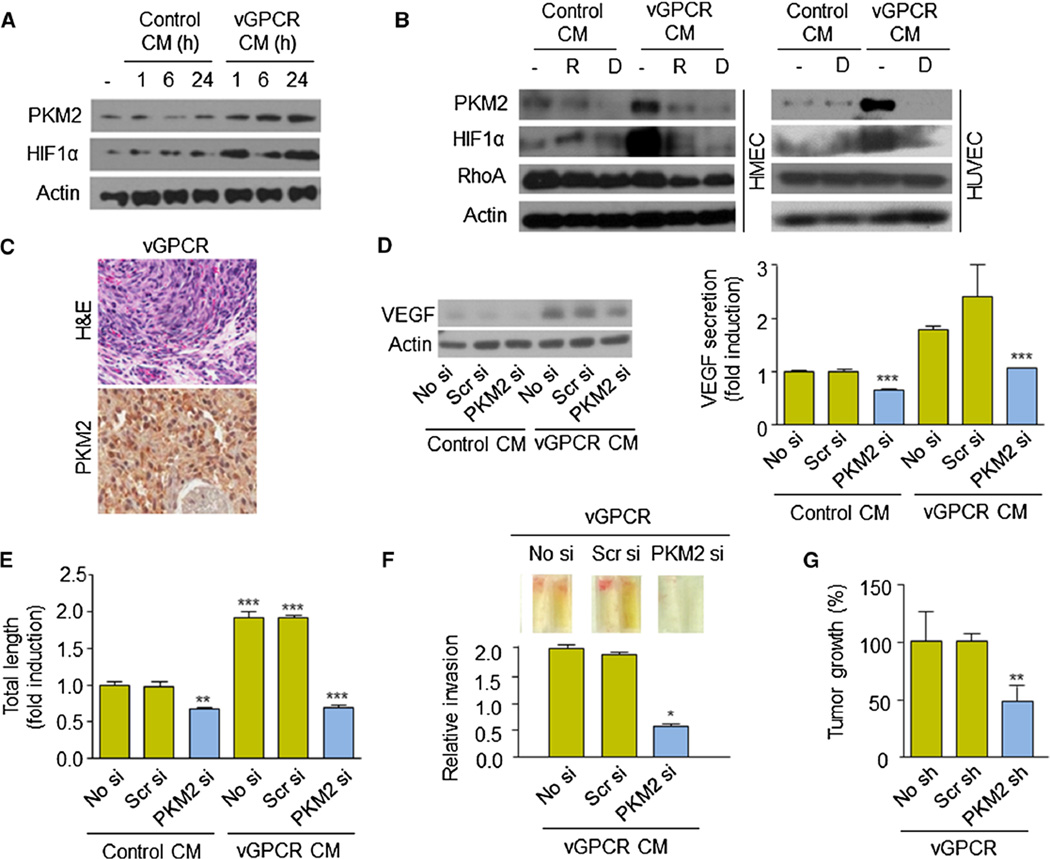
KSHV encodes multiple genes that have been previously implicated in the molecular pathogenesis of KS and that upregulate HIF-1 activity, including the viral G protein-coupled receptor (vGPCR), viral IFN regulatory factor 3 (IRF3) and the KSHV latency-associated nuclear antigen 1 (LANA1) [4, 7, 27]. Since PKM2 promotes the angiogenicity of KSHV-infected cells by both autocrine and paracrine mechanisms, we decided to further investigate whether PKM2 could be a downstream effector in the signaling by vGPCR. This constitutively active viral receptor, which has only limited expression in a small number of cells within the KS tumor, is sufficient to cause KS-like tumor formation in vivo through the secretion of a wide array of inflammatory and angiogenic factors, several of them up-regulated through HIF-1 transcriptional activation in vGPCR-expressing and neighboring cells [4, 10, 28–30]. Interestingly, we observed an increase in PKM2 expression when we treated endothelial cells with conditioned media from cells over-expressing vGPCR (Fig. 6a), which was inhibited when cells were treated with the mTOR inhibitor, rapamycin (R), or the HIF-inhibitor, digoxin (D) (Fig. 6b). PKM2 expression was observed in cells from tumor allografts obtained upon injection of nude mice with endothelial cells expressing vGPCR (Fig. 6c) [30].
Fig. 6.
Inhibition of PKM2 inhibits vGPCR oncogenesis. a PKM2 analysis upon treatment of HUVEC cells with control or vGPCR conditional media (CM) over time. b PKM2 analysis upon treatment of HMEC or HUVEC cells with control or vGPCR conditional media (CM). Cells were treated with vehicle (−), rapamycin (50 nM, 24 h, R), digoxin (100 nM, 24 h, D). Levels of RhoA serve as negative control. c Immunohistochemistry of PKM2 in vGPCR tumor allograft [30]. d, f Effect of PKM2 knockdown (no siRNA (No si), scrambled siRNA (Scr si) or PKM2 siRNA (PKM2 si) on VEGF expression and secretion (d), endothelial cell differentiation (tube formation assay) (e), and in vivo blood vessel development (DIVAA assay) (f). g PKM2 knockdown blocks vGPCR-induced allograft growth in nude mice. Mice were injected in the flank with (1:10) mixed populations of SVEC stably expressing vGPCR and cells expressing no shRNA (No sh), scrambled shRNA (Scr sh) or PKM2 shRNA (PKM2 sh). Percentage of tumor growth is shown
We then decided to explore whether the increase in PKM2 levels could affect the vGPCR paracrine up-regulation of the secretion of HIF angiogenic factors, such as VEGF. Interestingly, expression of PKM2 siRNA was able to reduce the levels of VEGF expression and secretion induced by treatment of HUVEC with supernatants derived from vGPCR-expressing cells (Fig. 6d). Indeed, inhibition of PKM2 reduced the angiogenic potential of vGPCR secretions (Fig. 6e, f). Finally, to assess whether PKM2 was necessary for vGPCR tumorigenesis in vivo, we established stable murine endothelial (SVEC) cell lines expressing vGPCR [30] or expressing a specific shRNA for PKM2 or control scrambled shRNA. These cells were used to establish (mixed-cell) allografts in a 1:10 ratio (resembling limited vGPCR expression in KS tumors) in athymic nu/nu mice, as previously described [29, 30]. Figure 6f shows that tumor growth in vivo was significantly reduced when PKM2 expression was blocked in the injected endothelial cells. Collectively, these results suggest that PKM2 is a critical player in the promotion of vGPCR angiogenesis and tumorigenesis.
Discussion
The investigation of the molecular pathogenesis of KS as well as its clinical management has been challenging due to the multiple factors involved in the development of this tumor [2]. Infection with KSHV is considered a necessary but not sufficient condition for the growth of KS lesions, which also depends on genetic, environmental and immunologic factors, as well as on an immunosuppressed state of the KSHV-infected individual. One of the hallmarks in KS development is the presence of a profound cytokine dysregulation, which is linked to inflammation and aberrant neovascularization in KS [3]. The release of KS cytokines and inflammatory factors within the tumor is associated in part to the activation of TSC/mTOR/HIF, a signaling pathway that has shown to be a driving force in the progression of multiple human cancers [31, 32]. Increased expression of HIF-1α and HIF-2α is observed in human KS lesions and has been associated with the upregulation of VEGF, ANGPT2, and ANGPTL4 and the promotion of KS tumor maintenance [4, 11]. HIFs also upregulate latent and lytic KSHV replication and have shown to play a central role in KSHV life cycle [33]. Indeed, HIF-1 inhibitors have been shown to block tumor growth in an animal model of KS [4].
The contribution of HIFs to tumor proliferation is not limited to the promotion of angiogenesis but includes many other biological functions such as stem cell maintenance, metabolic reprograming, epithelial-mesenchymal transition, invasion, and metastasis [19]. Here, we have found that this transcriptional activator also contributes to Kaposi’s sarcomagenesis by promoting the Warburg effect [13, 14]. KSHV-infected cells had been shown to shift their glucose metabolism away from oxidative phosphorylation to aerobic glycolysis, a metabolic feature required to allow and maintain latent KSHV infection [23] (Fig. 1). Here, we have found that HIF is an important player in the establishment of that metabolic switch. Interestingly, transcriptional reprograming via HIF1 activation has recently been reported to be involved in the metabolic transformation of KSHV-infected endothelial cells induced by KSHV microRNAs [34]. Moreover, digoxin was able to block tumor growth in a xenograft model for KS [4]. Collectively, these findings underscore the role of HIF as a critical regulator of KSHV oncogenesis and a molecular target in the development of mechanism-based therapies for KS patients.
We have seen that the contribution of HIF to the Warburg effect in KSHV-infected cells is, in part, through the elevation of the expression of PKM2, one of the isoforms of the kinase involved in the final step reaction of glycolysis [25]. PKM2 was found to be up-regulated upon KSHV infection and was critical for the metabolic rewiring from oxidative phosphorylation toward aerobic glycolysis to maintain the increased anabolic requirements of KSHV-infected cells. However, PKM2 was not necessary to regulate mitochondria levels in these cells, suggesting that the decrease in the number of these organelles observed upon KSHV infection must be caused by alternative routes. Interestingly, we have observed that PKM2 also promotes KS tumor growth by other intriguing mechanism: the activation of HIF-dependent transcription leading to the upregulation of HIF pro-angiogenic effectors, including VEGF, and the promotion of dysregulated angiogenesis [19]. In this regard, we observed that PKM2 was able to regulate the expression of VEGF by cytokines secreted by vGPCR, a KSHV-encoded receptor that promotes KSHV tumorigenesis in endothelial cells through paracrine mechanisms [29]. PKM2 upregulation is expected to be a significant event in KS sarcomagenesis as other KSHV genes, including LANA1 (which is expressed in latently infected tumor cells), also activate HIF1. The role of PKM2 in the metabolic reprograming and angiogenic dysregulation observed in KS suggests that PKM2 may be a potential therapeutic target for the treatment of patients with this disease.
PKM2 has become an attractive molecular target for the design of cancer therapy due to its pleotropic effects in the control of multiple biological functions linked to cell growth and proliferation. There is evidence showing that knockdown of PKM2 by siRNA induces apoptosis in multiple cancer cell lines, and the delivery of PKM2 siRNA in vivo alone or combined with chemotherapy drugs causes substantial tumor regression of established xenografts [25]. Genetic replacement of PKM2 with PKM1 and treatment with pharmacological agents that increase PKM2 activity have also shown to reduce tumor growth [35, 36]. Still, recent investigations also suggest that the efficacy of targeting PKM2 as an anticancer approach may depend on the cell of origin as well as on specific nutritional and metabolic characteristics of each tumor [25].
With respect to KS, treatment options for KS are diverse and differ with the specific form of the disease presented by the patient. Chemotherapeutic drugs include vincristine, bleomycin, doxorubicin, and etoposide, as single agents or in combination, but they have well-known toxicities that limit their prolonged use [37]. Clinical trials on anti-VEGF treatments or immunomodulating agents are in progress and show efficacy in a variable portion of treated patients [38, 39]. Interestingly, the recent identification of mTOR as a hallmark of KS development has encouraged the use of rapalogs (sirolimus and others) for KS management with promising results [40]. However, novel molecular-based mechanisms for the treatment of this disease are still in need. Whether HIF or PKM2 inhibitors could serve as antitumor drugs for this tumor warrants further investigation.
Materials and methods
Constructs and reagents
GFP and KSHV vGPCR gene expression vectors were constructed as previously described [29]. Recombinant KSHV (rKSHV.219) was a gift from Dr. Vieira [22]. Production of the viral particles was performed as previously described [22]. Expression vectors (pcDNA3.1-V5-His) encoding PKM1, PKM2, or PKM2 K270M or (pCEFL myc) HIF1αΔODD have been described elsewhere [21, 41]. siRNAs oligos (HIF1β, PKM2) were purchased from Qiagen. VEGF ELISA kit was obtained from R&D system. Control and vGPCR condition media (CM) was obtained as previously reported [11].
Cell lines
Telomerase-immortalized human umbilical vein endothelial cells (HUVEC), immortalized human microvascular endothelial cell (HMEC), immortalized murine endothelial cells (SVEC) and SVEC stable cell lines have been previously described [11]. KSHV cells were generated by puromycin selection of HUVEC infected with rKSHV.219 [22].
Western blot, immunohistochemistry, immunocytochemistry, and microscopy
Western blot, immunohistochemistry, immunocytochemistry, and fluorescence microscopy were performed as described [11]. Antibodies were purchased as follows: LANA1 from Leica, PKM2, HIF1α and HIF1β from Cell Signaling, HIF2α from Novus, PKM1 from Abgen, VEGF from Abcam, and Actin from Santa Cruz. Confocal microscopy was conducted by using a Zeiss LSM 510 Meta microscope.
Real-time PCR
Total RNA was isolated using the GenElute Mammalian Total RNA Miniprep kit (Sigma), and reverse transcription was performed using SuperScript III First-Strand Synthesis System (Invitrogen).GAPDH was used for normalization. Sequence of primers is 5′-CGGGCCAAGAGTGTGCTAAA-3′ and 5′-TGACGATACCGGAGCCAATG-3′ for human GLUT1, 5′-AGCTCTGTGGCGCAGGCATG-3′ and 5′-TCGGACAGGCCACAGCAGTG-3′ for human HK2, 5′-ACCAGGACAGCCAATACAAG-3′ and 5′-CCTCGGTCACTCATCTTCAC-3′ for human PDK1, 5′-ATCTTGACCTACGTGGCTTGGA-3′ and 5′-CCATACAGGCACACTGGAATCTC-3′ for human LDH A, 5′-AACTCAGATTGGATATGGGATTGG-3′ and 5′-AGAGCAGCAGAGATGGAAGG-3′ for human BNIP3, 5′-GCACAATGGTCTCACGTTCTC-3′ and 5′-CCGTTCGAACTGTCTCACCA-3′ for human ANGPT2, 5′-CGTGGGGACCTTAACTGTG-3′ and 5′-TTCCATGTTTTCCAGAAGATAC-3′ for human ANGPTL4, 5′-GGGCAGAATCATCACGAAGT-3′ and 5′-TGGTGATGTTGGACTCCTCA-3′ for human VEGF, 5′-AGAAGGCTGGGGCTCATTTG-3′ and 5′-AGGGGCCATCCACAGTCTTC-3′ for human GAPDH.
Endothelial cell migration and matrigel assays
Endothelial cell migration was evaluated in a modified Boyden chamber assay using transwell filters inserts with 8-µm pore size (Neuro Probe, Inc.), as previously described [11]. The formation of capillary-like endothelial tubes was determined by plating endothelial cells on growth factor-reduced Matrigel (BD) and following the procedure described in [11].
Directed in vivo angiogenesis assay (DIVAA)
The DIVAA assay was performed according to manufacturer’s protocol (Trevigen). Briefly, 6-week-old female nude mice were anesthetized with tribromoethanol (i.p., 250 mg/kg). A 1-cm incision was made on both dorsal–lateral surfaces of the animal, and two BME-filled angioreactors were implanted on each side. Each angioreactor contained a total of 25 µl BME pre-mixed with 20 µg/ml heparin and 2 µl of test conditioned medium or medium containing recombinant protein (control). Angioreactors were removed 11 days post-implantation. The matrigel was digested with 300 µl of CellSperse solution for 1 h (37 °C), and endothelial cells were labeled with FITC-lectin. Fluorescence was measured in 96-well plates (excitation 485 nm, emission 510 nm). Values for cell invasion are calculated as the ratio of relative fluorescent units (RFUs) of the test sample with respect to control sample.
Oxygen consumption and lactate measurements
Three thousand cells/ml in DMEM medium with glucose 4.5 g/l were placed in a stirred, sealed chamber at room temperature. Oxygen concentration was monitored with a fiber-optic oxygen sensor (Instech Laboratories, Inc.). The oxygen consumption rate was calculated from the slope of the oxygen concentration–time curve. A l-Lactate assay kit (Cayman Chemicals) was used to determine extracellular lactate concentration.
Mitochondrial DNA and mitochondrial mass measurements
Mitochondrial DNA content was measured by quantitative PCR analysis as previously described [42]. For mitochondrial mass measurements, cells were stained with (25 ng/ml) nonyl acridine orange (NAO, Invitrogen) for 10 min at 37 °C. Cells were collected and resuspended in PBS with 5 % BSA. Samples were analyzed by flow cytometry analysis.
Luciferase reporter assays
Cells were transfected with the pGL-VEGF/K reporter, containing the luciferase gene under the control of the VEGF promoter, and the control reporter pCEFL Renilla luciferase enhanced EGFP [28] along the indicated expression vectors or siRNAs. FLuc and RLuc activities were determined using the Dual-Luciferase Assay System (Promega).
Tumorigenesis assays
All procedures involving animals were approved by the Institutional Animal Care and Use Committee. For tumor allograft formation, SVEC cells stably expressing vGPCR [30] and SVEC stably infected with lentiviruses expressing scrambled shRNA or PKM2 shRNA (or no shRNA) were injected (in a 1:10 ratio) into the flank of 8-week-old athymic (nu/nu) nude female mice as previously described [30]. The animals were monitored twice weekly for tumor formation. The longest length (L) and shortest width (W) of the tumor were measured using a caliper at different time points throughout the experiment. Tumor volume was then converted into tumor weight using the formula LW2/2, as described previously [30].
Statistical analysis
Results are representative of least three independent experiments. Statistical analysis was performed with ANOVA test by GraphPad InStat 3 software (GraphPad, ***, p < 0.001; **, p < 0.01; *, p < 0.05).
References
- 1.Moore PS, Chang Y. Molecular virology of Kaposi’s sarcoma-associated herpesvirus. Philos Trans R Soc Lond B Biol Sci. 2001;356(1408):499–516. doi: 10.1098/rstb.2000.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mesri EA, Cesarman E, Boshoff C. Kaposi’s sarcoma and its associated herpesvirus. Nat Rev Cancer. 2010;10(10):707–719. doi: 10.1038/nrc2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jham BC, Montaner S. The Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor: lessons on dysregulated angiogenesis from a viral oncogene. J Cell Biochem. 2010;110(1):1–9. doi: 10.1002/jcb.22524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jham BCMT, Hu J, Chaisuparat R, Friedman ER, Pandolfi PP, Schneider A, Sodhi A, Montaner S. Amplification of the angiogenic signal through the activation of the TSC/mTOR/HIF axis by the KSHV vGPCR in Kaposi’s sarcoma. PLoS One. 2011;6(4):e19103. doi: 10.1371/journal.pone.0019103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll PA, Kenerson HL, Yeung RS, Lagunoff M. Latent Kaposi’s sarcoma-associated herpesvirus infection of endothelial cells activates hypoxia-induced factors. J Virol. 2006;80(21):10802–10812. doi: 10.1128/JVI.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catrina SB, Botusan IR, Rantanen A, Catrina AI, Pyakurel P, Savu O, Axelson M, Biberfeld P, Poellinger L, Brismar K. Hypoxia-inducible factor-1alpha and hypoxia-inducible factor-2 alpha are expressed in kaposi sarcoma and modulated by insulin-like growth factor-I. Clin Cancer Res. 2006;12(15):4506–4514. doi: 10.1158/1078-0432.CCR-05-2473. [DOI] [PubMed] [Google Scholar]
- 7.Shin YC, Joo CH, Gack MU, Lee HR, Jung JU. Kaposi’s sarcoma-associated herpesvirus viral IFN regulatory factor 3 stabilizes hypoxia-inducible factor-1 alpha to induce vascular endothelial growth factor expression. Cancer Res. 2008;68(6):1751–1759. doi: 10.1158/0008-5472.CAN-07-2766. [DOI] [PubMed] [Google Scholar]
- 8.Cai QL, Knight JS, Verma SC, Zald P, Robertson ES. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS Pathog. 2006;2(10):e116. doi: 10.1371/journal.ppat.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29(5):625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sodhi A, Chaisuparat R, Hu J, Ramsdell AK, Manning BD, Sausville EA, Sawai ET, Molinolo A, Gutkind JS, Montaner S. The TSC2/mTOR pathway drives endothelial cell transformation induced by the Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor. Cancer Cell. 2006;10(2):133–143. doi: 10.1016/j.ccr.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 11.Ma T, Jham BC, Hu J, Friedman ER, Basile JR, Molinolo A, Sodhi A, Montaner S. Viral G protein-coupled receptor upregulates Angiopoietin-like 4 promoting angiogenesis and vascular permeability in Kaposi’s sarcoma. Proc Natl Acad Sci U S A. 2010;107(32):14363–14368. doi: 10.1073/pnas.1001065107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20(1):51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vander Heiden MG, Lunt SY, Dayton TL, Fiske BP, Israelsen WJ, Mattaini KR, Vokes NI, Stephanopoulos G, Cantley LC, Metallo CM, Locasale JW. Metabolic pathway alterations that support cell proliferation. Cold Spring Harb Symp Quant Biol. 2011;76:325–334. doi: 10.1101/sqb.2012.76.010900. [DOI] [PubMed] [Google Scholar]
- 16.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15(4):300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G, Rabinowitz JD, Asara JM, Cantley LC. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329(5998):1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaneton B, Gottlieb E. Rocking cell metabolism: revised functions of the key glycolytic regulator PKM2 in cancer. Trends Biochem Sci. 2012;37(8):309–316. doi: 10.1016/j.tibs.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Luo W, Semenza GL. Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol Metab. 2012;23(11):560–566. doi: 10.1016/j.tem.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamada M, Suematsu M, Saya H. Pyruvate kinase M2: multiple faces for conferring benefits on cancer cells. Clin Cancer Res. 2012;18(20):5554–5561. doi: 10.1158/1078-0432.CCR-12-0859. [DOI] [PubMed] [Google Scholar]
- 21.Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145(5):732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vieira J, O’Hearn PM. Use of the red fluorescent protein as a marker of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. Virology. 2004;325(2):225–240. doi: 10.1016/j.virol.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 23.Delgado T, Carroll PA, Punjabi AS, Margineantu D, Hockenbery DM, Lagunoff M. Induction of the Warburg effect by Kaposi’s sarcoma herpesvirus is required for the maintenance of latently infected endothelial cells. Proc Natl Acad Sci U S A. 2010;107(23):10696–10701. doi: 10.1073/pnas.1004882107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11(5):407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Li Z, Yang P, Li Z. The multifaceted regulation and functions of PKM2 in tumor progression. Biochim Biophys Acta. 2014;2:285–296. doi: 10.1016/j.bbcan.2014.07.008. (1846) [DOI] [PubMed] [Google Scholar]
- 26.Guedez L, Rivera AM, Salloum R, Miller ML, Diegmueller JJ, Bungay PM, Stetler-Stevenson WG. Quantitative assessment of angiogenic responses by the directed in vivo angiogenesis assay. Am J Pathol. 2003;162(5):1431–1439. doi: 10.1016/S0002-9440(10)64276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai Q, Murakami M, Si H, Robertson ES. A potential alpha-helix motif in the amino terminus of LANA encoded by Kaposi’s sarcoma-associated herpesvirus is critical for nuclear accumulation of HIF-1alpha in normoxia. J Virol. 2007;81(19):10413–10423. doi: 10.1128/JVI.00611-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sodhi A, Montaner S, Patel V, Zohar M, Bais C, Mesri EA, Gutkind JS. The Kaposi’s sarcoma-associated herpes virus G protein-coupled receptor up-regulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1alpha. Cancer Res. 2000;60(17):4873–4880. [PubMed] [Google Scholar]
- 29.Montaner S, Sodhi A, Molinolo A, Bugge TH, Sawai ET, He Y, Li Y, Ray PE, Gutkind JS. Endothelial infection with KSHV genes in vivo reveals that vGPCR initiates Kaposi’s sarcomagenesis and can promote the tumorigenic potential of viral latent genes. Cancer Cell. 2003;3(1):23–36. doi: 10.1016/s1535-6108(02)00237-4. [DOI] [PubMed] [Google Scholar]
- 30.Montaner S, Sodhi A, Ramsdell AK, Martin D, Hu J, Sawai ET, Gutkind JS. The Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor as a therapeutic target for the treatment of Kaposi’s sarcoma. Cancer Res. 2006;66(1):168–174. doi: 10.1158/0008-5472.CAN-05-1026. [DOI] [PubMed] [Google Scholar]
- 31.Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012;33(4):207–214. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. 2013;126(Pt 8):1713–1719. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veeranna RP, Haque M, Davis DA, Yang M, Yarchoan R. Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen induction by hypoxia and hypoxia-inducible factors. J Virol. 2012;86(2):1097–1108. doi: 10.1128/JVI.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yogev O, Lagos D, Enver T, Boshoff C. Kaposi’s sarcoma herpesvirus microRNAs induce metabolic transformation of infected cells. PLoS Pathog. 2014;10(9):e1004400. doi: 10.1371/journal.ppat.1004400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 36.Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, Yang H, Mattaini KR, Metallo CM, Fiske BP, Courtney KD, Malstrom S, Khan TM, Kung C, Skoumbourdis AP, Veith H, Southall N, Walsh MJ, Brimacombe KR, Leister W, Lunt SY, Johnson ZR, Yen KE, Kunii K, Davidson SM, Christofk HR, Austin CP, Inglese J, Harris MH, Asara JM, Stephanopoulos G, Salituro FG, Jin S, Dang L, Auld DS, Park HW, Cantley LC, Thomas CJ, Vander Heiden MG. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8(10):839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dittmer DP, Richards KL, Damania B. Treatment of Kaposi sarcoma-associated herpesvirus-associated cancers. Front Microbiol. 2012;3:141. doi: 10.3389/fmicb.2012.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yarchoan R, Pluda JM, Wyvill KM, Aleman K, Rodriguez-Chavez IR, Tosato G, Catanzaro AT, Steinberg SM, Little RF. Treatment of AIDS-related Kaposi’s sarcoma with interleukin-12: rationale and preliminary evidence of clinical activity. Crit Rev Immunol. 2007;27(5):401–414. doi: 10.1615/critrevimmunol.v27.i5.10. [DOI] [PubMed] [Google Scholar]
- 39.Uldrick TS, Polizzotto MN, Yarchoan R. Recent advances in Kaposi sarcoma herpesvirus-associated multicentric Castleman disease. Curr Opin Oncol. 2012;24(5):495–505. doi: 10.1097/CCO.0b013e328355e0f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krown SE, Roy D, Lee JY, Dezube BJ, Reid EG, Venkataramanan R, Han K, Cesarman E, Dittmer DP. Rapamycin with antiretroviral therapy in AIDS-associated Kaposi sarcoma: an AIDS Malignancy Consortium study. J Acquir Immune Defic Syndr. 2012;59(5):447–454. doi: 10.1097/QAI.0b013e31823e7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 42.Venegas V, Wang J, Dimmock D, Wong LJ. Real-time quantitative PCR analysis of mitochondrial DNA content. In: Haines JL, et al., editors. Current protocols in human genetics/editorial board. Unit 19. Chap 19. 2011. p. 17. [DOI] [PubMed] [Google Scholar]